Abstract
The single nucleotide polymorphisms (SNPs) rs449647, rs769446 and rs405509 in the promoter region of the APOE gene have been variously suggested to be ɛ4-independent risk factors for Alzheimer's disease (AD). A previous Italian study found that the rs449647 was significantly associated with late-onset AD. The aim of this study was to verify whether these APOE promoter SNPs are genetic risk factors for AD and to investigate their interaction with the common APOE polymorphism. A total of 169 clinically diagnosed AD patients and 99 cognitively intact age-matched controls were included in the study. Significant associations with AD independent from sex, age and APOE/ɛ4 status were found for rs449647 A/A and rs405509 G/G genotypes (positive), and rs449647 A/T and rs405509 T/T genotypes (negative). Haplotype frequency estimation at the APOE locus showed significant associations for the ATG4, ATT4 and ACG3 (positive) and ATT2, ATT3 and TCG3 (negative) haplotypes. Therefore this study confirms the role of the rs449647 A/A genotype as risk factor for AD in Italy and suggests that promoter genotypes and APOE haplotypes might have a complex function in AD-associated genetic risk factors.
Keywords: APOE promoter, haplotypes, Alzheimer's disease, genetic risk factors
Introduction
Alzheimer's disease (AD), in its sporadic form, is a paradigmatic model of complex diseases resulting from the interaction between genetic and nongenetic factors such as environmental ones.1, 2 Mendelian inheritance of pathogenic mutations has been described only for a small amount of AD cases (less than 5%).1 To find an association with sporadic AD more than 100 biological or genetic factors have been analyzed.1, 2 In spite of several studies carried out in large AD samples of ethnically different subgroups, the ɛ4 allele of the APOE gene remains the only established genetic risk factor for AD.1 The pathogenetic mechanism by which ɛ4 might promote the development of AD is still unclear. Several evidences indicate a possible effect of the ɛ4 allele on β-amyloid (Aβ) formation and deposition and a consequent role in the ‘amyloid cascade'.1 This effect, however, could explain only less than half the cases of AD because of the presence of AD patients ɛ4 noncarriers; in fact, there is a general agreement about the ɛ4 allele as a variable neither necessary nor sufficient to develop AD.3, 4 In search for other genetic risk factors potentially associated with AD pathogenesis, three common single nucleotide polymorphisms (SNPs) located in the promoter region of the APOE gene have been described.5 This region, spanning from position –1017 to +406 of the APOE genetic locus (19q13.31), showed three major SNPs, rs449647 (A−491 → T), rs769446 (C−427 → T) and rs405509 (G−219 → T) with a potential functional role.1, 6, 7, 8 The promoter region could modulate the transcriptional activity of APOE coding region,7, 8, 9, 10, 11 and this modified expression of APOE may be the pathogenetic trigger depending on the promoter polymorphisms in AD.11 However, comprehensive analyses of these studies reported contradictory results1, 8 and data on this polymorphic region regarding an association with AD in the Italian population are rare.12 The aim of this study was to assess whether the association among AD and the three SNPs of the promoter region of APOE gene might be considered specific risk factors per se independently of APOE/ɛ4 status.
Materials and methods
Patients and controls
The study was designed as a hospital-based case–control study. It was conducted according to the Declaration of Helsinki Principles and the guidelines for Good Clinical Practice, and was approved by the respective local ethics committee. Written informed consent was obtained from the patients or from their legal guardians before participation into the study. A total of 268 unrelated Caucasians (enrolled from central and southern Italy) were consecutively evaluated at the Neuropsychological Unit of the Catholic University School of Medicine in Rome, Italy. Among these subjects, 169 were diagnosed as probable AD according to NINDS-ADRDA criteria and 99 as cognitively intact (MMSE score >28/30). Clinical diagnosis of sporadic probable AD was confirmed at the follow up after 12 months. In cognitively intact subjects no personal or familial psychiatric or cognitive impairment history, and no alcohol or drug abuse were reported. We considered these subjects as controls (CTRL). Moreover, all subjects affected by cerebrovascular diseases were excluded from the study because they were considered at risk to develop a vascular form of dementia. Clinical and demographic characteristics of patients and controls are reported in Table 1. No extra European as well as Jewish subjects were included in the study.
Table 1. Subjects characteristics.
| AD | Controls | |
|---|---|---|
| Number of subjects | 169 | 99 |
| Sex | ||
| Male/Female | 53/116 | 50/49 |
| Male (%) | 31.36 | 50.51 |
| Mean age (years±SD) | 63.38±7.39 | 66.21±7.31 |
| Early onseta | ||
| Number of subjects | 53 | 43 |
| Mean age (years±SD) | 59.81±3.73 | 59.72±3.32 |
| Late onsetb | ||
| Number of subjects | 116 | 56 |
| Mean age (years±SD) | 72.29±4.93 | 71.20±5.35 |
Abbreviation: AD, Alzheimer's disease.
Age<65 years for both cases and controls.
Age≥65 years for both cases and controls.
Genetic analyses
Genomic DNA was manually purified from 4 ml of frozen blood samples by organic protein extraction and ethanol precipitation according to standard methods. The APOE promoter genotypes were determined by PCR and agarose gel electrophoresis. The rs449647 (A−491 → T) and rs769446 (T−427 → C) genotypes were determined by a previously described method based on a nested PCR.5 A first amplification of 374 bp fragment was obtained using a two-base mismatched forward primer 5′-CATGTTGGCCAGGCTGGTtTtAA-3 and the reverse primer 5′-GGAAGGAGGTGGGGCATAGA-3′ at the following reaction conditions: 94°C for 5 min followed by 30 cycles at 95°C for 30 s, 50°C for 30 s and 72°C for 30 s, with an 8 min of final extension. The second amplification of the 374 bp PCR product, producing a 304 bp fragment, was performed with the same forward mismatched primer and the reverse primer 5′-CCCAGTAATACAGACACCCTCC-3′ at the following conditions: 94°C for 5 min followed by 30 cycles at 95°C for 30 s, 63°C for 30 s and 72°C for 30 s with an 8 min of final extension. Restriction analysis with DraI or AluI permits to identify the rs449647 and rs769446 genotypes on 3% agarose gel. The rs405509 (−219 GT) genotype was determined by a previously described method,13 by an amplification of a 220 bp fragment using the following forward primer 5′-AGAATGGAGGAGGGTGTCCG-3′ and reverse primer 5′-ACTTGTCCAATTATAGGGCTCC-3′. PCR conditions were 94°C for 5 min followed by 35 cycles at 95°C for 30 1 min, 58°C for 1 min and 72°C for 1 min, with an 8 min of final extension. Restriction analysis with HpaII allows to determine the three possible genotypes on 3% standard agarose gel. The rs429358 (C3937 → T) and rs7412 (C4075 → T) genotypes forming the APOE coding region polymorphism were analyzed as recently described.14
Statistical analysis
Agreement of the observed genotype frequencies with the expected Hardy–Weinberg (HW) frequencies was verified for each SNP in both study groups, including the two SNPs generating the common APOE polymorphism. Relative allele frequencies were estimated by the gene-counting method.15 Genotype frequencies in 3 × 2 cross tables were compared by means of Pearson's χ2-test. Comparison of genotype and allele frequencies in 2 × 2 cross tables was made by means of Fisher's exact test according to the two-way contingency table analysis of the Interactive Statistical Calculation Pages (available at URL: http://statpages.org/ctab2x2.html). The odds ratios (ORs) and relative risk (RR) were also calculated. Correlations between the promoter polymorphisms were tested by using the Spearman's correlation rank-sum test. Binary logistic regression analysis using sex and age at onset as covariates was also used to estimate the adjusted significances. Both these analyses were made with the SPSS version 10.1.3 (SPSS Inc., Chicago, IL, USA) statistical software package. The synergy between the investigated SNPs was estimated according to Leandro et al16 assuming an additive model. Briefly, the exceeding of relative excess risk was verified according to the formula RR(AB)−1>RR(A)–1+RR(B)–1, with A and B indicating the genotype to be evaluated. In the stratification of the APOE promoter polymorphism, according to the common APOE polymorphism, we assumed as ɛ2+ genotypes all genotypes containing at least one ɛ2 allele, that is genotypes ɛ2/ɛ2, ɛ2/ɛ3 and ɛ2/ɛ4. Similarly, ɛ4+ genotypes were considered the genotypes ɛ2/ɛ4, ɛ3/ɛ4 and ɛ4/ɛ4. The power of the study, including an estimation of the effect size (h), was also calculated. The haplotype analysis was made with the PHASE software version 2.1.1 assuming default parameters.17, 18 The overall phase probability at the APOE locus was calculated as the product of the probability at each polymorphism. The linkage disequilibrium (LD) analysis was made with the Haploview software version 4.1 (downloaded from URL: http://www.broad.mit.edu/haploview/haploview). If not already specified, all the statistical analyses were made with the R software for statistical computing, version 2.7.2 (downloaded from URL: http://www.r-project.org/). In all statistical analyses a two-tail P-values, setting statistical significance at P<0.05, were considered. ORs and the 95% confidence intervals (95% CI) were also reported.
Results
The APOE promoter polymorphisms
Genotypes and estimated allele frequencies of the three promoter SNPs are reported in Table 2. No differences to the expected HW frequencies were found in both AD and controls for rs449647 (P=0.093 and 0.787, respectively), rs769446 (P=0.208 and 0.182, respectively) and rs405509 (P=0.111 and 0.412, respectively). In the analysis of rs449647 a significant difference was found between AD and controls (P=0.005). The frequency of the wild-type A/A was higher in AD than in controls (76.33 vs 57.58%; OR=2.376, 95% CI 1.397–4.043), whereas the frequency of the A/T heterozygote was lower in AD than in controls (20.12 vs 37.37%; OR=0.422, 95% CI 0.243–0.733). Thus, the frequency of the A allele resulted higher in AD than in controls (0.864 vs 0.763%; OR=1.976, 95% CI 1.261–3.097), whereas the frequency of the T allele resulted lower in AD than in controls (0.136 vs 0.237%; OR=0.506, 95% CI 0.323–0.793). In the analysis of rs769446 no difference was found between AD and controls (P=0.345). In the analysis of rs405509 a significant difference was found between AD and controls (P<0.001). The frequency of the wild-type G/G was higher in AD than in controls (42.60 vs 29.29%; OR=1.792, 95% CI 1.058–3.034), whereas the frequency of the T/T homozygotes was lower in AD than in controls (7.69 vs 25.25%; OR=0.247, 95% CI 0.121–0.505). Thus, the frequency of the G allele was higher in AD than in controls (0.675 vs 0.520%; OR=1.912, 95% CI 1.335–2.738), whereas the frequency of the T allele was lower in AD than in controls (0.325 vs 0.480%; OR=0.523, 95% CI 0.365–0.749). Sex- and age-adjusted estimates for the association of the APOE promoter and APOE polymorphism genotypes with AD were estimated; this analysis of the adjustment for sex and age at onset did not change the strength of the associations (data not shown).
Table 2. Major SNPs in the promoter region of APOE gene.
| AD (n=169) | Controls (n=99) | ||||||
|---|---|---|---|---|---|---|---|
| DNA change (SNP ID) | Genotype/allele | N | Frequency | N | Frequency | OR | 95% CI |
| A−491 → T (rs449647) | A/A | 129 | 76.33% | 57 | 57.58% | 2.376 | 1.397–4.043 |
| A/T | 34 | 20.12% | 37 | 37.37% | 0.422 | 0.243–0.733 | |
| T/T | 6 | 3.55% | 5 | 5.05% | 0.692 | 0.218–2.196 | |
| A | 292 | 0.864 | 151 | 0.763 | 1.976 | 1.261–3.097 | |
| T | 46 | 0.136 | 47 | 0.237 | 0.506 | 0.323–0.793 | |
| T−427 → C (rs769446) | T/T | 129 | 76.33% | 83 | 83.84% | 0.622 | 0.330–1.174 |
| T/C | 35 | 20.71% | 14 | 14.14% | 1.586 | 0.812–3.092 | |
| C/C | 5 | 2.96% | 2 | 2.02% | 1.479 | 0.323–6.721 | |
| T | 293 | 0.867 | 180 | 0.909 | 0.651 | 0.368–1.153 | |
| C | 45 | 0.133 | 18 | 0.133 | 1.536 | 0.867–2.718 | |
| G−219 → T (rs405509) | G/G | 72 | 42.60% | 29 | 29.29% | 1.792 | 1.058–3.034 |
| G/T | 84 | 49.70% | 45 | 45.45% | 1.186 | 0.722–1.947 | |
| T/T | 13 | 7.69% | 25 | 25.25% | 0.247 | 0.121–0.505 | |
| G | 228 | 0.675 | 103 | 0.520 | 1.912 | 1.335–2.738 | |
| T | 110 | 0.325 | 95 | 0.480 | 0.523 | 0.365–0.749 | |
Abbreviations: AD, Alzheimer's disease; SNP, single nucleotide polymorphisms.
Significant values are in boldface.
Spearman's correlation rank-sum test: rs449647 and rs769446, P=0.096; rs449647 and rs405509, P=0.946; rs769446 and rs405509; P=0.308.
Synergistic effect: A/A (rs449647) and G/G (rs405509), relative excess risk 0.365 vs −0.074; A/T (rs449647) and T/T (rs405509), relative excess risk −0.783 vs −0.797.
Genotypes and estimated allele frequencies.
The APOE promoter polymorphisms according to the common APOE alleles
As expected, significant differences were found in the APOE genotype distribution between AD and controls. The frequency of the ɛ2+ genotypes was lower in AD than in controls (7.56 vs 17.00 %; OR=0.399, 95% CI 0.187–0.851). Similarly, the frequency of the ɛ3/ɛ3 genotype was lower in AD than in controls (44.19 vs 73.00%; OR=0.293; 95% CI 0.172–0.498). Conversely, the frequency of the ɛ4+ genotypes was higher in AD than in controls (48.26 vs 10.00%; OR=8.389, 95% CI 4.133–17.009).
Genotype frequencies of the APOE promoter polymorphisms according to the ɛ2, the ɛ3/ɛ3 and ɛ4 status are reported in Table 3. Genotype frequencies of rs449647 according to the APOE status are reported in Table 3a. In ɛ2+ subjects the frequency of A/A genotype was lower in AD than in controls (4.14 vs 12.12%; OR=0.215, 95% CI 0.082–0.566). No differences were found for the A/T and T/T genotypes. In ɛ3/ɛ3 subjects the frequency of the A/A genotype was lower in AD than in controls (34.32 vs 40.40%; OR=0.347, 95% CI 0.180–0.672). Similarly, the frequency of the A/T genotype was lower in AD than in controls (80.88 vs 29.29%; OR=0.218, 95% CI 0.079–0.605). No differences were found for the T/T genotype. In ɛ4+ subjects the frequency of the A/A genotype was higher in AD than in controls (38.46 vs 6.06%; OR=8.633, 95% CI 3.536–20.978). Similarly, the frequency of the A/T genotype was higher in AD than in controls (9.47 vs 4.04%; OR=7.333, 95% CI 2.204–24.046). Notably, the T/T genotype was absent in controls.
Table 3. Genotype frequencies of the most common rs449647 (A), rs769446 (B) and rs44405509 (C) according to the APOE genotypes.
| AD (n=169) | Controls (n=99) | |||||
|---|---|---|---|---|---|---|
| N | (%) | N | (%) | OR | 95% CI | |
| (a) | ||||||
| ɛ2+ | ||||||
| A/A | 7 | (4.14) | 12 | (12.12) | 0.215 | 0.082–0.566 |
| A/T | 5 | (2.96) | 4 | (4.04) | 1.422 | 0.374–5.390 |
| T/T | 1 | (0.59) | 1 | (1.01) | 0.800 | 0.062–10.286 |
| ɛ3/ɛ3 | ||||||
| A/A | 58 | (34.32) | 40 | (40.40) | 0.347 | 0.180–0.672 |
| A/T | 15 | (8.88) | 29 | (29.29) | 0.218 | 0.079–0.605 |
| T/T | 3 | (1.78) | 4 | (4.04) | 0.250 | 0.024–3.035 |
| ɛ4+ | ||||||
| A/A | 65 | (38.46) | 6 | (6.06) | 8.633 | 3.536–20.978 |
| A/T | 16 | (9.47) | 4 | (4.04) | 7.333 | 2.204–24.046 |
| T/T | 2 | (1.18) | — | — | — | — |
| (b) | ||||||
| ɛ2+ | ||||||
| T/T | 11 | (7.69) | 14 | (14.14) | 0.459 | 0.201–1.051 |
| T/C | 2 | (6.51) | 2 | (2.02) | 0.364 | 0.057–2.309 |
| C/C | — | — | 1 | (1.01) | — | — |
| ɛ3/ɛ3 | ||||||
| T/T | 59 | (34.91) | 62 | (62.63) | 0.285 | 0.157–0.521 |
| T/C | 14 | (8.28) | 10 | (10.10) | 0.267 | 0.073–0.982 |
| C/C | 3 | (1.78) | 1 | (1.01) | 1.500 | 0.093–25.586 |
| ɛ4+ | ||||||
| T/T | 61 | 36.09) | 7 | (7.07) | 9.739 | 4.243–22.227 |
| T/C | 20 | (11.83) | 2 | (2.02) | 8.000 | 1.696–36.325 |
| C/C | 2 | (1.18) | 1 | (1.01) | 0.667 | 0.039–10.802 |
| (c) | ||||||
| ɛ2+ | ||||||
| G/G | 5 | (2.96) | 3 | (3.03) | 0.647 | 0.157–2.626 |
| G/T | 7 | (4.14) | 13 | (13.13) | 0.224 | 0.084–0.599 |
| T/T | 1 | (0.59) | 1 | (1.01) | 2.000 | 0.192–20.852 |
| ɛ3/ɛ3 | ||||||
| G/G | 36 | (21.30) | 24 | (924.24) | 0.208 | 0.074–0.590 |
| G/T | 35 | (20.71) | 27 | (27.27) | 0.476 | 0.229–0.990 |
| T/T | 5 | (2.96) | 22 | (22.22) | 0.085 | 0.018–0.418 |
| ɛ4+ | ||||||
| G/G | 33 | (19.53) | 2 | (2.02) | 15.654 | 3.851–62.639 |
| G/T | 42 | (24.85) | 6 | (6.06) | 6.500 | 2.540–16.527 |
| T/T | 8 | (4.73) | 2 | (2.02) | 18.400 | 3.217–101.166 |
Abbreviation: AD, Alzheimer's disease.
AD group: ɛ2/ɛ3, 5.92% (n=10); ɛ2/ɛ4, 1.78% (n=3); ɛ3/ɛ3, 44.97% (n=76); ɛ3/ɛ4, 40.83% (n=69); ɛ4/ɛ4, 6.51% (n=11). HWE P=0.931. Control group: ɛ2/ɛ3, 16.16% (n=16); ɛ2/ɛ4, 1.01% (n=1); ɛ3/ɛ3, 73.74% (n=73); ɛ3/ɛ4, 9.09% (n=9). HWE P=0.999.
Significant values are in boldface.
Genotype frequencies of rs769446 according to the APOE status are reported in Table 3b. In ɛ2+ subjects no differences were found in the frequency distribution of the A/T or the T/C genotypes between cases and controls. No C/C genotypes were found in AD. In ɛ3/ɛ3 subjects, the frequency of the T/T genotype was lower in AD than in controls (34.91 vs 62.63%; OR=0.285, 95% CI 0.157–0.521). Similarly, the frequency of the T/C genotype was lower in AD than in controls (8.28 vs 10.10%; OR=0.267, 95% CI 0.073–0.982). In ɛ4+ subjects the frequency of the T/T genotype was higher in AD than in controls (36.09 vs 7.07%; OR=9.739, 95% CI 4.243–22.227). Similarly, the frequency of the T/C genotype was higher in AD than in controls (11.83 vs 2.02%; OR=8.000, 95% CI 1.696–36.325).
Genotype frequencies of rs44405509 according to the APOE status are reported in Table 3c. In ɛ2+ subjects a lower frequency of the G/T genotype was found in AD than in controls (4.14 vs 13.13%; OR=0.224, 95% CI 0.084–0.599), whereas no differences were found for the G/G or T/T genotypes. In ɛ3/ɛ3 subjects, a lower frequency of the G/G genotype was found in AD than in controls (21.30 vs 24.24; OR=0.208, 95% CI 0.074–0.590). Similarly, a lower frequency was found for both G/T and T/T genotypes in AD than in controls (20.71 vs 27.27%; OR=0.476, 95% CI 0.229–0.990 and 2.96 vs 22.22%; OR=0.085, 95% CI 0.018–0.418, respectively). In ɛ4+ subjects a higher frequency in AD than in controls was found for G/G (19.53 vs 2.02%; OR=15.654, 95% CI 3.851–62.639), G/T (24.85 vs 6.06%; OR=6.500, 95% CI 2.540–16.257) and T/T (4.73 vs 2.02%; OR=18.400, 95% CI 3.217–101.166).
Haplotype analysis
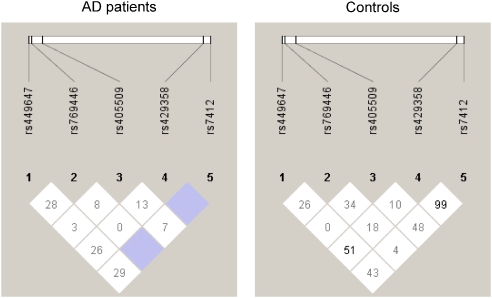
Estimation of the haplotype frequencies in the 3 kb block spanning the APOE gene locus is summarized in Table 4. The ATG3 haplotype resulted the most represented in both AD and controls (37.87 and 34.85%, respectively), followed in decreasing order by the ATT3 haplotype (11.24 vs 26.26%, respectively). In AD or in controls all the other haplotypes showed a frequency less than 10%. Notably, the total frequency of the haplotypes showing both the −491 A and the −427 T alleles, that is, the first six haplotypes in Table 4 (AT− haplotypes), represented more than 70% of the total frequency of the estimated haplotypes (73.66 and 72.23%, respectively for AD and controls). In the analysis, the frequency of the ATG4 haplotype (10.36 vs 1.01%; OR=11.320, 95% CI 2.971–43.025) as well as ATT4 haplotype (11.83 vs 3.03%; OR=4.295, 95% CI 1.829–10.069) was higher in AD than in controls. Notably, the frequency of the ACG3 haplotype was also higher in AD than in controls (10.95 vs 1.01%; OR=12.047, 95% CI 3.167–45.709). Conversely, the frequency of the ATT2 haplotype (1.18 vs 5.56%; OR=0.204, 95% CI 0.068–0.615), as well as the ATT3 haplotype (11.24% vs 26.26%; OR=0.356, 95% CI 0.224–0.564) and the TTG3 haplotype (3.55 vs 12.12%; OR=0.267, 95% CI 0.132–0.540) was lower in AD than in controls. A schematic representation of the LD coefficient D′ among the five SNPs, that is, the three SNPs in the promoter region and the two in the coding region of the APOE gene, is shown in Figure 1. As expected, according to the genetics of the APOE polymorphism,19 a strong LD was observed in AD between markers rs429358 and rs7412 (D′=1.0). Similarly, an LD between the same markers was suggested in controls (D′=0.9). We also observed in AD, and not in controls, a strong LD between markers rs769446 and rs7412 (D′=1.0). Conversely, the presence of LD was suggested in controls and not in AD between the markers rs449647 and rs429358 (D′=0.5).
Table 4. Haplotype frequencies in the 3 kb block spanning the APOE locus.
| AD (n=338) | Controls (n=198) | |||||
|---|---|---|---|---|---|---|
| Haplotype | N | (%) | N | (%) | OR | 95% CI |
| ATG2 | 4 | (1.18) | 3 | (1.52) | 0.778 | 0.193–3.140 |
| ATG3 | 128 | (37.87) | 69 | (34.85) | 0.140 | 0.791–1.641 |
| ATG4 | 35 | (10.36) | 2 | (1.01) | 11.320 | 2.971–43.025 |
| ATT2 | 4 | (1.18) | 11 | (5.56) | 0.204 | 0.068–0.615 |
| ATT3 | 38 | (11.24) | 52 | (26.26) | 0.356 | 0.224–0.564 |
| ATT4 | 40 | (11.83) | 6 | (3.03) | 4.295 | 1.829–10.069 |
| ACG3 | 37 | (10.95) | 2 | (1.01) | 12.047 | 3.167–45.709 |
| ACG4 | — | — | 2 | (1.01) | — | — |
| ACT2 | — | — | 1 | (0.51) | — | — |
| ACT3 | 1 | (0.30) | 3 | (1.52) | 0.193 | 0.027–1.359 |
| ACT4 | 5 | (1.48) | — | — | — | — |
| TTG2 | 4 | (1.18) | 1 | (0.51) | 2.359 | 0.351–15.780 |
| TTG3 | 12 | (3.55) | 24 | (12.12) | 0.267 | 0.132–0.540 |
| TTG4 | 6 | (1.78) | — | — | — | — |
| TTT2 | 1 | (0.30) | — | — | — | — |
| TTT3 | 13 | (3.85) | 12 | (6.06) | 0.620 | 0.282–1.362 |
| TTT4 | 8 | (2.37) | — | — | — | — |
| TCG3 | 2 | (0.59) | — | — | — | — |
| TCT2 | — | — | 1 | (0.51) | — | — |
| TCT3 | — | — | 9 | (4.55) | — | — |
Abbreviation: AD, Alzheimer's disease.
Significant values are in boldface.
Figure 1.
Schematic representation of linkage disequilibrium (D′ coefficient) among the markers spanning a 3 kb block at the APOE gene locus.
The phase probability, for each marker and for the 3 kb block spanning the APOE locus, for the ɛ2- and ɛ4-containing haplotypes common to both AD and controls is summarized in Table 5. No difference in phase probability estimation was found between AD and controls for the ɛ2- and ɛ4-containing haplotypes (data not shown).
Table 5. Phase probability at the APOE locus.
| AD | CTRS | Single phase probabilitya | Overall phase probability |
|---|---|---|---|
| ATG2/ATG3 | ATG2/ATG3 | k k k 1.00 | 1.00 |
| ATG3/ATG4 | ATG3/ATG4 | k k k 1.00 | 1.00 |
| ATG3/ATT2 | ATG3/ATT2 | k k 0.50 1.00 | 0.50 |
| ATG3/ATT4 | ATG3/ATT4 | k k 0.50 1.00 | 0.50 |
| ATT3/ATT4 | ATT3/ATT4 | k k k 1.00 | 1.00 |
Abbreviation: AD, Alzheimer's disease.
k indicates a known phase.
An estimation of the power of the analysis of this study based on the association of the rs449647 A/A genotype showed an h=0.402 with a power of 0.889. To be even more accurate in the estimate of the power of the study it would be possible to make the calculation on the less common genotype where an association resulted (T/T of the rs405509). As a consequence the estimate would be even more relevant than the previous one resulting an h=0.491 and a power of 0.972.
Discussion
The main purpose of this study was to analyze the role of the major APOE promoter polymorphisms in AD susceptibility and their interactions with the APOE polymorphism.
This study shows a statistically significant increased frequency of the A/A genotype and A allele of the rs449647 in AD. This result is consistent with that of a previous Italian study,12 reporting an increased A/A genotype and A allele frequencies in AD. We confirmed this association also when the estimated rs449647 frequencies were adjusted for sex and age at onset. This association has been reported by previous studies9, 12, 20, 21, 22, 23, 24, 25 and resulted in agreement with similar data collected from several independent samples genotyped for the apoE coding region and APOE promoter polymorphisms.26 Whereas our data support these reported positive associations between rs449647 and AD, other studies failed to show similar results.27, 28, 29 It is not still clear how rs449647 exerts its effect on AD risk. A possible explanation might be its potential role to modify the APOE transcriptional activity.10, 30 Several studies reported that APOE expression is important for brain amyloid loading and deposition in AD and in elderly healthy individuals, also independently from APOE genotype.11, 31, 32 It might be possible that the risk for AD may be modulated by the apoE protein or apoE mRNA levels as well as by the common apoE protein isoforms.33, 34 Recent studies30, 31 supported the notion that rs449647 might be related to variations in apoE brain levels and reported an increased level of apoE in the brains of AD carrying the A/A genotype as compared to controls with the same genotype. Significantly higher plasma apoE levels, independent from the ɛ4 status, in AD subjects carrying the A/A genotype have been reported.32 On the contrary, another recent study, performed as a part of the Rotterdam study,24 suggested a lowering effect of the rs449647 A/A genotype on plasma apoE levels in AD. Thus, the biological effect of the rs449647 polymorphism remains controversial.
The analysis of the distribution of genotypic and estimated allelic frequencies of rs769446 did not show any difference between AD and controls. These data are in agreement with previous studies.9, 13 One of the earlier investigations on the possible functional effects of the ApoE promoter polymorphisms5 showed that only the –491 and the –219 polymorphic sites produce variations on the transcriptional activity of the APOE coding region, whereas the T to C substitution at the rs769446 site had no significant effect on APOE promoter activity. On the other hand, a study carried out on a Spanish population reported an association between rs769446 polymorphism and AD showing an increased risk for AD in subjects bearing the A−491 and C−427 haplotypes.11 The results of this study suggested a possible effect of the C−427 allele on the transcriptional activity of the APOE gene with a consequent higher level of apoE in plasma and brain. At variance, a recent study on a French population35 showed a positive association between T−427 allele and AD risk; in the same study the −427 allele has been found in the haplotype conferring an increased risk for AD. However, as observed in these two studies, the positive association between rs769446 and AD shows contrasting results on the possible allele (T or C) conferring a higher AD risk. The analysis of the distribution of the genotypic and estimated allelic frequencies of rs405509 showed an overrepresentation of G/G genotype and the G allele in AD patients according to Myllykangas et al.36 At variance, our results are in contrast with those carried out on French population9, 22 reporting an increased frequency of the T−219 allele in AD patients. The same results were obtained in elderly AD subjects as reported in a recent meta-analysis26 and in other studies on population-based cohort.37, 38 In line with these conflicting results, it is not clear yet how the G−219 → T polymorphism exerts its possible biological effect in AD.5, 40
To evaluate the possible interaction between the promoter variants and the common APOE polymorphism, we also classified the promoter polymorphisms according to the ɛ2+, ɛ3/ɛ3 and ɛ4+ APOE genotypes. For rs449647 the overrepresentation of the A/A genotype and for rs405509 the overrepresentation of the G/G genotype observed in AD were confirmed only in presence of the ɛ4+ genotypes. For rs769446 significant differences were observed, meanly in the presence of ɛ3/ɛ3 and ɛ4+ genotypes. However, these associations may not be explained by LD (Figure 1).
The haplotype analysis showed an overrepresentation of the ATG family haplotype in AD (P=0.007) and confirmed previous results describing the haplotypes of the ATG family as the most frequent haplotypes, and one ATG- haplotype at risk for AD.35 At variance, we did not confirm an overrepresentation of the ATT- haplotype in AD,35 (P=0.010). Haplotype analysis also showed a significant increased frequencies of the two major haplotypes of the -ɛ4 family (ATG4 and ATT4) in AD. The strongest association to AD observed for the ATG4 haplotype, compared to the ATT4, might be explained by the presence of the G−219 allele. The overall association of these two haplotypes of the ɛ4 family, however, slightly changed the association of the ɛ4 allele with AD. On the contrary, two haplotypes of the -ɛ3 family (ATT3 and TTG3) showed a significant decreased frequency in AD. The overall association of these haplotypes of the -ɛ3 family, however, did not significantly change the association of the ɛ3 allele. Notably, we observed an -ɛ3 haplotype (ACG3) that was overrepresented in AD than in controls. This result might suggest a possible interaction between A−491 and C−427 alleles, independently from APOE4. A similar observation was previously reported by Artiga et al11 on Spanish population.
Despite several studies on the APOE promoter polymorphisms in AD, the haplotype analysis is not common and, when estimated, it has been often restricted to one or two promoter polymorphisms.27, 39 A Finnish study,27 estimating a two-point haplotype (rs449647 and the common exon 4 polymorphism) indicated the A4 as the higher-risk haplotype for AD and this haplotype frequency was the same of the ɛ4 allele in AD group confirming an LD between the two alleles. This result is in agreement with another earlier study,39 which found the highest AD risk for the A4 haplotype and the lowest for the T3 ones. On the contrary, a recent study on Colombian population using an haplogroup analysis of the APOE promoter polymorphisms confirmed their independent contribution as genetic risk factors for AD.40 Different possible explanations of the contrasting results on allelic and haplotype distribution of the APOE promoter polymorphisms have been reported. The first might be the presence of an LD among the different alleles of the APOE promoter polymorphisms and the APOE coding region. The real risk-conferring allele such as that reported for the A−491 might bear persistently another allele in the haplogroup or haplotype. A second possible explanation, considering the absence of an LD in our sample, might be that for a specific combination of alleles or genotypes of the APOE promoter polymorphisms it is necessary to perform the complex interaction between these biological factors that led to a modified transcriptional activity of the APOE coding region.
To investigate the possible cis-acting interaction of quantitative promoter effects on qualitative effects of the APOE coding variants we estimated the phase probability for the investigated haplotypes. The phase probabilities of the genotypes common to both AD and controls are summarized in Table 5. In fact, it has been reported that the T−491 allele caused a decrease in APOE promoter activity, whereas the T−219 caused an increased promoter activity.11, 31 Thus, haplotypes containing these alleles may have a cis-acting effects, in particular on the expression of the ɛ4 allele.
The contrasting findings reported by previous studies on APOE promoter polymorphisms and AD probably depend on background heterogeneity and sample selection criteria, both important parameters for the APOE polymorphisms evaluation. These parameters include ethnic origins and the age range of the investigated samples. Therefore our study confirms the conclusions reached by other studies on Caucasians, especially on Spanish population samples, about a function of the promoter polymorphisms in AD, suggesting that the same genetic background might be the more plausible explanation for these similar results. However, future studies on larger cohort of AD might further increase the power of the analysis as well as confirm the possible risk/protective effects associated with these polymorphisms/haplotypes and their complex interaction.
Acknowledgments
This study was partially supported by a grant from the Italian Ministry of Health (4AN/F9-2003-06) to CM and IRCCS Research Program, Ricerca Corrente 2006-2008, Linea n. 2 ‘Malattie di rilevanza sociale'.
References
- St George-Hyslop PH, Petit A. Molecular biology and genetics of Alzheimer's disease. C R Biol. 2005;328:119–130. doi: 10.1016/j.crvi.2004.10.013. [DOI] [PubMed] [Google Scholar]
- Zawia NH, Basha MR. Environmental risk factors and the developmental basis for Alzheimer's disease. Rev Neurosci. 2005;16:325–337. doi: 10.1515/revneuro.2005.16.4.325. [DOI] [PubMed] [Google Scholar]
- American College of Medical Genetics/American Society of Human Genetics Working Group on ApoE and Alzheimer Disease Statement on use of apolipoprotein E testing for Alzheimer disease. JAMA. 1995;274:1627–1629. [PubMed] [Google Scholar]
- National Institute on Aging/Alzheimer's Association Working Group Apolipoprotein E genotyping in Alzheimer's disease. Lancet. 1996;347:1091–1095. [PubMed] [Google Scholar]
- Artiga MJ, Bullido MJ, Sastre I, et al. Allelic polymorphisms in the transcriptional regulatory region of apolipoprotein E gene. FEBS Lett. 1998;421:105–108. doi: 10.1016/s0014-5793(97)01543-3. [DOI] [PubMed] [Google Scholar]
- Bullido MJ, Valdivieso F. Apolipoprotein E gene promoter polymorphisms in Alzheimer's disease. Microsc Res Tech. 2000;50:261–267. doi: 10.1002/1097-0029(20000815)50:4<261::AID-JEMT2>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- Alvarez-Arcaya A, Combarros O, Llorca J, Sanchez-Guerra M, Berciano J, Fernandez-Luna JL. The −491 TT apolipoprotein E promoter polymorphism is associated with reduced risk for sporadic Alzheimer's disease. Neurosci Lett. 2001;304:204–208. doi: 10.1016/s0304-3940(01)01790-6. [DOI] [PubMed] [Google Scholar]
- Bray NJ, Jehu L, Moskvina V, et al. Allelic expression of APOE in human brain: effects of epsilon status and promoter haplotypes. Hum Mol Genet. 2004;13:2885–2892. doi: 10.1093/hmg/ddh299. [DOI] [PubMed] [Google Scholar]
- Lambert JC, Berr C, Pasquier F, et al. Pronounced impact of Th1/E47cs mutation compared with −491 AT mutation on neural APOE gene expression and risk of developing Alzheimer's disease. Hum Mol Genet. 1998;7:1511–1516. doi: 10.1093/hmg/7.9.1511. [DOI] [PubMed] [Google Scholar]
- Laws SM, Hone E, Gandy S, Martins RN. Expanding the association between the APOE gene and the risk of Alzheimer's disease: possible roles for APOE promoter polymorphisms and alterations in APOE transcription. J Neurochem. 2003;84:1215–1236. doi: 10.1046/j.1471-4159.2003.01615.x. [DOI] [PubMed] [Google Scholar]
- Artiga MJ, Bullido MJ, Frank A, et al. Risk for Alzheimer's disease correlates with transcriptional activity of the APOE gene. Hum Mol Genet. 1998;7:1887–1892. doi: 10.1093/hmg/7.12.1887. [DOI] [PubMed] [Google Scholar]
- Casadei VM, Ferri C, Veglia F, et al. APOE −491 promoter polymorphism is a risk factor for late-onset Alzheimer's disease. Neurology. 1999;53:1888–1889. doi: 10.1212/wnl.53.8.1888. [DOI] [PubMed] [Google Scholar]
- Wang JC, Kwon JM, Shah P, Morris JC, Goate A. Effect of APOE genotype and promoter polymorphism on risk of Alzheimer's disease. Neurology. 2000;55:1644–1649. doi: 10.1212/wnl.55.11.1644. [DOI] [PubMed] [Google Scholar]
- Seripa D, Signori E, Gravina C, Matera MG, Rinaldi M, Fazio VM. Simple and effective determination of apolipoprotein E genotypes by positive/negative polymerase chain reaction products. Diagn Mol Path. 2006;15:180–185. doi: 10.1097/01.pdm.0000213451.99655.1d. [DOI] [PubMed] [Google Scholar]
- Gerdes LA, Klausen IB, Sihm I, Færgeman O. Apolipoprotein E polymorphism in a Danish population compared to findings in 45 other study populations around the world. Genet Epidemiol. 1992;9:155–167. doi: 10.1002/gepi.1370090302. [DOI] [PubMed] [Google Scholar]
- Leandro G, Duca P. The role of hepatitis B and C viruses in hepatocellular carcinoma in a hepatitis B endemic area: a case–control study. Cancer. 1993;71:510–511. doi: 10.1002/1097-0142(19930115)71:2<510::aid-cncr2820710236>3.0.co;2-n. [DOI] [PubMed] [Google Scholar]
- Stephens M, Smith N, Donnelly P. A new statistical method for haplotype reconstruction from population data. Am J Hum Genet. 2001;68:978–989. doi: 10.1086/319501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens M, Donnelly P. A comparison of Bayesian methods for haplotype reconstruction from population genotype data. Am J Hum Genet. 2003;73:1162–1169. doi: 10.1086/379378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seripa D, Matera MG, Daniele A, et al. The missing ApoE allele. Ann Hum Genet. 2007;71:496–500. doi: 10.1111/j.1469-1809.2006.00344.x. [DOI] [PubMed] [Google Scholar]
- Ahmed ARH, MacGowan SH, Culpan D, Jones RW, Wilcock GK. The −491 A/T polymorphism of the apolipoprotein E gene is associated with the ApoEe4 allele and Alzheimer's disease. Neurosci Lett. 1999;263:217–219. doi: 10.1016/s0304-3940(99)00126-3. [DOI] [PubMed] [Google Scholar]
- Lambert JC, Mann D, Goumidi L, et al. Effect of the APOE promoter polymorphisms on cerebral amyloid peptide deposition in Alzheimer's disease. Lancet. 2001;357:608–609. doi: 10.1016/S0140-6736(00)04063-0. [DOI] [PubMed] [Google Scholar]
- Lambert JC, Pasquier F, Cottel D, Frigard B, Amouyel P, Chartier-Harlin MC. A new polymorphism in the APOE promoter associated with risk of developing Alzheimer's disease. Hum Mol Genet. 1998;7:533–540. doi: 10.1093/hmg/7.3.533. [DOI] [PubMed] [Google Scholar]
- Nicodemus KK, Stenger JE, Schmechel DE, et al. Comprehensive association analysis of APOE regulatory region polymorphisms in Alzheimer disease. Neurogenetics. 2004;5:201–208. doi: 10.1007/s10048-004-0189-9. [DOI] [PubMed] [Google Scholar]
- Roks G, Cruts M, Houwing-Duistermaat JJ, et al. Effect of the APOE −491A/T promoter polymorphism on apolipoprotein E levels and risk of Alzheimer disease: the Rotterdam Study. Am J Med Genet. 2002;114:570–573. doi: 10.1002/ajmg.10407. [DOI] [PubMed] [Google Scholar]
- Yang JD, Feng GY, Zhang J, Cheung J, St Clair D, He L. Apolipoprotein E −491 promoter polymorphism is an independent risk factor for Alzheimer's disease in the Chinese population. Neurosci Lett. 2003;350:25–28. doi: 10.1016/s0304-3940(03)00815-2. [DOI] [PubMed] [Google Scholar]
- Lambert JC, Araria-Goumidi L, Myllykangas L, et al. Contribution of APOE promoter polymorphisms to Alzheimer's disease risk. Neurology. 2002;59:59–66. doi: 10.1212/wnl.59.1.59. [DOI] [PubMed] [Google Scholar]
- Helisalmi S, Hiltunen M, Valonen P, et al. Promoter polymorphism (−491 A/T) in the APOE gene of Finnish Alzheimer's disease patients and control individuals. J Neurol. 1999;246:821–824. doi: 10.1007/s004150050461. [DOI] [PubMed] [Google Scholar]
- Song YQ, Rogaeva E, Premkumar S, et al. Absence of association between Alzheimer disease and the −491 regulatory region polymorphism of APOE. Neuroscience Lett. 1998;250:189–192. doi: 10.1016/s0304-3940(98)00470-4. [DOI] [PubMed] [Google Scholar]
- Toji H, Maruyama H, Sasaki K, Nakamura S, Kawakami H. Apolipoprotein E promoter polymorphism and sporadic Alzheimer's disease in a Japanese population. Neurosci Lett. 1999;259:56–58. doi: 10.1016/s0304-3940(98)00855-6. [DOI] [PubMed] [Google Scholar]
- Laws SM, Hone E, Taddei K, et al. Variation at the APOE −491 promoter locus is associated with altered brain levels of apolipoprotein E. Mol Psychiatry. 2002;7:886–890. doi: 10.1038/sj.mp.4001097. [DOI] [PubMed] [Google Scholar]
- Laws SM, Taddei K, Martins G, et al. The −491 AA polymorphism in the APOE gene is associated with increased plasma apoE levels in Alzheimer's disease. Neuroreport. 1999;10:879–882. doi: 10.1097/00001756-199903170-00038. [DOI] [PubMed] [Google Scholar]
- Pahnke J, Walker LC, Schroeder E, et al. Cerebral beta-amyloid deposition is augmented by the –491 AA promoter polymorphism in non-demented elderly individuals bearing the apolipoprotein E ɛ4 allele. Acta Neuropathol. 2003;105:25–29. doi: 10.1007/s00401-002-0602-0. [DOI] [PubMed] [Google Scholar]
- Holtzman DM, Bales KR, Tenkova T, et al. Apolipoprotein E isoform-dependent amyloid deposition and neuritic degeneration in a mouse model of Alzheimer's disease. Proc Natl Acad Sci USA. 2000;97:2892–2897. doi: 10.1073/pnas.050004797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holtzman DM, Bales KR, Wu S, et al. Expression of human apolipoprotein E reduces amyloid-beta deposition in a mouse model of Alzheimer's disease. J Clin Invest. 1999;103:R15–R21. doi: 10.1172/JCI6179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zurutuza L, Verpillat P, Raux G, et al. APOE promoter polymorphisms do not confer independent risk for Alzheimer's disease in a French population. Eur J Hum Genet. 2000;8:713–716. doi: 10.1038/sj.ejhg.5200513. [DOI] [PubMed] [Google Scholar]
- Myllykangas L, Polvikoski T, Reunanen K, et al. ApoE e3-haplotype modulates Alzheimer beta-amyloid deposition in the brain. Am J Med Genet. 2002;114:288–291. doi: 10.1002/ajmg.10202. [DOI] [PubMed] [Google Scholar]
- Heijmans BT, Slagboom PE, Gussekloo J, et al. Association of APOE varepsilon2/varepsilon3/varepsilon4 and promoter gene variants with dementia but not cardiovascular mortality in old age. Am J Med Genet. 2002;107:201–208. doi: 10.1002/ajmg.10142. [DOI] [PubMed] [Google Scholar]
- Lambert JC, Berr C, Cottel D, Amouyel P, Helbecque N. APOE promoter polymorphisms and dementia in the elderly. Neurosci Lett. 2004;365:116–119. doi: 10.1016/j.neulet.2004.04.063. [DOI] [PubMed] [Google Scholar]
- Town T, Paris D, Fallin D, et al. The –491A/T apolipoprotein E promoter polymorphism association with Alzheimer's disease: independent risk and linkage disequilibrium with the known APOE polymorphism. Neurosci Lett. 1998;252:95–98. doi: 10.1016/s0304-3940(98)00567-9. [DOI] [PubMed] [Google Scholar]
- Parra-Bonilla G, Arboleda G, Yunis J, et al. Haplogroup analysis of the risk associated with APOE promoter polymorphism (−219T/G, −491A/T and –427T/C) in Colombian Alzheimer's disease patients. Neurosci Lett. 2003;349:159–162. doi: 10.1016/s0304-3940(03)00816-4. [DOI] [PubMed] [Google Scholar]