Abstract
Interstitial deletions of 6q are rare. We report a detailed clinical and molecular characterization of four patients with interstitial deletion involving 6q25. All of our patients presented with microcephaly, developmental delay, dysmorphic features and hearing loss, whereas two of them had agenesis of the corpus callosum. We determined the size, extent and genomic content of the deletions using high-density array-comparative genomic hybridization (a-CGH), and found that a common segment spanning 3.52 Mb within the 6q25.2–q25.3 region was deleted in all four cases. We hypothesize that a subset of genes in the commonly deleted region are dosage sensitive and that haploinsufficieny of these genes impairs normal development of the brain and hearing.
Keywords: 6q deletion; hearing loss; microcephaly, developmental delay; agenesis of the corpus callosum; array-CGH
Introduction
Since the first reported case in 1975,1 over 65 cases of deletions involving the long arm of chromosome 6 have been reported.2, 3, 4, 5, 6, 7 Though the clinical phenotype is quite variable, Hopkin et al2 attempted to correlate the phenotype with genotype, by dividing patients into three groups. Those in Group A with del(6)(q11q16) had a high incidence of hernias, up-slanting palpebral fissures, thin lips, a lower frequency of microcephaly, micrognathia and heart malformations. Patients categorized to Group B with del(6)(q15q25) had intrauterine growth retardation, abnormal respiration, hypertelorism and upper limb malformations, whereas those in group C with del(6)(q25) presented with retinal abnormalities, cleft palate and genital hypoplasia. However, there was significant overlap of features among the groups and some features such as developmental delay (100%), ear anomalies (90%), hypotonia (82%) and postnatal growth retardation (68%) were common to all groups. Hearing loss has been reported only in a minority of publications.1, 2, 6, 8, 9
Most of the reported deletions were identified and characterized based on routine karyotyping with no further attempt to delineate the breakpoints. We determined the size, extent and genomic content of four cases involving interstitial deletion of 6q25. All four deletions have a common deleted segment in 6q25.2–q25.3 with the smallest region of overlap (SRO) of 3.52 Mb. All of the patients presented with microcephaly, developmental delay, dysmorphic features and hearing loss. We hypothesize that a subset of genes deleted in the SRO are dosage sensitive and are important for CNS and hearing development.
Clinical reports
Patient 1
The proband is a male child born to nonconsanguineous Hispanic parents. Family history was negative for known chromosomal anomalies or birth defects. Pregnancy was uncomplicated and delivery was by Cesarean section that was performed for fetal distress. Birth weight was 3.6 kg (25th centile), whereas the length and FOC are not available. Apgar scores were 7 and 9 at 1 and 5 min, respectively. The perinatal period was characterized by feeding difficulties. At the 6-month examination, his motor development was that of a 3-month old. A peripheral G-banded karyotyping performed at 6 months was normal. When examined by a genetics consultant at 33 months of age, his FOC (46.2 cm) was at the 2nd centile (z-score −2.0), whereas the height (89.2 cm) and weight (13.1 kg) were at the 9th and 27th centile, respectively. Examination was remarkable for plagiocephaly, down-slanting palpebral fissures, epicanthic folds, midface hypoplasia, low set ears, long philtrum and micrognathia (Figure 1a). He had global developmental delay; rolled over at 1 year, walked at 23 months and had only two words at 33 months. Audiology evaluation showed severe sensorineural hearing loss on the right. Brain imaging could not be performed.
Figure 1.
Photographs of patients 1 (a), 2 (b) and 4 (c) showing facial characteristics. Note that hypertelorism, posteriorly rotated auricles, broad nasal root and midface hypoplasia are common to all. In addition, patient 1 has epicanthic folds, down-slanting palpebral fissures and retrognathia; patient 2 has a bulbous nose and thin upper lip, whereas patient 4 has malformed, low-set auricles and a thin upper lip.
Patient 2
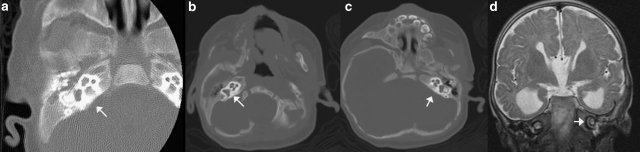
This female child was born at 37 weeks of gestation to nonconsanguineous Caucasian parents. The mother had an abnormal triple screen and underwent amniocentesis that revealed a normal female chromosome constitution, 46,XX. Growth parameters at birth were remarkable for weight of 2.2 kg (0.3 centile, z-score −2.8) and FOC of 31 cm (0.1 centile, z-score −3.9), whereas length 48 cm (5th centile) was normal. Soon after birth, she had respiratory compromise requiring ventilation with the aid of bag and mask. The perinatal period was also complicated by subarachnoid hemorrhage that was managed medically without any surgical intervention. The proband had feeding difficulties until 3 months. Her medical problems also included two episodes of febrile seizures and hypotonia. At the 9-month evaluation, she was noted to have a bulbous nose, thin upper lip and a pit on the right helix (Figure 1b). Audiology evaluation revealed bilateral moderate-to-severe sensorineural hearing loss. MRI of the brain revealed plagiocephaly and no structural abnormalities of the brain. CT scan of the temporal bones showed normal cochlear structure. (Figure 2a) Ophthalmological examination revealed esotropia of the left eye. She had global developmental delay; she sat unsupported at 1 year, crawled at 14 months and could not walk without support at 18 months (age at last evaluation). Her expressive language was delayed, and at 18 months, she did not have any words.
Figure 2.
Imaging showing normal two and a half turns of the cochlea in patients 2 (a), 3 (b, c) and 4 (d). The normal cochlear anatomy implies that the hearing loss could be secondary to abnormal sensory epithelium or its connecting pathways.
Patient 3
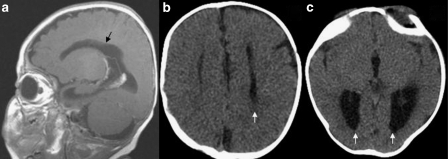
The proband was born to nonconsanguineous Hispanic parents at 39 weeks of gestation by Cesarean section performed for nonprogression of labor. Family history was noncontributory. Pregnancy was uncomplicated; however, prenatal ultrasounds revealed the possibility of hydrocephalus and agenesis of the corpus callosum (ACC). Growth parameters at birth for weight (2.62 kg, 3rd centile) and FOC (32 cm; 0.1 centile, z-score of −3.0) were below normal, but the length (48 cm, 5th centile) was within normal limits. The perinatal period was complicated by feeding difficulties that mandated the placement of a feeding tube. Examination at 2 months of age revealed a FOC of 33 cm (0.1 centile, z-score of −5.0), length of 52 cm (0.5 centile, z-score −2.6) and a weight of 3.45 kg (0.3 centile, z-score −2.7). Dysmorphic features included low set and posteriorly rotated ears, midface hypoplasia, a high arched palate, bifid uvula, small hands with mild fifth finger clinodactyly and small feet. CT scan of the head at 2 months of age revealed the absence of corpus callosum with colpocephaly (Figure 3b and c), a marked increase in the size of the lateral ventricles, and premature fusion of both lambdoid as well as the posterior aspect of the superior sagittal sutures giving rise to plagiocephaly. Auditory brainstem response (ABR) showed mild conductive loss in the left ear and a normal auditory acuity in the right ear. CT scan showed normal cochlear anatomy (Figure 2b and c). At the 10-month examination, her weight (5.38 kg, 0.1 centile, z-score −4.0), length (64 cm, 0.3 centile, z-score −2.7) and FOC (39.5, 0.1 centile, z-score −4.3) were well below the normal limits. She had significant delay and had not attained any motor or language milestones. Peripheral blood karyotyping performed elsewhere was reported as 46,XX,del(6)q25.2–q25.3.
Figure 3.
(a) MRI showing the absence of corpus callosum in patient 4. CT scan showing parallel orientation of lateral ventricles (b) and colpocephaly (c) suggestive of agenesis of corpus callosum in patient 3.
Patient 4
This male patient was born to a 30-year-old–gravida 2, para1–woman with no significant medical history. The parents are nonconsanguineous, and family history was noncontributory. Pregnancy was uncomplicated until 31 weeks of gestational age when prenatal ultrasound revealed enlarged cerebral ventricles. The proband was born through vaginal delivery at 38 weeks of gestation. The growth parameters were remarkable for a weight of 2.8 kg (3rd centile), length of 47 cm (1.1 centile, z-score −2.3) and an FOC of 31 cm (0.4 centile, z-score −2.7). Apgar scores were 6 and 8 at 1 and 5 min, respectively. The perinatal period was unremarkable. At the 2-month examination, his FOC of 34.5 cm (0.1 centile, z-score −4.2) and length of 53 cm (0.1 centile, z-score −3.0) were well below the normal, whereas the weight (4.44 kg) was at the 3rd centile. Dysmorphic features including mild midface hypoplasia, low set posteriorly rotated ears with abnormal helices and high arched palate were evident (Figure 1c). The hands were held in a clenched position, but there were no overlapping fingers and palmar creases were normal. The lower limbs had a mild valgus deformity at the knee and rocker bottom feet. There was mild hypertonia in all four extremities with normal deep tendon reflexes. He had penoscrotal webbing that was surgically corrected with scrotoplasty at 6 months. Ophthalmological examination was abnormal and showed intermittent esotropia, lash ptosis and strabismic amblyopia that was treated with eye patching. MRI of the brain showed agenesis of corpus callosum with accompanying colpocephaly and a diffuse delay in myelination (Figure 3a). ABR was consistent with a moderate-to-severe sensorineural hearing loss in both ears. The cochlear anatomy was unremarkable (Figure 2d). Echocardiogram showed a fenestrated septum secundum ASD with minimal left-to-right shunting. At 10 months, he continued to be microcephalic with an FOC of 40.4 (0.1 centile, z-score −4.5), whereas his weight (8 kg) and length (69.5 cm) were at the 5th and 6th centiles, respectively. He had global delay and could not sit, crawl or rollover. G-banded karyotyping performed on peripheral blood leukocytes was consistent with a terminal deletion of 6q; 46XY,del(6)(q25.3).
Material and methods
Human subjects
Following an initial result of the chromosome microarray analysis (CMA),10 informed consent was obtained from the parents for further analysis by whole-genome array-based oligonucleotide comparative genomic hybridization (CGH). The research protocol was approved by the Institutional Review Board (IRB) for Baylor College of Medicine and affiliated hospitals.
Clinical chromosome microarray analysis
CMA was initially performed on all four cases. The microarrays were designed and manufactured in the Medical Genetics Laboratory as described previously.10 The procedures for DNA digestion, labeling and hybridization, as well as data analysis were performed as described previously.11
FISH analysis
Confirmatory FISH analyses with the BAC clones were performed using standard procedures. Briefly, the BAC clone of interest was grown in broth media with 20 μg/ml of chloramphenicol. DNA was extracted from bacterial artificial chromosome clones (Eppendorf Plasmid Mini Prep kit, Hamburg, Germany) and directly labeled with SpectrumOrangeTM dUTP by nick-translation (Vysis, Downer Grove, IL, USA) according to the manufacturers' instructions.
High-resolution oligonucleotide array
The Agilent 244K Whole Human Genome Oligo Microarray Kit (Agilent Technologies Inc., Santa Clara, CA, USA) contains 238 459 arrayed 60-mer oligonucleotides, representing a compiled view of the human genome at an average resolution of 6.4 kb. The procedures for DNA digestion, labeling and hybridization were performed according to the manufacturer's instructions with some modifications.12
Results
Clinical features
The clinical phenotype of the patients is shown in Table 1. Microcephaly, developmental delay and dysmorphic features, such as hypertelorism, abnormal root of the nose, midface hypoplasia and ear abnormalities were common to all (Figure 1). Plagiocephaly and growth restriction were seen in three of the four patients. Moderate-to-severe sensorineural loss was found in patients 1, 2 and 4, whereas patient 3 had mild conductive hearing loss. ACC was seen in patients 3 and 4.
Table 1. Clinical features observed in our cohort compared to those seen in studies with similar cytogenetically determined deletions.
| Patient 1 | Patient 2 | Patient 3 | Patient 4 | Pirola et al | Narahara et al | Sukumar et al case 2 | Sukumar et al case 3 | Hopkin et al case 3 | Oliveira-Duarte et al | Rivas et al | Meng et al case 1 | Meng et al case 2 | Barthoshesky et al | Valtat et al case 3 | Valtat et al case 4 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sex | M | F | F | M | F | M | M | M | M | F | F | M | F | M | F | M |
| Cytogenetic band deleted | 6q25.2–q25.3 | 6q25.2–q26 | 6q24.3–q25.3 | 6q25.2–q27 | q25.1–q25.3 | q25.1–q25.3 | 6q25.1–q25.3 | 6q25.1–q26 | 6q25-qter | 6q25-qter | 6q25-qter | 6q24.3-qter | 6q25.3-qter | 6q25-qter | 6q25-q27 | 6q25-qter |
| Age at report | 4 years | 18 months | 11 months | 15 months | 9 months | 7 months | 17 years | Birth | NA | NA | 5 years | 4 months | 2 years | NA | Birth | 10 years |
| Growth restriction | − | − | + | + | + | + | + | + | − | − | + | + | + | + | + | − |
| Microcephaly | + | + | + | + | − | + | + | + | − | + | + | + | + | + | + | − |
| Abnormal shape of head | + | + | + | − | + | + | − | − | − | + | + | − | + | NA | NA | NA |
| Facial features | ||||||||||||||||
| Epicanthic folds | + | − | − | − | − | + | + | − | − | + | − | − | + | + | + | + |
| Downslanting palpebral fissure | + | − | − | + | − | − | + | − | − | + | − | + | − | + | NA | + |
| Upslanting palpebral fissure | − | − | − | − | − | + | − | − | − | NA | − | − | − | − | NA | |
| Hypertelorism | + | + | + | + | − | − | − | − | + | NA | − | + | − | + | NA | + |
| Nasal root abnormality | + | + | + | + | − | + | − | − | + | + | + | + | + | NA | NA | |
| Low set ears | + | − | + | + | + | − | − | − | + | − | NA | + | + | + | NA | NA |
| Posteriorly rotated auricles | + | + | + | + | − | − | − | − | + | − | + | − | − | + | NA | NA |
| Abnormal auricles or pits | − | + | − | + | − | − | + | + | + | + | + | − | − | + | + | − |
| Midface hypoplasia | + | + | + | + | − | − | − | − | − | − | − | − | − | NA | NA | |
| Long philtrum | + | − | − | + | − | − | − | − | − | + | + | − | − | + | NA | − |
| Thin upper lip | − | + | − | + | − | − | − | − | − | + | − | + | + | − | − | + |
| Palatal abnormality | − | − | + | + | + | − | − | − | NA | + | NA | + | + | + | + | − |
| Hands | ||||||||||||||||
| Transverse crease | − | − | − | − | − | − | − | − | NA | NA | + | − | + | + | − | |
| Clinodactyly | − | − | + | − | − | − | − | − | − | NA | NA | NA | NA | − | NA | NA |
| Congenital heart disease | − | − | − | + | − | + | − | − | + | − | − | + | + | + | + | − |
| Nervous system | ||||||||||||||||
| Developmental delay | + | + | + | + | + | + | + | NA | + | + | + | NA | + | + | NA | + |
| Seizures | − | + | − | − | − | − | + | − | − | + | NA | − | − | NA | NA | NA |
| Structural brain abnormalities | NA | − | + | + | + | NA | − | + | + | + | + | − | + | NA | NA | NA |
| Agenesis of corpus callosum | NA | − | + | + | + | NA | − | + | − | − | − | + | − | NA | NA | NA |
| Hypotonia | − | + | + | − | NA | − | NA | NA | + | + | + | + | + | NA | NA | NA |
| Hypertonia | − | − | − | + | NA | − | NA | NA | − | − | − | − | − | NA | NA | NA |
| Retinal or eye abnormalities | − | + | − | + | − | − | − | + | + | + | + | + | + | NA | NA | NA |
| Hearing loss | + | + | + | + | NA | NA | − | + | + | NA | NA | NA | NA | NA | NA | NA |
| Genitourinary abnormality | − | − | − | + | − | − | − | + | + | + | NA | − | − | + | NA | NA |
| Sacral and/or anorectal anomalies | − | − | − | − | + | − | − | − | + | − | NA | + | − | NA | NA | NA |
The first four studies involve interstitial deletions around the SRO seen in our cases, whereas the others involve terminal deletions encompassing the SRO. + Denotes that a characteristic is present, − denotes the absence of the characteristic and NA implies that data were not available.
Clinical chromosome microarray analysis
We initially performed array-CGH (a-CGH) analysis on DNA extracted from blood leukocytes on the clinical microarray platform routinely used in our institution. The interrogating BAC clones deleted are shown in Table 2.
Table 2. Molecular mapping of the deletions.
| Patient | Chromosomal segment | BAC or PAC clones corresponding to the deleted region | Confirmation of deletion | Proximal and distal breakpoint regions on the 244K aCGH | Deletion Size (Mb) as per 244K aCGH | Genes deleted |
|---|---|---|---|---|---|---|
| 1 | 6q25.2–q25.3 | RP11-266C7, RP11-115G17 | ish del(6)(q25.3q25.3) (RP11-115G17-) | 155085617–155094102, 158870729–158876467 | 3.77 Mb | TIAM2 → TULP4 |
| 2 | 6q25.2–q26 | RP11-134L4, RP11-266C7, RP11-115G17, RP156L9 | ish del(6)(q25.3q25.3) (RP11-115G17-) | 154841486–154847023, 161614264–161623426 | 6.7 Mb | RBM16 → MAP3K4 |
| 3 | 6q24.3–q25.3 | RP11-134L4, RP11-266C7, RP11-115G17, RP156L9 | ish del(6)(q25.3q25.3) (RP11-115G17-) | 149951406–149959837, 160265928–160276072 | 10.3 Mb | LATS1 → PNLDC1 |
| 4 | 6q25.2–q27 | RP11-266C7, RP11-115G17, RP156L9, RP1-56L9, RP1-45F6, RP1-51J12, RP11-256A6, RP11-104N13, RP346016, RP11-168L15, RP3-366N23, RP3-495K2 | del(6)(q25.3q27) | 155336861–155349955, 169163256–169178124 | 13.81 Mb | TIAM2 → SMOC2 |
The clinical chromosome microarray analysis was performed using either BAC-based10, 11 or BAC-emulated oligo arrays.13 The breakpoints and the deletion sizes were estimated by 244 oligonucleotide a-CGH. The genes in the deleted region were determined using the UCSC genome browser (March 2006 assembly).
High-resolution oligonucleotide array
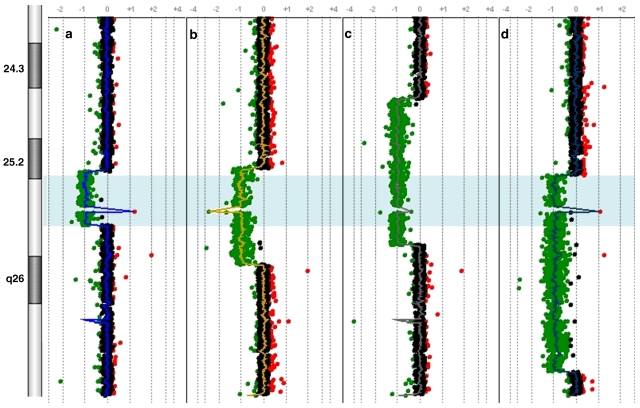
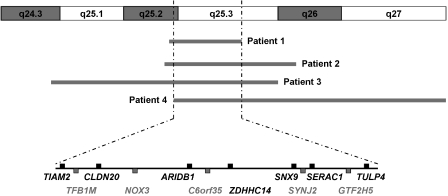
To determine the precise size extent and genomic content of the deletions, the patient samples were analyzed using an Agilent 244K oligonucleotide array. The breakpoints are as depicted in Table 2. The deletions varied in size from 3.77 to 13.81 Mb (Figure 4). The commonly deleted genomic interval (SRO) corresponds to chromosomal region 6q25.2–25.3 and encompasses an area of 3.52 Mb that contains 15 genes mapping from TIAM2 (T-cell lymphoma invasion and metastasis 2) to TULP4 (Figure 5).
Figure 4.
Mapping of breakpoints and common deleted region (blue; a, b, c and d). Each point represents an oligonucleotide probe. The normalized data for each probe is represented along a vertical line that indicates its relative position on 6q. Loss of copy number is indicated by deviation to the left of the center (depicted in green), whereas gain of copy number is indicated by deviation to the right of the center (depicted in red). The deletions of four patients span 3.77, 6.7, 10.3 and 13.81 Mb. The commonly deleted region (shaded) spans 3.52 Mb from 155349955 to 158870729 bp. The spike in the 6q25.3 region represents the copy number variation (CNV) of the only one oligonucleotide (60 mer, positioned at 157888702–157888761 bp) in the flanking 420-Kb genomic region. The oligonucleotide is located in a known CNV region and is most likely indicative of polymorphism. Note that there is grouping of proximal breakpoints in patients 1, 2 and 4. A full-colour version of this figure is available at the EJHG journal online.
Figure 5.
Protein-coding genes in the smallest region of overlap (SRO). All the genes except NOX3 are expressed in the brain.
Discussion
Interstitial deletions of the long arm of chromosome 6 are rare.2, 3, 4, 5, 6, 7, 14, 15 We report and characterize four patients with de novo interstitial deletions of 6q, including two submicroscopic deletions. The deletions were of different sizes with the SRO (3.52 Mb) corresponding to the chromosomal segment 6q25.2–q25.3.
The clinical features of our cases were compared with four other cases with similar interstitial deletions that had breakpoints determined cytogenetically to map between 6q24 and 6q27 and encompassing the SRO of our cases (Table 1).4, 6, 14, 16 This approach may help further delineate the phenotype associated with the deletion of 6q25.2–q25.3. The studies with terminal deletions of 6q with breakpoints at or distal to 6q24.3, the most proximal breakpoint of deletions in our cases, and containing the SRO seen in our cases are also summarized.5, 17, 18, 19, 20, 21 Studies with 6q monosomy because of translocations, deletions with proximal breakpoints centromeric to 6q24 or those not deleted for the 6q25.2–q25.3 were excluded from comparison.
Developmental delay in varying degrees was found in all the cases across all the above-mentioned studies. Growth restriction was found only in two of our patients (patients 3 and 4), whereas it was observed more commonly in patients with larger, cytogenetically visible deletions. This discrepancy may be because of the fact that the extent of deletions in the other studies may have been underestimated by the limited genomic resolution of G-banding analysis. Most of the dysmorphic features seen in our patients were nonspecific and have been reported in other studies as well (Table 1). The shared specific findings in our cases appear to be the association of microcephaly, ACC and hearing loss with deletion of the 6q25 region.
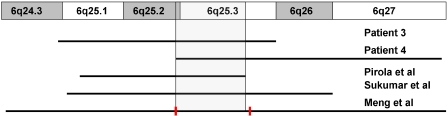
Microcephaly was present in all of our patients as well in all studies with interstitial or terminal deletions involving 6q25 except two cases.2, 17 (Table 1). In the case of Hopkin et al,2 the proband had macrocephaly because of hydrocephalus, whereas in Valtat et al,17 the FOC was 1.5 SD below the mean. Pirola et al16 reported a normal head circumference (50th centile) at 9 months of age in their case, but the birth FOC was 2 SD below the mean. ACC was seen in two of our patients (3 and 4), and has been reported in other 6q-deletion patients.5, 6, 15, 16, 22 By comparing these studies, we delimited a critical region of 3.3 Mb for ACC (Figure 6). This interval falls within the SRO of 3.52 Mb seen in our cases. However, there may be incomplete penetrance for this abnormality, as some cases with deletions of this critical region (eg, patient 2) have normal corpus callosal development. Hearing loss was seen in all of our patients. Hopkin et al2 describe sensorineural hearing loss in one of their cases, whereas Sukumar et al6 mention that the hearing of one of the patients was abnormal, but do not provide results of objective testing. The other reports do not specifically mention the hearing evaluation. There are two known regions in the vicinity 6q25 that have been implicated in deafness. One locus at 6q26–q27 identified by linkage analysis in a consanguineous family is associated with autosomal recessive congenital sensorineural deafness, whereas the other region at 6q23.2 harbors the EYA4 gene implicated in later onset of autosomal dominant hearing loss.23, 24
Figure 6.
The distal deletions of 6q associated with agenesis of the corpus callosum (ACC) share a common minimal region (gray) that is within the smallest region of overlap (SRO) seen in our cases (red bars). This indicates that a gene important for normal corpus callosal development lies in the 6q25.2–q25.3 region. A full-colour version of this figure is available at the EJHG journal online.
Our data suggest that microcephaly, ACC and hearing loss seen in interstitial or terminal deletions involving 6q25 may map to a 3.52-Mb region at 6q25.2–q25.3. This region harbors 12 protein-coding genes (Figure 5); among them, the ones that may be pertinent to the clinical phenotype are TIAM2, NOX3 and SYNJ2.
TIAM2 encodes a guanine nucleotide exchange factor. A highly similar protein (Stef) is implicated in normal neural development in a murine model.25 The pattern of expression of Stef in a stage and region-specific manner in the mouse brain corresponded to neuronal morphological changes.
SYNJ2 is a phosphoinositol 5-phosphatase that is involved in vesicular trafficking and actin dynamics. In glioblastoma cell lines, SYNJ2 is implicated in the regulation of the formation of invadopodia and lamellipodia leading to tumor cell invasion and migration.26 It is, however, not known whether SYNJ2 has a role in normal migration of neurons during development. It is tempting to hypothesize that TIAM2 and SYNJ2 may be involved in normal development of the brain and that haploinsufficieny of either or both causes microcephaly, ACC and developmental delay seen in 6q25.3 deletions.
NOX3 is an NADPH oxidase that is implicated in normal development of otoconia in mice.27 Homozygous mutations lead to lack of otoconia and significant vestibular dysfunction, whereas the heterozygous mice had normal auditory and vestibular functions. The other inner ear structures including the sensory epithelium and the hearing of the affected mice were normal. It is, however, possible that haploinsufficieny of NOX3 may have auditory as well as vestibular effects in humans.
Genomic rearrangements describe mutational changes in the genome such as duplication, deletion, insertion and inversion that are different from the traditional Watson–Crick base pair alterations. Nonrecurrent genomic rearrangements can be mediated by nonhomologous end joining (NHEJ)28 or fork stalling and template switching (FoSTeS).29 Given that there was grouping of the proximal breakpoints in three of our patients (Figure 4), it is possible that an underlying genomic architecture important to the rearrangement is present in the vicinity of the deleted regions.
In summary, interstitial deletions involving 6q25.2–q25.3 described herein presented with microcephaly, ACC, developmental delay, hearing loss and dysmorphic features. The deletions can remain undetected by routine karyotyping techniques as evidenced by normal karyotyping results in two of our patients. We hence recommend a-CGH for diagnosis of such deletions. From a clinical standpoint, we suggest that MRI of the brain and a formal hearing evaluation should be considered in all patients with deletions involving 6q25.
Acknowledgments
We thank the participating families for their kind cooperation. This work was supported in part by fellowship grants from the Osteogenesis Imperfecta Foundation (SNSC) and the National Urea Cycle Foundation and Children's National Medical Center (AE).
References
- Milosevic J, Kalicanin P. Long arm deletion of chromosome no. 6 in a mentally retarded boy with multiple physical malformations. J Ment Defic Res. 1975;19:139–144. [PubMed] [Google Scholar]
- Hopkin RJ, Schorry E, Bofinger M, et al. New insights into the phenotypes of 6q deletions. Am J Med Genet. 1997;70:377–386. [PubMed] [Google Scholar]
- Matkins SV, Meyer JE, Berry AC. A child with partial monosomy 6q secondary to a maternal direct insertional event. J Med Genet. 1987;24:227–229. doi: 10.1136/jmg.24.4.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLeod DR, Fowlow SB, Robertson A, Samcoe D, Burgess I, Hoo JJ. Chromosome 6q deletions: a report of two additional cases and a review of the literature. Am J Med Genet. 1990;35:79–84. doi: 10.1002/ajmg.1320350115. [DOI] [PubMed] [Google Scholar]
- Meng J, Fujita H, Nagahara N, Kashiwai A, Yoshioka Y, Funato M. Two patients with chromosome 6q terminal deletions with breakpoints at q24.3 and q25.3. Am J Med Genet. 1992;43:747–750. doi: 10.1002/ajmg.1320430419. [DOI] [PubMed] [Google Scholar]
- Sukumar S, Wang S, Hoang K, et al. Subtle overlapping deletions in the terminal region of chromosome 6q24.2–q26: three cases studied using FISH. Am J Med Genet. 1999;87:17–22. [PubMed] [Google Scholar]
- Titomanlio L, Giurgea I, Baumann C, et al. A locus for sacral/anorectal malformations maps to 6q25.3 in a 0.3 Mb interval region. Eur J Hum Genet. 2006;14:971–974. doi: 10.1038/sj.ejhg.5201635. [DOI] [PubMed] [Google Scholar]
- Pandya A, Braverman N, Pyeritz RE, Ying KL, Kline AD, Falk RE. Interstitial deletion of the long arm of chromosome 6 associated with unusual limb anomalies: report of two new patients and review of the literature. Am J Med Genet. 1995;59:38–43. doi: 10.1002/ajmg.1320590109. [DOI] [PubMed] [Google Scholar]
- Schuster M, Lohscheller J, Kummer P, Eysholdt U, Rosanowski F. Severe sensory hearing loss in del(6q)-syndrome. Int J Pediatr Otorhinolaryngol. 2003;67:1263–1266. doi: 10.1016/j.ijporl.2003.07.014. [DOI] [PubMed] [Google Scholar]
- Cheung SW, Shaw CA, Yu W, et al. Development and validation of a CGH microarray for clinical cytogenetic diagnosis. Genet Med. 2005;7:422–432. doi: 10.1097/01.gim.0000170992.63691.32. [DOI] [PubMed] [Google Scholar]
- Lu X, Shaw CA, Patel A, et al. Clinical implementation of chromosomal microarray analysis: summary of 2513 postnatal cases. PLoS ONE. 2007;2:e327. doi: 10.1371/journal.pone.0000327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Probst FJ, Roeder ER, Enciso VB, et al. Chromosomal microarray analysis (CMA) detects a large X chromosome deletion including FMR1, FMR2, and IDS in a female patient with mental retardation. Am J Med Genet A. 2007;143:1358–1365. doi: 10.1002/ajmg.a.31781. [DOI] [PubMed] [Google Scholar]
- Ou Z, Kang SH, Shaw CA, et al. Bacterial artificial chromosome-emulation oligonucleotide arrays for targeted clinical array-comparative genomic hybridization analyses. Genet Med. 2008;10:278–289. doi: 10.1097/GIM.0b013e31816b4420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narahara K, Tsuji K, Yokoyama Y, et al. Specification of small distal 6q deletions in two patients by gene dosage and in situ hybridization study of plasminogen and alpha-L-fucosidase 2. Am J Med Genet. 1991;40:348–353. doi: 10.1002/ajmg.1320400322. [DOI] [PubMed] [Google Scholar]
- Rubtsov N, Senger G, Kuzcera H, et al. Interstitial deletion of chromosome 6q: precise definition of the breakpoints by microdissection, DNA amplification, and reverse painting. Hum Genet. 1996;97:705–709. doi: 10.1007/BF02346176. [DOI] [PubMed] [Google Scholar]
- Pirola B, Bortotto L, Giglio S, et al. Agenesis of the corpus callosum with Probst bundles owing to haploinsufficiency for a gene in an 8 cM region of 6q25. J Med Genet. 1998;35:1031–1033. doi: 10.1136/jmg.35.12.1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valtat C, Galliano D, Mettey R, Toutain A, Moraine C. Monosomy 6q: report on four new cases. Clin Genet. 1992;41:159–166. doi: 10.1111/j.1399-0004.1992.tb03655.x. [DOI] [PubMed] [Google Scholar]
- Oliveira-Duarte MH, Martelli-Soares LR, Sarquis-Cintra T, Machado ML, Lison MP. Distal monosomy of the long arm of chromosome 6 (6q25–6qter) inherited by maternal translocation t(6q;17q) Ann Genet. 1990;33:56–59. [PubMed] [Google Scholar]
- Rivas F, Ruiz C, Rivera H, Moller M, Serrano-Lucas JI, Cantu JM. De novo del(6)(q25) associated with macular degeneration. Ann Genet. 1986;29:42–44. [PubMed] [Google Scholar]
- Bartoshesky L, Lewis MB, Pashayan HM. Developmental abnormalities associated with long arm deletion of chromosome No. 6. Clin Genet. 1978;13:68–71. doi: 10.1111/j.1399-0004.1978.tb04129.x. [DOI] [PubMed] [Google Scholar]
- Stevens CA, Fineman RM, Breg WR, Silken AB. Report of two cases of distal deletion of the long arm of chromosome 6. Am J Med Genet. 1988;29:807–814. doi: 10.1002/ajmg.1320290410. [DOI] [PubMed] [Google Scholar]
- Shen-Schwarz S, Hill LM, Surti U, Marchese S. Deletion of terminal portion of 6q: report of a case with unusual malformations. Am J Med Genet. 1989;32:81–86. doi: 10.1002/ajmg.1320320117. [DOI] [PubMed] [Google Scholar]
- Ansar M, Ramzan M, Pham TL, et al. Localization of a novel autosomal recessive non-syndromic hearing impairment locus (DFNB38) to 6q26–q27 in a consanguineous kindred from Pakistan. Hum Hered. 2003;55:71–74. doi: 10.1159/000071813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hildebrand MS, Coman D, Yang T, et al. A novel splice site mutation in EYA4 causes DFNA10 hearing loss. Am J Med Genet A. 2007;143A:1599–1604. doi: 10.1002/ajmg.a.31860. [DOI] [PubMed] [Google Scholar]
- Yoshizawa M, Hoshino M, Sone M, Nabeshima Y. Expression of stef, an activator of Rac1, correlates with the stages of neuronal morphological development in the mouse brain. Mech Dev. 2002;113:65–68. doi: 10.1016/s0925-4773(01)00650-5. [DOI] [PubMed] [Google Scholar]
- Chuang YY, Tran NL, Rusk N, Nakada M, Berens ME, Symons M. Role of synaptojanin 2 in glioma cell migration and invasion. Cancer Res. 2004;64:8271–8275. doi: 10.1158/0008-5472.CAN-04-2097. [DOI] [PubMed] [Google Scholar]
- Paffenholz R, Bergstrom RA, Pasutto F, et al. Vestibular defects in head-tilt mice result from mutations in Nox3, encoding an NADPH oxidase. Genes Dev. 2004;18:486–491. doi: 10.1101/gad.1172504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupski JR, Stankiewicz P. Genomic disorders: molecular mechanisms for rearrangements and conveyed phenotypes. PLoS Genet. 2005;1:e49. doi: 10.1371/journal.pgen.0010049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JA, Carvalho CM, Lupski JR. A DNA replication mechanism for generating nonrecurrent rearrangements associated with genomic disorders. Cell. 2007;131:1235–1247. doi: 10.1016/j.cell.2007.11.037. [DOI] [PubMed] [Google Scholar]