Abstract
The origin of the Etruscans (the present day Tuscany, Italy), one of the most enigmatic non-Indo-European civilizations, is under intense controversy. We found novel genetic evidences on the mitochondrial DNA (mtDNA) establishing a genetic link between Anatolia and the ancient Etruria. By way of complete mtDNA genome sequencing of a novel autochthonous Tuscan branch of haplogroup U7 (namely U7a2a), we have estimated an historical time frame for the arrival of Anatolian lineages to Tuscany ranging from 1.1±0.1 to 2.3±0.4 kya B.P.
Keywords: mtDNA, haplogroup, complete genome, Tuscany, Etruscans
Introduction
The origin of the Etruscans, one of the most ancient and enigmatic non-Indo-European civilizations, is being the target of a controversial debate. A recent study identified among modern Tuscans a rather high prevalence of Near Eastern mtDNA haplogroups and an exclusive haplotype sharing between them and Near Eastern populations.1 The finding has been interpreted as evidence in support of the classical theory that Etruscans may have come from the East through the Mediterranean Sea (Herodotus, Historiae, Vol I, p 94), which currently find little support by archaeologists and historians.2 In favor of the Eastern Mediterranean origin of the Etruscan civilization, the finding that the extent of mtDNA variation observed in Tuscan cattle breeds is similar to that observed in the Near East and much higher than that observed in the rest of Italy and Europe.3 The two facts could be compliant with other hypotheses. Thus, studies on fossil DNA in Italy have identified ancient pre-Neolithic bovine – aurochs – whose types are closer to modern bovine than West European aurochs: this contradicts the bovine migration theory and suggests either in loco domestication or population continuity across Italy–Balkans–Anatolia during the Palaeolithic4 (however, see the recent study by Achilli et al5 for a high-resolution study on mitochondrial DNA (mtDNA) of aurochs and domestic cattle). Furthermore, currently available analysis of archaeological Etruscan remains seems to indicate genetic continuity with Tuscans, with closer, but not specific, affinity with Anatolia6 (however, see Bandelt7 and Malyarchuk and Rogozin8).
To further test these hypotheses, we have analyzed a total of 258 Tuscan samples using mtDNA single nucleotide polymorphisms (SNPs), which allow the classification of Near Eastern typical haplogroups (HV lineages that are non-H and non-HV0, R0a, U7 and U3). mtDNA complete genome sequencing of a novel and autochthonous U7 sub-haplogroup has allowed, for the first time, to provide a time frame for this event.
Materials and Methods
Samples
We have undertaken a sample collection campaign covering 10 areas in Tuscany,9 whose geographical location extends to a wide area covering continental Etruria and the Elba Island (whose ferrous beds exploitation has set a landmark in driving ancient Etruscan craftsmanship): Arezzo (N=11), Chiusi (N=36), Collevecchio (N=24), Elba Island (N=53), Magliano Sabina (N=49), Monte Fiascone (N=17), Pitigliano (N=16), Tarquinia (N=15), Tuscania (N=26) and Vulci (N=11).
Genotyping of mtDNA SNPs
We have used the minisequencing technique for screening a total of 258 samples,10 all for a set of 24 mtDNA SNPs that allow to classify mtDNA sequences into major European haplogroups plus those that are more likely to be of Near East origin (Supplementary Table S1). Those mtDNAs with a SNP profile compatible with a typical Near Eastern haplogroups, that is, HV lineages that are non-H and non-HV0, R0a, U7 and U3, were further sequenced for the first hypervariable segment (HVS-I); only a small fraction of them (∼10 of the total sample size) were finally confirmed to belong to the mentioned haplogroups. Some other samples showing ambiguous haplogroup affiliation (eg, members of the broad macro-haplogroup N*) or that could reveal some phylogeographic information at the control region level (eg, haplogroups I, W, X) were also sequenced for the HVS-I. In total, 63 samples out of 258 were sequenced for the HVS-I.
Automatic sequencing
PCR amplification was carried out in a 9700 Thermocycler (AB). The temperature profile for 32 cycles of amplification was 95°C for 10 s, 60°C for 30 s and 72°C for 30 s. Sequencing primers were described earlier by Wilson et al.11 PCR product purification and sequencing were performed according to Salas et al.12
Nine samples from the Isle of Elba were sequenced for the complete mtDNA genome. The primers used for PCR amplification and sequencing were those reported by Torroni et al13 with minor modifications. More technical details concerning the PCR and sequencing reaction can be provided under request.
A posteriori sequence quality was evaluated following the methods described earlier.14, 15, 16, 17
Databases
We have compiled a total of 15 631 HVS-I sequences into a database that contains 13 155 West European profiles (including 1099 from different areas of Italy) and 2476 from Near East.
Coalescence age
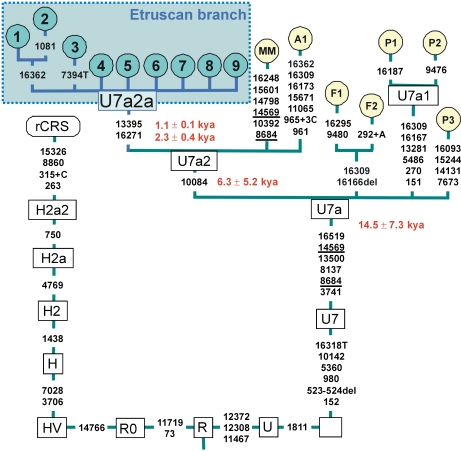
Estimation of the time to the most recent common ancestor of each cluster and SDs was carried out according to Saillard et al18 and using an evolutionary rate estimate of 1.26 × 0.08 × 10−8 base substitutions (other than a deletion or insertion) per nucleotide per year in the coding region (between 577 and 16 023), corresponding to 5140 years per substitution in the entire coding region.19 The coalescence age needed to accumulate the variation within U7a2a was estimated using both control and coding region information, which corresponds in Figure 1 with the first and the second term, respectively.
Figure 1.
Phylogeny of complete genomes belonging to haplogroup U7. Source of the data: nos. 1–9, present study (GenBank acc. no. EU445683-EU445691; MM=no. AndalAF_11 from Andalucia, Southern Spain (acc. no. AF382011); F1, no. 146 from Finland (acc. no. AY339547); F2, no. 147 from Finland (acc. no. AY339548); P1, no. B19 from India (acc. no. AY714013); P2, no. B81 from India (acc. no. AY714014); P3, no. C22 from India (acc. no. AY714004); A1, no. 13 from Pakistan (A Achilli, personal communication; acc. no. AY882391). The coalescence age needed to accumulate the variation within U7a2a was estimated using both control (top) and coding (bottom) region information.
Results and discussion
A total of 63 mtDNAs were sequenced for the HVS-I as described in Materials and Methods. The resulting haplotypes were searched across a European and Near Eastern database of more than 15 500 sequences. Overall, 34 out of the 63 sequenced Tuscan individuals (21 haplotypes) have a counterpart in Near East. Five HVS-I haplotypes (eight individuals who constitute ∼3% of the individuals in the total sample) singled out of the 63 sequenced individuals were not present in a large European database containing over 12 500 and including more than 1000 Italian sequences from outside Tuscany. Interestingly, some of those ‘Near Eastern sequences' emerging from our Tuscan sample did match with the Tuscan haplotypes described by Achilli et al.1
On the basis of combined information of SNPs and HVS-I sequence data, we confirmed that ∼10% (26 individuals out of 258) of our Tuscans actually belong to one of the typical Near Eastern haplogroups (see Supplementary Table S1), and have also a match with Near East populations. All of these Near East haplotypes are diverse (with the exception of those belonging to U7, see below) and fall at the tips of the phylogeny, suggesting a recent arrival to the region.
The typical Near Eastern U7 haplogroup occurs at relatively high frequency in the Elba Island (∼17%; 9 mtDNAs out of 53), and all of these U7 mtDNAs share the same HVS-I motif (T16271C-A16318T-T16519C), indicating that this lineage could represent a Near Eastern founder in the Isle. The T16271C-A16318T motif matches only two additional sequences in a worldwide database of more than >70 000 profiles; interestingly, both correspond to DNA samples collected in the ‘Etruscan area' (in Lucca; author's personal communication; GenBank acc. no.: DQ081609 and DQ081665;20). Complete genome sequencing of these nine U7 mtDNAs allowed the identification of a new sub-clade, U7a2a, characterized by transitions A13395G and T16271C. U7a2a is a sub-branch of U7a2; to our knowledge, only two other U7a2 complete genomes lacking the diagnostic motif of U7a2a have been reported in the literature, one was identified in a Pakistani and the other in an Andalusian (see Figure 1). The amount of variation accumulated within U7a2a Etruscan cluster (assuming a single founder) can be dated in the range 1.1±0.1 to 2.3±0.4 kya B.P., consistent with a recent arrival of this haplogroup to this Isle and compatible with the Etrurian culture (9th–1st century BC).
The investigation of a large and representative sample set and the analysis of complete mtDNA genomes support the hypothesis that Tuscany still preserves the fingerprint of a historical connection with the Near East. However, it should be stressed that this represents just a minor component of the Tuscan genetic make-up and suggests that historically different layers were superimposed over the Mesolithic gene pool of the Peninsula.
Note added in proof
Analysis performed by coalescent simulations21 suggested a model with little or no continuity between Ancient Etruscans and Modern Tuscans. However, the ancient dataset was extremely small and only a larger sample size would set the issue of diachronic continuity in Tuscany.
Acknowledgments
We would like to thanks Antonio Torroni for his useful comments and suggestions on the article and Alessandro Achilli for his help in submitting the complete genome fasta files to GenBank. The grant of the Xunta de Galicia (PGIDIT06PXIB208079PR), and the grant from the Fundación de Investigación Médica Mutua Madrileña, given to AS, partially supported this project.
Footnotes
Supplementary Information accompanies the paper on European Journal of Human Genetics website (http://www.nature.com/ejhg)
Supplementary Material
References
- Achilli A, Olivieri A, Pala M, et al. Mitochondrial DNA variation of modern Tuscans supports the near eastern origin of Etruscans. Am J Hum Genet. 2007;80:759–768. doi: 10.1086/512822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pallottino M. Storia della Prima Italia. Rusconi: Italy, Milano; 1999. [Google Scholar]
- Pellecchia M, Negrini R, Colli L, et al. The mystery of Etruscan origins: novel clues from Bos taurus mitochondrial DNA. Proc Biol Sci. 2007;274:1175–1179. doi: 10.1098/rspb.2006.0258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beja-Pereira A, Caramelli D, Lalueza-Fox C, et al. The origin of European cattle: evidence from modern and ancient DNA. Proc Natl Acad Sci USA. 2006;103:8113–8118. doi: 10.1073/pnas.0509210103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Achilli A, Olivieri A, Pellecchia M, et al. Mitochondrial genomes of extinct aurochs survive in domestic cattle. Curr Biol. 2008;18:R157–R158. doi: 10.1016/j.cub.2008.01.019. [DOI] [PubMed] [Google Scholar]
- Vernesi C, Caramelli D, Dupanloup I, et al. The Etruscans: a population-genetic study. Am J Hum Genet. 2004;74:694–704. doi: 10.1086/383284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bandelt H-J.Etruscan artifacts Am J Hum Genet 200475919–920.author reply 923–917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malyarchuk BA, Rogozin IB.On the Etruscan mitochondrial DNA contribution to modern humans Am J Hum Genet 200475920–923.author reply 923–927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capelli C, Brisighelli F, Scarnicci F, et al. Y chromosome genetic variation in the Italian peninsula is clinal and supports an admixture model for the Mesolithic-Neolithic encounter. Mol Phylogenet Evol. 2007;44:228–239. doi: 10.1016/j.ympev.2006.11.030. [DOI] [PubMed] [Google Scholar]
- Alvarez-Iglesias V, Jaime JC, Carracedo Á, Salas A. Coding region mitochondrial DNA SNPs: targeting East Asian and Native American haplogroups. Forensic Sci Int Genet. 2007;1:44–55. doi: 10.1016/j.fsigen.2006.09.001. [DOI] [PubMed] [Google Scholar]
- Wilson MR, Polanskey D, Butler J, DiZinno JA, Replogle J, Budowle B. Extraction, PCR amplification and sequencing of mitochondrial DNA from human hair shafts. Biotechniques. 1995;18:662–669. [PubMed] [Google Scholar]
- Salas A, Comas D, Lareu MV, Bertranpetit J, Carracedo Á. mtDNA analysis of the Galician population: a genetic edge of European variation. Eur J Hum Genet. 1998;6:365–375. doi: 10.1038/sj.ejhg.5200202. [DOI] [PubMed] [Google Scholar]
- Torroni A, Rengo C, Guida V, et al. Do the four clades of the mtDNA haplogroup L2 evolve at different rates. Am J Hum Genet. 2001;69:1348–1356. doi: 10.1086/324511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bandelt H-J, Quintana-Murci L, Salas A, Macaulay V. The fingerprint of phantom mutations in mitochondrial DNA data. Am J Hum Genet. 2002;71:1150–1160. doi: 10.1086/344397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bandelt H-J, Salas A, Bravi CM. Problems in FBI mtDNA database. Science. 2004;305:1402–1404. doi: 10.1126/science.305.5689.1402b. [DOI] [PubMed] [Google Scholar]
- Salas A, Bandelt H-J, Macaulay V, Richards MB. Phylogeographic investigations: the role of trees in forensic genetics. Forensic Sci Int. 2007;168:1–13. doi: 10.1016/j.forsciint.2006.05.037. [DOI] [PubMed] [Google Scholar]
- Salas A, Carracedo Á, Macaulay V, Richards M, Bandelt H-J. A practical guide to mitochondrial DNA error prevention in clinical, forensic, and population genetics. Biochem Biophys Res Commun. 2005;335:891–899. doi: 10.1016/j.bbrc.2005.07.161. [DOI] [PubMed] [Google Scholar]
- Saillard J, Forster P, Lynnerup N, Bandelt H-J, Nørby S. mtDNA variation among Greenland Eskimos: the edge of the Beringian expansion. Am J Hum Genet. 2000;67:718–726. doi: 10.1086/303038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishmar D, Ruiz-Pesini E, Golik P, et al. Natural selection shaped regional mtDNA variation in humans. Proc Natl Acad Sci USA. 2003;100:171–176. doi: 10.1073/pnas.0136972100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falchi A, Giovannoni L, Calo CM, et al. Genetic history of some western Mediterranean human isolates through mtDNA HVR1 polymorphisms. J Hum Genet. 2006;51:9–14. doi: 10.1007/s10038-005-0324-y. [DOI] [PubMed] [Google Scholar]
- Belle EM, Ramakrishnan U, Mountain JL, Barbujani G. Serial coalescent simulations suggest a weak genealogical relationship between Etruscans and modern Tuscans. Proc Natl Acad Sci USA. 2006;103:8012–8017. doi: 10.1073/pnas.0509718103. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.