Abstract
Several lines of evidence link glucose-6-phosphate dehydrogenase (G6PD) deficiency to protection from severe malaria. Early reports suggested most G6PD deficiency in sub-Saharan Africa was because of the 202A/376G G6PD A− allele, and recent association studies of G6PD deficiency have employed genotyping as a convenient way to determine enzyme status. However, further work has suggested that other G6PD deficiency alleles are relatively common in some regions of West Africa. To investigate the consequences of unrecognized allelic heterogeneity on association studies, in particular studies of G6PD deficiency and malaria, we carried out a case–control analysis of 2488 Gambian children with severe malaria and 3875 controls. No significant association was found between severe malaria and the 202A/376G G6PD A− allele when analyzed alone, but pooling 202A/376G with other deficiency alleles revealed the signal of protection (male odds ratio (OR) 0.77, 95% CI 0.62–0.95, P=0.016; female OR 0.71, 95% CI 0.56–0.89, P=0.004). We have identified the 968C mutation as the most common G6PD A− allele in The Gambia. Our results highlight some of the consequences of allelic heterogeneity, particularly the increased type I error. They also suggest that G6PD-deficient male hemizygotes and female heterozygotes are protected from severe malaria.
Keywords: genetic association study, G6PD deficiency, Plasmodium falciparum, allelic heterogeneity
Introduction
Geographical, epidemiological and in vitro evidence support the hypothesis that glucose-6-phosphate dehydrogenase (G6PD) deficiency confers protection from disease caused by the Plasmodium falciparum parasite.1 G6PD is a key component in the pentose phosphate pathway, employed by erythrocytes to handle oxidative damage. After invasion of host erythrocytes, malaria parasites digest haemoglobin to provide growing space and to obtain nutrients. This process releases toxic by-products, inducing oxidative stress on the cell. G6PD is encoded by a 16.2 kb gene found on the X chromosome. Approximately 160 genetic variants causing clinical deficiency of G6PD have been characterized.2 The geographical distribution of these deficiency alleles closely reflects populations exposed historically to endemic malaria.3, 4
In 1995, a case–control study of 2039 African children from The Gambia and Kenya reported that female heterozygotes and G6PD-deficient male hemizygotes were protected from severe malaria.5 Recently, another study of 3197 Malian children (2765 with uncomplicated malaria and 432 with severe malaria) reported that hemizygous males, and not heterozygous females, were protected against severe disease.6 Earlier studies indicated generally that male hemizygotes and female homozygotes were protected from malaria,3, 7, 8, 9 although a few authors have suggested that it is the female heterozygotes who receive protection.10, 11, 12 A number of methodological issues have been highlighted to explain these different findings, including the small sample size of earlier studies, variation in phenotype definition, choice of controls, village surveys vs hospital-based studies, age or immune status of subjects and study location.6, 13, 14
The 202A/376G G6PD A− allele is considered to be the most common G6PD deficiency allele in sub-Saharan Africa. This allele has two non-synonymous single nucleotide polymorphisms (SNPs) at positions 202 and 376 of the coding sequence,15 and exhibits around 12% of normal enzyme activity compared with that of the wild-type G6PD B allele.5, 16 Haplotypes with only a derived allele at position 376, and not at 202, mark the G6PD A allele, which has around 85% of normal enzyme activity. Other G6PD deficiency alleles are known to exist in sub-Saharan Africa, but are thought to make up only 5% of the total (an estimate based on an early study of 20 African Americans).1, 16 Owing to the relative ease of high-throughput genotyping, modern disease association studies have employed DNA genotyping, focusing on the G202A/A376G SNP combination.5, 6 However, a recent study from Senegal in West Africa found two other common deficiency alleles in the Sereer population G6PD Santamaria (542T/376G, 1% frequency) and G6PD Betica-Selma (968C/376G, 10%).17
The rates of G6PD deficiency in West Africa range around 10–20% or more,18 and yet the 202A allele frequency is frequently substantially lower (Senegal (Sereer), 1.0% frequency;17 Sierre Leone (Mende), 3.9%;4 Ghana (Fante), 5.7%;4 The Gambia (mixed), 5.9%;5 Ghana (mixed), 10.5%;4 Sierre Leone (Temne), 10.5%;4 Mali (Dogon and Malinke), 15.3%;6 Ghana (Ga), 18.9%;4 Nigeria (Yoruba), 21.3%4). These discrepancies raise the possibility that alternative G6PD deficiency alleles may be relatively common in parts of West Africa, and may sometimes outnumber the 202A/376G G6PD A− allele. We set out to investigate whether the allelic heterogeneity of G6PD deficiency could complicate genetic association studies of severe malaria susceptibility. We carried out a population-based association analysis of 2488 Gambian children with severe malaria and 3875 controls. We genotyped G6PD G202A and A376G along with the three other G6PD variants, studied in Senegal.17 The Gambia is geographically close to Senegal in West Africa, contains members of the Sereer ethnic group and is one of the three populations studied previously in large genetic association studies.5
Methods
Patient samples were collected as part of the ongoing epidemiological studies regarding severe malaria at the Royal Victoria Hospital, Banjul, The Gambia. Cases were children (mostly <5 years of age) presenting with an evidence of P. falciparum on blood film and clinical features of severe malaria, such as cerebral malaria (CM), severe malarial anaemia (SMA), or another severe complications, such as respiratory distress or death.19, 20 CM was defined as a significant impairment of consciousness (Blantyre coma score ≤3) not attributable to convulsions, hypoglycaemia, meningitis or another cause.21 SMA was defined as haemoglobin concentration <5 g/dl or packed cell volume <15%. The distribution of phenotypes among the cases was: CM (63.8%), SMA (19.1%), both CM and SMA (11.3%) and others (mostly respiratory distress) (5.8%). Controls were cord blood samples obtained from birth clinics in the same regions as the cases. All DNA samples were collected and genotyped after obtaining approval from the relevant research ethics committees and informed consent from participants. The majority (84.2%) of participants are from four self-reported ethnic groups (Mandinka, 34.7%; Fula, 18.0%; Jola, 17.0%; Wollof, 14.5%; others, 15.8%, including Sereer, 3.1%), consistent with the ethno-demographics of The Gambia.
Genomic DNA samples underwent whole genome amplification through either Primer Extension Pre-amplification (PEP)22 or Multiple Displacement Amplification (MDA),23 before genotyping on a Sequenom MassArray genotyping platform.24, 25 Details of the genotyped SNPs are documented in Figure 1 and Table 1. The G6PD G202A and A376G SNPs were genotyped in all 6363 samples. The additional three SNPs (A542T, G680T and T968C) were genotyped in a subset (1999 cases and 620 controls). Sequenom genotyping for the sickle variant (HbS, rs334) was carried out for all samples as described previously.26 Genotype calling was carried out using an automated algorithm, followed by visual inspection to assess quality. As the proportion of male heterozygotes was low (0.2%), they were removed from the analysis. The rate of genotype call discordance in the 10% of samples replicated was low (0.2%). No deviations from the Hardy–Weinberg equilibrium (Supplementary Table 1) were observed in population controls (P>0.05). The SNP and haplotype association analysis was undertaken using logistic regression, with an adjustment for the potentially confounding effects of ethnic groups (Fula, Jola, Mandinka, Wollof, other) and HbS status. The association analysis of each ethnic group was not carried out individually because of small sample size. Haplotypes were reconstructed using an expectation-maximization algorithm. Statistical analysis was carried out using the R software package (http://www.r-project.org).
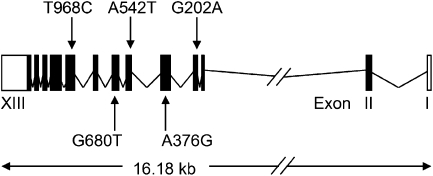
Figure 1.
G6PD exons and relative positions of coding variation studied. The G6PD gene is coded on the negative strand of the X chromosome. The gene is, therefore, drawn from Exon 1 (right) to Exon 13 (left). The position of the 5 single nucleotide polymorphisms studied are marked with arrows, and annotated by their cDNA position and alleles (see Table 1, for more details).
Table 1. Properties of the G6PD polymorphisms genotyped.
| Chromosome: coordinatea | Ref SNP identifier | Coding DNA positionb | Allelesc | Amino acid position | Residue change |
|---|---|---|---|---|---|
| X:153417411 | rs1050828 | 202 | G/A | 68 | V/M |
| X:153416686 | rs1050829 | 376 | A/G | 126 | N/D |
| X:153415849 | rs5030872 | 542 | A/T | 181 | D/V |
| X:153415534 | d | 680 | G/T | 227 | R/L |
| X:153414434 | d | 968 | T/C | 323 | L/P |
NCBI 36 assembly of the human genome (November 2005).
The originally described cDNA position. These cDNA positions vary according to the G6PD transcript and genome build. We refer to these designations for continuity in this paper, and include here the rs numbers, if assigned, and their chromosomal coordinates. (http://www.ensembl.org/Homo_sapiens/geneview?gene=ENSG00000160211). RefSNP accession ID (rs number).
Negative (coding) strand, ancestral/derived); 202A/376G is a G6PD A− allele, 376A is the G6PD B allele, 542T/376G is the G6PD Santamaria allele, 680T/376G is a G6PD A− allele and 968C/376G is the G6PD Betica-Selma allele.
Not in dbSNP.
Results
We genotyped the G6PD SNPs, G202A and A376G, in 2488 children with severe malaria and 3875 cord blood controls from The Gambia. We began by analyzing the G6PD G202A and A376G SNPs individually (Table 2). The frequency of 202A was lower than reported previously (all controls, 2.8%; Fula, 3.4%; Jola, 1.2%; Mandinka, 2.9%; Wollof, 3.3%; other ethnic groups, 3.0%).5 Case–control analysis indicated limited evidence for 202A protecting from severe malaria (males A vs G odds ratio (OR) 0.82, 95% CI 0.59–1.14, P=0.234; females AG vs GG OR 0.76, 95% CI 0.53–1.10, P=0.144; females AA vs GG OR 3.14, 95% CI 0.49–20.16, P=0.228) (Table 2). In contrast, the 376G allele (frequency 31.9%, marker for G6PD A, an allele with near normal enzyme activity) appeared to be associated with protection in Gambian males (OR 0.89, 95% CI 0.79–0.99, P=0.04). This result is likely to be due to the complex haplotypic structure of the G6PD locus. Specifically, among subjects who have 376G, some are G6PD A (near normal enzyme activity), whereas others (by virtue of having another mutation in cis) are G6PD A− (with severely reduced enzyme activity). In the Gambians, the 376G variant is present on haplotypes with at least three non-synonymous SNPs capable of causing enzyme deficiency. No associations were seen during multi-SNP analysis of G202A and A376G (Table 3).
Table 2. Minor allele frequencies and allelic/genotypic-based tests of association for the G6PD polymorphisms.
| MAF | Allelic-based tests of association | Genotype-based tests of associationa | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Polymorphismb | Allele | Control | SM | Alleles | OR | 95% CI | P-value | Genotypes | OR | 95% CI | P-value | ||
| (3875) | (2488) | ||||||||||||
| [1897] | [1294] | ||||||||||||
| HbS | S | 0.075 | 0.013 | S vs A | 0.16 | 0.12 | 0.21 | <0.001 | AS vs AA | 0.12 | 0.08 | 0.19 | <0.001 |
| G202A – Male | A | 0.027 | 0.023 | A vs G | 0.82 | 0.59 | 1.14 | 0.234 | A vs G | 0.88 | 0.55 | 1.42 | 0.602 |
| G202A – Female | A | 0.029 | 0.022 | A vs G | 0.73 | 0.52 | 1.03 | 0.070 | AA vs GG | 3.14 | 0.49 | 20.16 | 0.228 |
| AG vs GG | 0.76 | 0.53 | 1.10 | 0.144 | |||||||||
| A376G – Male | G | 0.318 | 0.293 | G vs A | 0.89 | 0.79 | 0.99 | 0.039 | G vs A | 0.89 | 0.76 | 1.05 | 0.172 |
| A376G – Female | G | 0.319 | 0.298 | G vs A | 0.91 | 0.81 | 1.01 | 0.089 | AG vs AA | 0.98 | 0.84 | 1.15 | 0.809 |
| GG vs AA | 0.85 | 0.65 | 1.11 | 0.232 | |||||||||
| (620) | (1999) | ||||||||||||
| [336] | [1050] | ||||||||||||
| A542T– Male | T | 0.024 | 0.010 | T vs A | 0.39 | 0.20 | 0.77 | 0.006 | T vs A | 0.43 | 0.16 | 1.15 | 0.092 |
| A542T – Female | T | 0.019 | 0.005 | T vs A | 0.27 | 0.11 | 0.63 | 0.003 | AT vs AA | 0.26 | 0.10 | 0.67 | 0.005 |
| T968C – Male | C | 0.070 | 0.052 | C vs T | 0.72 | 0.50 | 1.03 | 0.070 | C vs T | 0.63 | 0.378 | 1.07 | 0.085 |
| T968C – Female | C | 0.086 | 0.056 | C vs T | 0.63 | 0.44 | 0.89 | 0.010 | CC vs TT | 0.62 | 0.13 | 3.01 | 0.551 |
| CT vs TT | 0.62 | 0.41 | 0.95 | 0.027 | |||||||||
( ), total sample size; [ ], number of males; MAF, minor allele frequency; CC, case–controls; OR, odds ratio; CI, confidence interval; SM, severe malaria.
Adjusted for HbS and ethnicity (Fula, Jola, Mandinka, Wollof, other).
202A is the G6PD A− allele and 376A is the G6PD B allele.
Table 3. Multi-SNP association analysisa of severe malaria and the G6PD phenotypes.
| Haplotype | Frequency | OR | 95% CI | P-value | ||
|---|---|---|---|---|---|---|
| 202/376 | ||||||
| Female | GA [B] | 0.689 | 1.00 | |||
| GG [A] | 0.285 | 0.94 | 0.84 | 1.06 | 0.293 | |
| AG [A−] | 0.026 | 0.80 | 0.57 | 1.13 | 0.215 | |
| Male | GA [B] | 0.693 | 1.00 | |||
| GG [A] | 0.281 | 0.95 | 0.87 | 1.03 | 0.224 | |
| AG [A−] | 0.025 | 0.89 | 0.69 | 1.13 | 0.340 | |
| Overall | GA [B] | 0.691 | 1.00 | |||
| GG [A] | 0.283 | 0.95 | 0.88 | 1.01 | 0.109 | |
| AG [A−] | 0.026 | 0.86 | 0.70 | 1.05 | 0.134 | |
| Female 202A/376G heterozygote | 0.73 | 0.51 | 1.05 | 0.093 | ||
| 202/376/542/968 | ||||||
| Female | GATT | 0.689 | 1.00 | |||
| GGTT | 0.203 | 1.09 | 0.92 | 1.29 | 0.331 | |
| GGTC | 0.069 | 0.73 | 0.53 | 1.02 | 0.063 | |
| GGAT | 0.013 | 0.29 | 0.12 | 0.69 | 0.005 | |
| AGTT | 0.026 | 0.81 | 0.57 | 1.14 | 0.222 | |
| Any deficient | 0.107 | 0.71 | 0.56 | 0.89 | 0.004 | |
| Male | GATT | 0.688 | 1.00 | |||
| GGTT | 0.217 | 0.89 | 0.75 | 1.04 | 0.148 | |
| GGTC | 0.057 | 0.73 | 0.56 | 0.96 | 0.023 | |
| GGAT | 0.014 | 0.58 | 0.36 | 0.95 | 0.032 | |
| AGTT | 0.023 | 1.02 | 0.63 | 1.65 | 0.927 | |
| Any deficient | 0.094 | 0.77 | 0.62 | 0.95 | 0.016 | |
| Overall | GATT | 0.689 | 1.00 | |||
| GGTT | 0.210 | 0.98 | 0.87 | 1.10 | 0.722 | |
| GGTC | 0.063 | 0.73 | 0.60 | 0.90 | 0.003 | |
| GGAT | 0.014 | 0.49 | 0.32 | 0.76 | 0.001 | |
| AGTT | 0.024 | 0.88 | 0.66 | 1.16 | 0.349 | |
| Any deficient | 0.101 | 0.74 | 0.63 | 0.86 | 0.0002 | |
| Female deficiency allele heterozygote | 0.62 | 0.42 | 0.92 | 0.018 | ||
OR, odds ratio; CI, confidence interval.
Adjusted for HbS.
Next, we repeated the analysis including the additional G6PD variants. Among the Gambian controls, the 968C allele was at a frequency of 7.8% and the 542T allele was at 2.2%. No 680T alleles were detected. Even with the smaller sample size (see Methods), our analysis found disease associations with 542T/376G and 968C/376G haplotypes in single- and multi-marker analysis. When we analyzed all the deficiency haplotypes together (pooling 202A/376G, 542T/376G and 968C/376G haplotypes) (Table 3), we see a significant protective effect (∼23% reduced risk) in males (P=0.016) and (∼29% reduced haplotype risk) in females (P=0.004). Female heterozygotes for a deficiency allele (202A/376G, 542T/376G or 968C/376G) appeared to be protected (heterozygotes vs homozygous wild-type OR 0.62, 95% CI 0.42–0.92, P=0.018). However, no protection for homozygote-deficient females was detected (homozygous deficient vs homozygous wild-type OR 0.99, 95% CI 0.72–1.38, P=0.978).
Interestingly, the 542T/376G G6PD Santamaria allele causes a more severe G6PD deficiency (∼2% residual enzyme activity),27, 28 and there was a trend towards relatively greater protection in males (OR 0.43, 95% CI 0.16–1.15) and females (heterozygotes vs homozygous wild-type OR 0.26, 95% CI 0.10–0.66) when compared with that of the 202A/376G G6PD A− allele (∼12% activity16 males OR 0.88, 95% CI 0.55–1.42; female heterozygotes vs homozygous wild-type OR 0.76, 95% CI 0.52–1.10) or the 968C/376G G6PD Betica-Selma allele (∼11% activity16; males OR 0.63, 95% CI 0.38–1.07; female heterozygotes vs homozygous wild-type OR 0.63, 95% CI 0.41–0.95).
The analysis of CM cases did not substantially alter the results and interpretations above. The HbS S allele frequency in the Gambian controls was 7.5% (Fula, 8.8%; Jola, 5.9%; Mandinka, 8.9%; Wollof, 7.0%; other, 5.4%), and we also confirmed the known protective effect (∼90% reduced risk) of the HbS AS genotype29 (Table 1).
Discussion
These results provide an empirical example of how unrecognized allelic heterogeneity can confound genetic association studies. Theoretical treatments and simulation studies have highlighted the potential for allelic heterogeneity to impair power in genetic association studies, particularly when functional variants go undetected.30, 31, 32 Our results confirm recent findings from Senegal,17 suggesting that the diversity of common G6PD deficiency alleles in parts of West Africa is probably greater than that considered previously. Furthermore, our data raises the possibility that unrecognized allelic heterogeneity may have complicated past studies of G6PD deficiency and severe malaria susceptibility, particularly those studies that have classified subjects according to their genotypes and not their G6PD enzyme activity. Although in a high-throughput setting, phenotypic assays of G6PD enzymatic activity are more difficult to undertake than sequence-based assays, it is preferable to use both. This would enable a more accurate assessment of the concordance between these two approaches.
Initial single- and multi-SNP analysis, ignoring additional deficiency alleles, suggested that the conventional 202A/376G G6PD A− allele was not associated with severe malaria. One reason for this result is limited statistical power due to low allele frequency. The 202A allele is at a frequency of only 2.8% in the Gambian controls, significantly lower than reported previously.5 The second explanation for the negative association in the single SNP analysis is that the 202A allele is being compared not only against the wild-type B allele but also against other deficiency variants. Depending on the protection offered by these other alleles, and hence their relative depletion among cases, this could mask the true effects of 202A. When other deficiency alleles are included in our analysis, we could clearly show an association between G6PD deficiency and severe malaria.
The issue, in which G6PD genotypes receive protection from malaria, has been the subject of much debate over the years. The majority of studies have reported that hemizygous males are protected.3, 5, 6, 7, 8, 9 This finding was replicated by our work. It seems plausible that females homozygous for deficiency alleles are also protected. However, the rarity of such individuals has made it difficult for even large association studies to show whether this is true.5, 6 We too found no association between homozygous-deficient females and severe malaria. With only around 0.5% of study subjects being female and homozygous, our statistical power is once again rather limited. Whether heterozygous females are protected from malaria has been controversial; only some studies have reported this finding.5, 10, 11, 12 We speculated initially that the association between female heterozygotes and protection, found by Ruwende et al, may have been due to misclassification of some compound heterozygotes (eg 202A/542T or 202A/968C genotypes) as simply G202A heterozygotes. However, having taken into account the additional alleles, heterozygous females still appear to be protected from severe malaria. Although it is possible that yet more unrecognized G6PD deficiency alleles exist in the Gambian population, our data suggest that both heterozygous females and hemizygous males are protected from severe malaria in The Gambia.
The exact mechanism for the protection offered by G6PD deficiency is still uncertain. Some, but not all, in vitro studies have suggested an impaired parasite growth in deficient cells (see Greene,13 for a review). More recent work has suggested other mechanisms may be involved, such as early phagocytosis of infected erythrocytes.33 Conceivably, heterozygotes might benefit from having a sub-population of G6PD-deficient erthythrocytes that are subjected to the same mechanisms as the G6PD-deficient cells of male hemizygotes. Some authors have also suggested an intrinsic benefit to heterozygosity, such as poor parasite adaptation to the mixture of G6PD-deficient and normal cells.12
G6PD deficiency was among the first human genetic traits found to influence complex disease, and it continues to provide an interesting paradigm.3 Gaining a greater understanding of exactly how G6PD variation modulates malaria susceptibility may suggest novel approaches in prevention and treatment. Future studies of G6PD deficiency in Africa, employing genetic epidemiology, need to consider establishing the region-specific repertoire of functional variation before embarking on focused genotyping. The large-scale resequencing of the G6PD gene in African populations is also likely to generate further fascinating insights into how P. falciparum has shaped our genome.
Acknowledgments
The authors thank the patients from the Gambian study population, as well as the many investigators involved in the original study for their contributions. The authors also thank Bronwyn MacInnis, Malcolm Molyneux, Shivang Shah and Tom Williams for useful comments on this paper. Funding for this work came from the Bill and Melinda Gates Foundation, Wellcome Trust, and Medical Research Council UK. AEF was funded by a Wellcome Trust Clinical Research Training Fellowship (GR074586AIA). MD was supported by a training fellowship from the International Atomic Energy Agency (IAEA).
Footnotes
Supplementary Information accompanies the paper on European Journal of Human Genetics website (http://www.nature.com/ejhg)
Supplementary Material
References
- Ruwende C, Hill A. Glucose-6-phosphate dehydrogenase deficiency and malaria. J Mol Med. 1998;76:581–588. doi: 10.1007/s001090050253. [DOI] [PubMed] [Google Scholar]
- Mason PJ, Bautista JM, Gilsanz F. G6PD deficiency: the genotype-phenotype association. Blood Rev. 2007;21:267–283. doi: 10.1016/j.blre.2007.05.002. [DOI] [PubMed] [Google Scholar]
- Allison AC, Clyde DF. Malaria in African children with deficient erythrocyte glucose-6-phosphate dehydrogenase. Br Med J. 1961;1:1346–1349. doi: 10.1136/bmj.1.5236.1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tishkoff SA, Varkonyi R, Cahinhinan N, et al. Haplotype diversity and linkage disequilibrium at human G6PD: recent origin of alleles that confer malarial resistance. Science. 2001;293:455–462. doi: 10.1126/science.1061573. [DOI] [PubMed] [Google Scholar]
- Ruwende C, Khoo SC, Snow RW, et al. Natural selection of hemi- and heterozygotes for G6PD deficiency in Africa by resistance to severe malaria. Nature. 1995;376:246–249. doi: 10.1038/376246a0. [DOI] [PubMed] [Google Scholar]
- Guindo A, Fairhurst RM, Doumbo OK, Wellems TE, Diallo DA. X-linked G6PD deficiency protects hemizygous males but not heterozygous females against severe malaria. PLoS Med. 2007;4:e66. doi: 10.1371/journal.pmed.0040066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler T. G-6-PD deficiency and malaria in Black Americans in Vietnam. Mil Med. 1973;138:153–155. [PubMed] [Google Scholar]
- Gilles HM, Fletcher KA, Hendrickse RG, Lindner R, Reddy S, Allan N. Glucose-6-phosphate-dehydrogenase deficiency, sickling, and malaria in African children in South Western Nigeria. Lancet. 1967;1:138–140. doi: 10.1016/s0140-6736(67)91037-9. [DOI] [PubMed] [Google Scholar]
- Kar S, Seth S, Seth PK. Prevalence of malaria in Ao Nagas and its association with G6PD and HbE. Hum Biol. 1992;64:187–197. [PubMed] [Google Scholar]
- Bienzle U, Ayeni O, Lucas AO, Luzzatto L. Glucose-6-phosphate dehydrogenase and malaria. Greater resistance of females heterozygous for enzyme deficiency and of males with non-deficient variant. Lancet. 1972;1:107–110. doi: 10.1016/s0140-6736(72)90676-9. [DOI] [PubMed] [Google Scholar]
- Luzzatto L. About hemoglobins, G6PD and parasites in red cells. Experientia. 1995;51:206–208. doi: 10.1007/BF01931096. [DOI] [PubMed] [Google Scholar]
- Usanga EA, Luzzatto L. Adaptation of Plasmodium falciparum to glucose 6-phosphate dehydrogenase-deficient host red cells by production of parasite-encoded enzyme. Nature. 1985;313:793–795. doi: 10.1038/313793a0. [DOI] [PubMed] [Google Scholar]
- Greene L.G6PD Deficiency as Protection Against Falciparum Malaria: An Epidemiological Critique of Population and Experimental Studies Yearbook of Physical Anthropology Wiley-Liss Inc.: Wilmington, DE, USA; 1993, vol 36, pp 153–178. [Google Scholar]
- Tripathy V, Reddy BM. Present status of understanding on the G6PD deficiency and natural selection. J Postgrad Med. 2007;53:193–202. doi: 10.4103/0022-3859.33867. [DOI] [PubMed] [Google Scholar]
- Hirono A, Beutler E. Molecular cloning and nucleotide sequence of cDNA for human glucose-6-phosphate dehydrogenase variant A(-) Proc Natl Acad Sci USA. 1988;85:3951–3954. doi: 10.1073/pnas.85.11.3951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beutler E, Kuhl W, Vives-Corrons JL, Prchal JT. Molecular heterogeneity of glucose-6-phosphate dehydrogenase A. Blood. 1989;74:2550–2555. [PubMed] [Google Scholar]
- De Araujo C, Migot-Nabias F, Guitard J, Pelleau S, Vulliamy T, Ducrocq R. The role of the G6PD AEth376G/968C allele in glucose-6-phosphate dehydrogenase deficiency in the Seerer population of Senegal. Haematologica. 2006;91:262–263. [PubMed] [Google Scholar]
- Scriver C.Glucose-6-Phosphate Dehydrogenase Deficiency7th edn, McGraw Hill: New York; 1995 [Google Scholar]
- Marsh K, Forster D, Waruiru C, et al. Indicators of life-threatening malaria in African children. N Engl J Med. 1995;332:1399–1404. doi: 10.1056/NEJM199505253322102. [DOI] [PubMed] [Google Scholar]
- World Health Organization, Division of Control of Tropical Diseases Severe and complicated malaria. Trans R Soc Trop Med Hyg. 1990;84 Suppl 2:1–65. [PubMed] [Google Scholar]
- Molyneux ME, Taylor TE, Wirima JJ, Borgstein A. Clinical features and prognostic indicators in paediatric cerebral malaria: a study of 131 comatose Malawian children. Q J Med. 1989;71:441–459. [PubMed] [Google Scholar]
- Zhang L, Cui X, Schmitt K, Hubert R, Navidi W, Arnheim N. Whole genome amplification from a single cell: implications for genetic analysis. Proc Natl Acad Sci USA. 1992;89:5847–5851. doi: 10.1073/pnas.89.13.5847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez JM, Portillo MC, Saiz-Jimenez C. Multiple displacement amplification as a pre-polymerase chain reaction (pre-PCR) to process difficult to amplify samples and low copy number sequences from natural environments. Environ Microbiol. 2005;7:1024–1028. doi: 10.1111/j.1462-2920.2005.00779.x. [DOI] [PubMed] [Google Scholar]
- Ross P, Hall L, Smirnov I, Haff L. High level multiplex genotyping by MALDI-TOF mass spectrometry. Nat Biotechnol. 1998;16:1347–1351. doi: 10.1038/4328. [DOI] [PubMed] [Google Scholar]
- Wilson JN, Rockett K, Jallow M, et al. Analysis of IL10 haplotypic associations with severe malaria. Genes Immun. 2005;6:462–466. doi: 10.1038/sj.gene.6364227. [DOI] [PubMed] [Google Scholar]
- Clark T, Diakite M, Auburn S, et al. Tumour necrosis factor and lymphotoxin alpha polymorphisms and severe malaria in African populations. J Infect Dis. 2009;199:569–575. doi: 10.1086/596320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beutler E, Kuhl W, Saenz GF, Rodriguez W. Mutation analysis of glucose-6-phosphate dehydrogenase (G6PD) variants in Costa Rica. Hum Genet. 1991;87:462–464. doi: 10.1007/BF00197169. [DOI] [PubMed] [Google Scholar]
- Saenz GF, Chaves M, Berrantes A, Elizondo J, Montero AG, Yoshida A. A glucose-6-phosphate dehydrogenase variant, Gd(-) Santamaria found in Costa Rica. Acta Haematol. 1984;72:37–40. doi: 10.1159/000206354. [DOI] [PubMed] [Google Scholar]
- Kwiatkowski DP. How malaria has affected the human genome and what human genetics can teach us about malaria. Am J Hum Genet. 2005;77:171–192. doi: 10.1086/432519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terwilliger JD, Hiekkalinna T. An utter refutation of the ‘Fundamental Theorem of the HapMap'. Eur J Hum Genet. 2006;14:426–437. doi: 10.1038/sj.ejhg.5201583. [DOI] [PubMed] [Google Scholar]
- Slager SL, Huang J, Vieland VJ. Effect of allelic heterogeneity on the power of the transmission disequilibrium test. Genet Epidemiol. 2000;18:143–156. doi: 10.1002/(SICI)1098-2272(200002)18:2<143::AID-GEPI4>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- Longmate JA. Complexity and power in case-control association studies. Am J Hum Genet. 2001;68:1229–1237. doi: 10.1086/320106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cappadoro M, Giribaldi G, O'Brien E, et al. Early phagocytosis of glucose-6-phosphate dehydrogenase (G6PD)-deficient erythrocytes parasitized by Plasmodium falciparum may explain malaria protection in G6PD deficiency. Blood. 1998;92:2527–2534. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.