Abstract
The chromosome break points of the t(8;21)(q21.3;q22.12) translocation associated with acute myeloid leukemia disrupt the RUNX1 gene (also known as AML1) and the RUNX1T1 gene (also known as CBFA2T3, MTG8 and ETO) and generate a RUNX1–RUNX1T1 fusion protein. Molecular characterization of the translocation break points in a t(5;8)(q32;q21.3) patient with mild-to-moderate mental retardation and congenital heart disease revealed that one of the break points was within the RUNX1T1 gene. Analysis of RUNX1T1 expression in human embryonic and fetal tissues suggests a role of RUNX1T1 in brain and heart development and support the notion that disruption of the RUNX1T1 gene is associated with the patient's phenotype.
Keywords: RUNX1T1, MTG8, ETO, acute myeloid leukemia (AML), brain development, heart development
Introduction
The proto-oncogene RUNX1T1 (runt-related transcription factor 1; translocated to, 1), formerly known as CBFA2T1, MTG8 and ETO, is involved in acute myeloid leukemia (AML) cases exhibiting recurrent t(8;21) translocations.1 In the t(8;21) AML cases the translocation break points disrupt both the RUNX1T1 gene located at 8q22 and the RUNX1 gene (runt-related transcription factor 1), formerly known as AML1, at 21q22.3. This results in a chimeric RUNX1–RUNX1T1 gene, which is expressed as a fusion protein.1, 2, 3, 4
RUNX1 encodes a transcription factor, which binds DNA through a conserved Runt domain and may act as an activator or repressor by an interaction with co-activators or co-repressors, respectively, and is shown to be essential for normal hematopoiesis.5
RUNX1T1 belongs to a highly conserved group of proteins which all contain four nervy homology regions,6, 7 but its biological role is not well understood. In murine embryonic stem cells expression of Runx1t1 is upregulated during hematopoietic differentiation8 and targeted mutation of Runx1t1 results in developmental defects in the gastrointestinal system.9 RUNX1T1 mRNA has been detected in several human tissues, with highest expression in brain and heart. Immunohistochemical analysis in mice has shown an expression primarily in the nuclei and to some extent in the cytoplasm of neural cells.10 RUNX1T1 forms multiprotein complexes with transcriptional co-repressors, DNA-binding transcription factors and histone deacetylaces.7 Interestingly, RUNX1T1 can interact with ATROPHIN-1 (ATN1) which is mutated in the neurodegenerative disorder dentatorubral-pallidoluysian atrophy, DRPLA (OMIM no. 125371). Co-transfection experiments in a neuronal cell model (Neuro-2a) suggested co-localization of Runx1t1 and Atn1 in discrete nuclear structures containing histone deacetylase complexes and that Runx1t1 can enhance the transcriptional repressor function of Atn1.11 All these studies indicate a function of RUNX1T1/Runx1t1 in the nervous system.
Here, we report a patient with mild-to-moderate mental retardation and congenital heart disease carrying a constitutional balanced t(5;8) translocation, which disrupts the RUNX1T1 gene. Expression analysis of the RUNX1T1 gene in human embryonic and fetal tissues suggested a role of RUNX1T1 in brain and heart development.
Materials and methods
Tissue samples
Human embryonic and fetal tissues were collected from spontaneous and legal abortions. Informed consent was obtained according to the Helsinki Declaration II. Embryonic or fetal age was based on measurement of crown-rump length (CRL). Immediately after dissection the samples intended for RNA extraction were snap frozen in liquid nitrogen or treated with RNAlater according to the manufacturer's instructions (Ambion, Austin, TX, USA). Samples for immunohistochemistry were dissected into appropriate tissue blocks and fixed for 12–24 h at 4°C in either 10% neutral-buffered formalin, 4% Formol-Calcium, Lillie's or Bouin's fixatives. The specimens were dehydrated with graded alcohols, cleared in xylene and paraffin-embedded. Serial sections, 3–5 μ thick, were cut in transverse, sagittal or horizontal planes and placed on silanized slides.
Cytogenetic analyses
Metaphase chromosomes were prepared from phytohemagglutinin-stimulated peripheral blood lymphocytes according to standard procedures and were analyzed by G-banding techniques. Array CGH (comparative genome hybridization) was performed using a submegabase resolution whole genome tiling path 32K BAC array as described earlier.12 In brief BAC DNA was amplified using linker–adapter ligation PCR and spotted on epoxy-coated slides (NUNC, Wiesbaden, Germany). Sonicated patient and reference DNAs were labeled by random priming (Bioprime Array CGH, Invitrogen, Carlsbad, CA, USA) with Cy3 and Cy5 (Amersham Biosciences, Piscataway, NJ, USA), respectively. Subsequent hybridizations were performed overnight in a SlideBooster (Advalytix, Munich, Germany) at 42°C. After high-stringency washes, slides were scanned using an Axon 4000B scanner and images were analyzed using Genepix 5.0 (Axon Instruments, Union City, CA, USA).
Fluorescent in situ hybridization (FISH) analysis
Translocation break points were mapped by FISH analysis using 200 ng BAC DNA according to standard procedures. The FISH signals were visualized using avidin-FITC detection system. Chromosomes were counterstained with DAPI (4,6-diamino-2-phenylindole) and the signals were investigated using a Leica DMRB epifluorescence microscope equipped with a Sensys 1400 CCD camera (Photometrics, Tucson, AZ, USA) and an IPLab Spectrum imaging software (Abbott laboratories, Abbott Park, IL, USA).
Parental analysis
Somatic cell hybrids were constructed from a lymphoblastoid cell line derived from the patient using mouse E2 cells as described previously.13 DNA extracted from the cell lines were genotyped for the presence of the human chromosomes 5 and 8 using the microsatellite markers D5S1462, D5S1453, D5S1719, D8S136, D8S1106, D8S1145 and D8S1477. FAM-labeled PCR products were analyzed using an ABI3130xl Genetic Analyzer and Genemapper software (Applied Biosystems, Foster City, CA, USA).
Real-time quantitative RT-PCR (QPCR)
Total RNA was isolated from tissues using TRIzol Reagent (Invitrogen, Taastrup, Denmark) and first strand cDNA synthesis was performed using SuperScript II (RNase H-) reverse transcriptase (Invitrogen) according to the manufacturer's instructions. QPCR analysis was carried out on an ABI 7500 Fast real-time PCR system using a LightCycler FastStart DNA MasterPLUS SYBR GreenI kit (Roche, Hvidovre, DK).
Primer sequences used for QPCR analysis of RUNX1T1 were 5′ GAAAGCCCACGACATGATCAC 3′ and 5′ CAGCCACTGCAGGTTTCACTC 3′. Primer sequences for housekeeping genes are available on request.
Immunohistochemistry
Sections were deparaffinized and rehydrated in xylene followed by a series of graded alcohols according to established procedures. The sections were treated with a fresh 0.5% solution of hydrogen peroxide in methanol for 15 min to quench endogenous peroxidase and then rinsed in TRIS-buffered saline (TBS, 5 m Tris-HCl, 146 m NaCl, pH 7.6). Non-specific binding was inhibited by incubation for 30 min with blocking buffer (ChemMate antibody diluent S2022, DakoCytomation, Glostrup, Denmark) at room temperature. The sections were then incubated overnight at 4°C with a polyclonal rabbit antibody which specifically recognizes human and rodent CBFA2T1 (RUNX1T1) by immunoblotting and immunohistochemistry (C5616, Sigma, 1:800) in blocking buffer (ChemMate antibody diluent S2022, DakoCytomation). The sections were washed with TBS and then incubated for 30 min with a peroxidase-labelled anti-rabbit polymer (DAKO EnVision™+ System/HRP K4011, DakoCytomation). The sections were washed with TBS, followed by an incubation for 10 min with 3,3′-diamino-benzidine chromogen solution. Positive staining was recognized as a brown color. The sections were dehydrated in graded alcohols followed by xylene and coverslipped with DPX-mounting media. Non-immune rabbit IgG1 (X0936) and staining without primary antibody were used as negative controls. Sections from early human fetal liver, which contain numerous hematopoietic stem cells served as positive controls. Control sections stained without antibody or with a non-immune rabbit IgG1, were blank, whereas the hematopoietic islands in the early developing liver were always positive.
Results
Case report
The prospositus is the only child of a healthy mother. Pregnancy started 6 months after termination of chemotherapy of the father's leukemia (Acute Lymphoblastic Leukemia). The father died at the age of 30 years before birth of his son. The father has 3 healthy sibs, who have no descendants. Miscarriages or stillbirths are unknown in the father's anamnesis. The patient was delivered spontaneously having normal measures. At the age of 1 year a ventricular septal defect (VSD) was surgically closed.
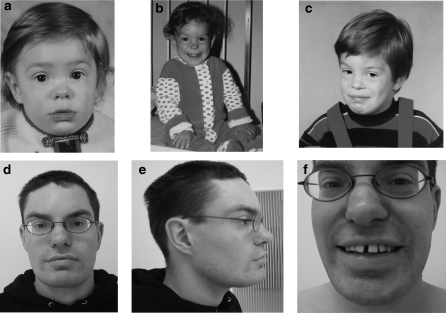
At the age of 28 years he is of normal height and weight. He has minor craniofacial dysmorphism (Figure 1). His face has a coarse appearance with full eye-brows and full lips, especially the lower lip which is also averted. The nose shows a broad tip and there is a deep transverse furrow on his chin. The forehead is high, receding and medially prominent. The posterior hairline is low set. He has dental anomalies (malocclusion and large wide-spaced medial incisors). Except from the craniofacial dysmorphism he has no further anomalies.
Figure 1.
Pictures of the patient at age of 1 year (a), 2 years (b), 4.5 years (c) and 27 years (d–f). He has minor craniofacial dysmorphism with dental anomalies and a coarse face with full eye-brows and full lips. The nose shows a broad tip, the forehead is high and the posterior hairline is low set.
The patient has mild/moderate mental retardation (the degree of which has never been evaluated by standardized tests) with infantile and temporarily auto aggressive behavior (self-mutilation of the finger nails). He has attended special assisted schooling, is able to read and write simple texts at a basic level and manage his practical life under his mother's guidance, but would not be able to lead an independent life. He has a full time job together with other mentally handicapped people in a “sheltered” supermarket run by an organization for the handicapped. The patient has no special health problems.
Cytogenetic analysis
A routine cytogenetic analysis of the patient suggested a balanced reciprocal translocation with the karyotype 46,XY,t(5;8)(q33;q22). The mother's karyotype was normal. Unfortunately, the father's karyotype was not determined before his death and no material was available for a post-mortem karyotyping. Array CGH analysis of the patient did not detect any potential disease associated copy number variants.
Parental analysis of the t(5;8) translocation
Four hybrid cell lines harboring only the normal human chromosomes 5 or 8, and the human derivative chromosomes der(5) or der(8), respectively, were established. DNA was extracted from the four hybrid cell lines, from a lymphoblastoid cell line derived from the patient and from a blood sample from the mother. These six samples were genotyped for microsatellite markers on the short arm of chromosomes 5 and 8, respectively. The genotype of microsatellite markers D5S1453, D8S136, D8S1130 and D8S1477 suggested that the normal chromosomes 5 and 8 were inherited from the mother and excluded the possibility of maternal inheritance of the derivative chromosomes (data not shown).
Mapping of translocation break points
The translocation break points were mapped using FISH. The 5q break point was investigated using 20 BAC clones covering an approximately 18.7 Mb region at 5q31.3–q33.3 (Supplementary Table 1). The break point was mapped to a gene-empty region at chromosome position 144 115 706–144 217 671 (UCSC Human Genome Browser, March 2006) at 5q32 represented by the overlapping region of two BAC clones spanning the break point.
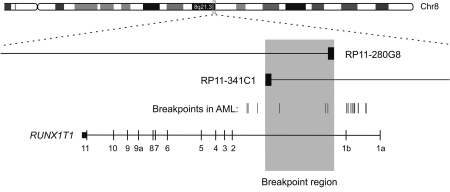
The 8q break point was investigated using 19 BAC clones covering an approximately 12 Mb region at 8q21.3–q22.3 (Supplementary Table 1). The break point was mapped to chromosome position 93 112 195–93 138 735 (UCSC Human Genome Browser, March 2006) at 8q21.3 represented by the overlapping region of two break point spanning BAC clones RP11-280G8 and RP11-341C1. The break point is within a region of 26.5 kb in intron 1b of the RUNX1T1 gene (Figure 2).
Figure 2.
Physical map of the break point region at 8q21.3. The chromosome 8 break point was within a 26.5 kb region (shaded gray) determined by the overlapping ends of the BAC clones RP11-280G8 and RP11-341C1. The break point thus mapped within intron 1b of RUNX1T1 near, and overlapping with some of the break points reported in AML (marked by vertical lines).14
Analysis of RUNX1T1 in embryonic and fetal tissues
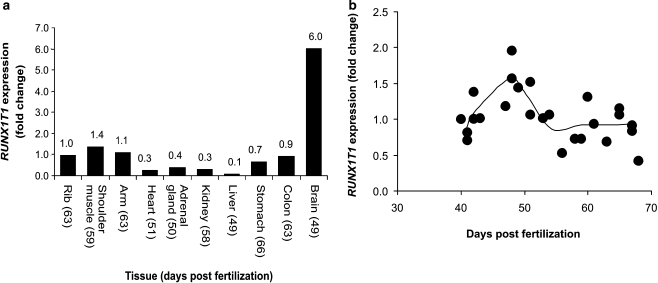
Expression of the RUNX1T1 gene was investigated by QPCR using a panel of RNA samples extracted from human embryonic and fetal tissues. Expression was detected in all tissues (Figure 3a). The lowest expression was observed in heart, liver, kidney and adrenal gland, while expression in muscle, stomach and colon was 2–3 times higher. The expression in brain tissues was almost 10-fold stronger than the average value (0.69) of the other investigated tissues. The temporal expression of RUNX1T1 during heart development was investigated in more detail using a panel of 25 hearts collected from human embryos and fetuses aged 40–68 days post fertilization. Some biological variation was observed, but the analysis suggested a gradual increase in the expression of RUNX1T1 in the developing heart until day 48 followed by a gradual decrease until day 54 where the expression level was constant at least until day 68 (Figure 3b).
Figure 3.
Expression of RUNX1T1 in human embryonic and fetal tissues. (a) Expression level in a panel of tissues. Expression level is shown as fold change compared to the expression in embryonic heart. The age of the embryo/fetus is indicated at each tissue. (b) Expression level in 25 human embryonic hearts. Expression level is shown as fold change compared to the expression in embryonic heart on day 40. The curve represents the average value of the data points in groups of three. Expression level was determined using QPCR. The CT values used for calculation of the relative expression were in the range of 19–26 cycles. The data was normalized using the average value of two housekeeping genes (B2M and HPRT).
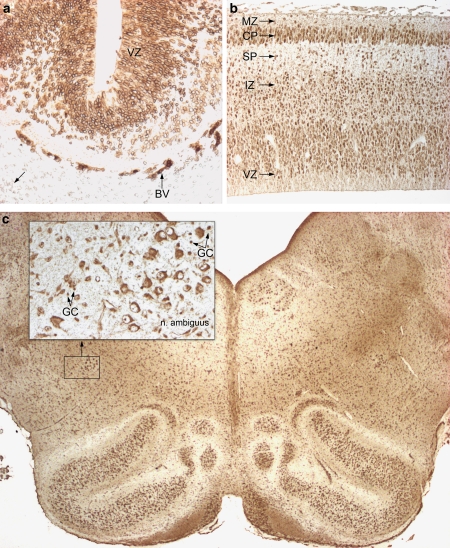
Immunohistochemical analysis of human embryonic and fetal tissue sections showed that RUNX1T1 is expressed in several tissues during development including the brain and the heart (Figures 4, 5, 6). Expression of RUNX1T1 was observed in several parts of the embryonic brain. At the earliest stage investigated (5-week-old embryos) a strong and distinct staining was seen in the perinuclear membrane of neural stem cells in the germinal matrix (ventricular zone) of the entire CNS. A weak staining was found in the nuclei per se. An example from the midbrain is shown in Figure 4a. Outside the CNS, the heart and the hemangiopoietic tissue as well as the developing gastrointestinal tract showed a similar pattern of reactivity, that is, a strong staining of the perinuclear cistern and a somewhat weaker staining of the nuclei (data not shown). Staining of the frontal cortex from an 8-week-old fetus showed a change in the pattern of reactivity. At this stage no apparent expression was observed in neuronal precursors of the ventricular zone, whereas an intermediate expression was found in the nuclei of migrating young neurons in the intermediate zone and subplate layer and a strong nuclear expression was found in neurons in the cortical plate (Figure 4b). Staining of the lower brain stem from a 16-week-old fetus showed strong cytoplasmic reactivity in the well-developed motor neurons innervating branchial arch-derived muscles (the nucleus ambiguous is shown in the inset in Figure 4c), whereas smaller nuclei of developing glial cells expressed a strong reactivity.
Figure 4.
Expression of RUNX1T1 in the developing human brain. (a) shows a sagittal section through the midbrain of a 5-week-old embryo. At this early stage of development all cell nuclei in the CNS exhibit a positive-staining reaction, which is particularly strong in the perinuclear space. The reactivity is most pronounced in the ventricular zone (VZ). Outside the CNS blood vessels (BV) are strongly reacting whereas the mesenchyme shows a weak reactivity (arrow). (b) depicts a coronal section through the forebrain of an 8-week-old fetus. The layers of the frontal cortex show a differential reactivity depending of differentiation: The radial glial cells/neuroblasts of the ventricular zone (VZ) express an overall weak reactivity. The migrating young neurons in the subventricular and intermediate zone (IZ) express strong nuclear reactivity, which is also seen in the subplate zone (SP). Differentiating neurons in the cortical plate (CP) show a strong nuclear but also a cytoplasmic reactivity. Nuclei of Cajal-Retzius cells in the marginal zone (MZ) are also distinctly labeled. (c) is from a horizontal section of the lower brain stem of a 16-week-old fetus. The inset shows mature neurons in the ambiguous nucleus with strong cytoplasmic reactivity, distinct nucleoli but a lack of expression in the nuclei. Early differentiating glial cells (GC) show a marked staining of their nuclei.
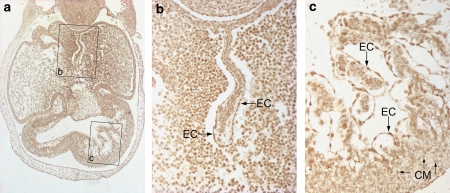
Figure 5.
Expression of RUNX1T1 in the developing human heart. This figure shows the overall reactivity for RUNXITI in the heart of a 6-week-old embryo (a). Note the positive-staining reaction in endocardial cells (EC) of the septum primum (b) and in the developing trabeculae (c). The nuclei of the cardiomyocytes (CM) are more weakly stained.
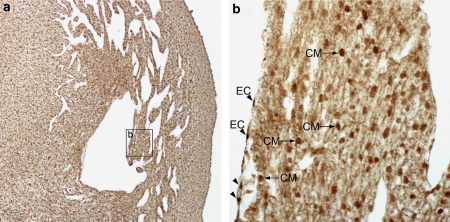
Figure 6.
Expression of RUNX1T1 in the developing human heart. (a) In the heart of a 9-week-old fetus the endocardial cells (EC) express a very strong cytoplasmic reactivity (b) whereas the nuclei of the cardiomyocytes (CM) are distinctly stained.
Staining of the heart from a 6-week-old embryo (Figure 5a) showed a particularly strong staining of both nuclei and cytoplasm of endocardial cells of the septum primum (Figure 5b) and of the developing trabeculae of the ventricular myocardium (Figure 5c). Nuclei of cardiomyocytes were also stained although not to the same degree (cf. CM in Figure 5c). At later stages of development, for example, in 9-week-old fetuses, endocardial cells expressed a strong cytoplasmic staining whereas the cardiomyocytes showed a distinct nuclear staining (Figures 6a and 6b).
Discussion
The 8q21 break point truncates the RUNX1T1 gene and the 5q31 break point maps within a gene-empty region, ruling out the possibility that a functional fusion protein may be produced by the translocation. Thus, we find it likely that the phenotype of the patient is caused by haploinsufficiency of RUNX1T1, although a position effect cannot be excluded.15
Deletions involving 8q21–q22 are rare. To our knowledge only three patients with deletions which presumably involve the RUNX1T1 gene has been reported.16, 17, 18 Psychomotor retardation was reported for all three patients and congenital heart disease (VSD and tetralogy of Fallot, respectively) was reported for two of the patients. However, the deletion in these cases was very large and the chromosomal break points were defined using karyotyping, thus comparison with our case is difficult.
Most frequently, break points related to the t(8;21) translocation in AML are clustering within one distinct region in intron 1a of RUNX1T1, suggesting a common causal mechanism.14 A less frequent break point region in AML patients has been identified in intron 1b of RUNX1T1, overlapping with the break point region in our patient. This co-localization may also argue for a common causal mechanism. Non-allelic homologous recombination mediated by low-copy repeats, or segmental duplications, has been identified as the causal mechanism in a significant proportion of recurrent chromosomal rearrangements.19
However, we did not find segmental duplications in the regions surrounding the break points in our patient suggesting that another mechanism is involved in the case presented here.
The phenotype of the proband and the location of the 8q21.3 break point within RUNX1T1 may suggest that this gene is involved in heart and brain development. To test the proposed involvement of RUNX1T1 in human heart and brain development, we analyzed the expression of RUNX1T1 in a panel of human embryonic and fetal tissues using QPCR. This analysis showed that the gene is expressed in several embryonic tissues. In embryonic brain tissue we observed a 10-fold higher expression compared to the average expression level of the other tissues analyzed. The abundant expression in the developing brain is in agreement with previous investigations.10, 20 Involvement of RUNX1T1 in brain development was further investigated using immunohistochemical analysis of human embryonic and fetal tissue sections.
In a 5-week-old embryo we observed strong and distinct staining in the perinuclear membrane of neural stem cells in the germinal matrix (ventricular zone) of the entire CNS. The same staining pattern was observed in the heart, hemangiopoietic tissue and the developing gastrointestinal tract. The nuclear envelope is involved in transcriptional regulation through chromatin modification21 and RUNX1T1 forms multiprotein complexes with transcriptional co-repressors and histone deacetylases.7 The distinct perinuclear expression pattern we observed therefore suggests that RUNX1T1 may be involved in transcriptional regulation in neural stem cells.
During development of the human cortex neural progenitors (neuroblasts) proliferate in the ventricular zone (VZ). The young neurons migrate from the VZ and establish the layers of the cortex in a sequential fashion.22, 23 Immunohistochemical analysis of the frontal cortex of an 8-week-old fetus showed no expression in neural progenitors of the VZ. Nuclear expression of RUNX1T1 was observed in young neurons in the intermediate zone and the subplate zone. The strongest nuclear expression was observed in neurons in the cortical plate. These data support the previously reported nuclear localization of RUNX1T1 in mouse neurons10 and suggest that RUNX1T1 is involved in differentiation of cortical neurons in human. Interestingly, a similar expression pattern of RUNX1T1 was observed during hematopoietic differentiation of murine embryonic stem (ES) cells.8 In this study, low levels of Runx1t1 expression were detected in undifferentiated ES cells and a gradual increase in expression was observed during differentiation. Thus, RUNX1T1 seems to play a role during stem cell differentiation, at least in hematopoietic and neuronal differentiation.
The cytoplasmic staining in neurons of the nucleus ambiguous in a 16-week-old fetus supports previous observations in hippocampal neurons10 and suggests that RUNX1T1 may have multiple roles during brain development.
QPCR analysis of 25 hearts dissected from human embryos at different developmental stages suggests that RUNX1T1 expression level in the heart peaks around day 48 post fertilization. These data suggest that the temporal expression of RUNX1T1 during heart development is tightly regulated and thus support that RUNX1T1 is involved in heart development.
Immunohistochemical detection of RUNX1T1 in cardiomyocytes and endothelial cells of a 9-week-old human fetal heart confirms that RUNX1T1 is expressed in the developing human heart and suggest that the protein may have multiple roles during cardiac development.
Septation of the cardiac ventricles starts at the end of the fourth week of embryonic development.24 In the middle of the seventh week the growth of the muscular ventricular septum halts before it meets the septum intermedium. Interestingly, this arrest of septum growth is concurrent with the temporal peak of RUNX1T1 expression we observed around day 48. Furthermore, immunohistochemical staining of the heart from a 6-week-old embryo showed a strong staining of endocardial cells of the developing trabeculae in the ventricular myocardium. Development of the muscular part of the ventricular septum is, at least in part, achieved by the compaction of trabeculae.25, 26, 27
Thus the temporal and spatial expression patterns of RUNX1T1 we observed in the developing heart support a role for RUNX1T1 in development of the cardiac ventricular septum, as suggested by the VSD observed in the translocation patient.
We conclude that mapping of the translocation break points within the RUNX1T1 gene in a t(5;8) patient with mental retardation and congenital heart disease and detection of differential RUNX1T1 expression in human embryonic brain and heart implicate a role for this gene in human brain and heart development.
Acknowledgments
This work was supported by The Danish Heart Association, The Novonordisk Foundation, Grosserer L. F. Foghts Fond, Ronald McDonald House Charities and Gangstedfonden. Wilhelm Johannsen Centre for Functional Genome Research is established by the Danish National Research Foundation. We thank Lillian Rasmussen, Kirsten Winther and Sussi Forchhammer for technical assistance.
Footnotes
Supplementary Information accompanies the paper on European Journal of Human Genetics website (http://www.nature.com/ejhg)
Supplementary Material
References
- Erickson P, Gao J, Chang KS, et al. Identification of breakpoints in t(8;21) acute myelogenous leukemia and isolation of a fusion transcript, AML1/ETO, with similarity to Drosophila segmentation gene, runt. Blood. 1992;80:1825–1831. [PubMed] [Google Scholar]
- Miyoshi H, Shimizu K, Kozu T, Maseki N, Kaneko Y, Ohki M. t(8;21) breakpoints on chromosome 21 in acute myeloid leukemia are clustered within a limited region of a single gene, AML1. Proc Natl Acad Sci USA. 1991;88:10431–10434. doi: 10.1073/pnas.88.23.10431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyoshi H, Kozu T, Shimizu K, et al. The t(8;21) translocation in acute myeloid leukemia results in production of an AML1-MTG8 fusion transcript. EMBO J. 1993;12:2715–2721. doi: 10.1002/j.1460-2075.1993.tb05933.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson LF, Boyapati A, Ahn EY, et al. Acute myeloid leukemia with the 8q22;21q22 translocation: secondary mutational events and alternative t(8;21) transcripts. Blood. 2007;110:799–805. doi: 10.1182/blood-2006-11-019265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamagata T, Maki K, Mitani K. Runx1/AML1 in normal and abnormal hematopoiesis. Int J Hematol. 2005;82:1–8. doi: 10.1532/IJH97.05075. [DOI] [PubMed] [Google Scholar]
- Davis JN, McGhee L, Meyers S. The ETO (MTG8) gene family. Gene. 2003;303:1–10. doi: 10.1016/s0378-1119(02)01172-1. [DOI] [PubMed] [Google Scholar]
- Rossetti S, Hoogeveen AT, Sacchi N. The MTG proteins: chromatin repression players with a passion for networking. Genomics. 2004;84:1–9. doi: 10.1016/j.ygeno.2004.02.011. [DOI] [PubMed] [Google Scholar]
- Okumura AJ, Peterson LF, Lo MC, Zhang DE. Expression of AML/Runx and ETO/MTG family members during hematopoietic differentiation of embryonic stem cells. Exp Hematol. 2007;35:978–988. doi: 10.1016/j.exphem.2007.03.002. [DOI] [PubMed] [Google Scholar]
- Calabi F, Pannell R, Pavloska G. Gene targeting reveals a crucial role for MTG8 in the gut. Mol Cell Biol. 2001;21:5658–5666. doi: 10.1128/MCB.21.16.5658-5666.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sacchi N, Tamanini F, Willemsen R, is-Donini S, Campiglio S, Hoogeveen AT. Subcellular localization of the oncoprotein MTG8 (CDR/ETO) in neural cells. Oncogene. 1998;16:2609–2615. doi: 10.1038/sj.onc.1201824. [DOI] [PubMed] [Google Scholar]
- Wood JD, Nucifora FC, Jr, Duan K, et al. Atrophin-1, the dentato-rubral and pallido-luysian atrophy gene product, interacts with ETO/MTG8 in the nuclear matrix and represses transcription. J Cell Biol. 2000;150:939–948. doi: 10.1083/jcb.150.5.939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erdogan F, Chen W, Kirchhoff M, et al. Impact of low copy repeats on the generation of balanced and unbalanced chromosomal aberrations in mental retardation. Cytogenet Genome Res. 2006;115:247–253. doi: 10.1159/000095921. [DOI] [PubMed] [Google Scholar]
- Yan H, Papadopoulos N, Marra G, et al. Conversion of diploidy to haploidy. Nature. 2000;403:723–724. doi: 10.1038/35001659. [DOI] [PubMed] [Google Scholar]
- Xiao Z, Greaves MF, Buffler P, et al. Molecular characterization of genomic AML1-ETO fusions in childhood leukemia. Leukemia. 2001;15:1906–1913. doi: 10.1038/sj.leu.2402318. [DOI] [PubMed] [Google Scholar]
- Kleinjan DA, van HV. Long-range control of gene expression: emerging mechanisms and disruption in disease. Am J Hum Genet. 2005;76:8–32. doi: 10.1086/426833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dallapiccola B, Santoro L, Trabace S, Ramenghi M, Mastroiacovo P, Gandini E. Deletion of the long arm of chromosome 8 resulting from a de novo translocation t(4;8) (q13;q213) Hum Genet. 1977;38:125–130. doi: 10.1007/BF00527393. [DOI] [PubMed] [Google Scholar]
- Donahue ML, Ryan RM. Interstitial deletion of 8q21–>22 associated with minor anomalies, congenital heart defect, and Dandy-Walker variant. Am J Med Genet. 1995;56:97–100. doi: 10.1002/ajmg.1320560122. [DOI] [PubMed] [Google Scholar]
- Fryburg JS, Golden WL. Interstitial deletion of 8q13.3–>22.1 associated with craniosynostosis. Am J Med Genet. 1993;45:638–641. doi: 10.1002/ajmg.1320450524. [DOI] [PubMed] [Google Scholar]
- Lupski JR, Stankiewicz P. Genomic disorders: molecular mechanisms for rearrangements and conveyed phenotypes. PLoS Genet. 2005;1:e49. doi: 10.1371/journal.pgen.0010049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolford JK, Prochazka M. Structure and expression of the human MTG8/ETO gene. Gene. 1998;212:103–109. doi: 10.1016/s0378-1119(98)00141-3. [DOI] [PubMed] [Google Scholar]
- Akhtar A, Gasser SM. The nuclear envelope and transcriptional control. Nat Rev Genet. 2007;8:507–517. doi: 10.1038/nrg2122. [DOI] [PubMed] [Google Scholar]
- Bystron I, Blakemore C, Rakic P. Development of the human cerebral cortex: Boulder Committee revisited. Nat Rev Neurosci. 2008;9:110–122. doi: 10.1038/nrn2252. [DOI] [PubMed] [Google Scholar]
- Molyneaux BJ, Arlotta P, Menezes JR, Macklis JD. Neuronal subtype specification in the cerebral cortex. Nat Rev Neurosci. 2007;8:427–437. doi: 10.1038/nrn2151. [DOI] [PubMed] [Google Scholar]
- Larsen WJ. Human Embryology. Philadelphia: Churchill Livingstone; 2001. [Google Scholar]
- Contreras-Ramos A, Sanchez-Gomez C, Garcia-Romero HL, Cimarosti LO. Normal Development of the Muscular Region of the Interventricular Septum – I. The Significance of the Ventricular Trabeculations. Anat Histol Embryol. 2008;37:344–351. doi: 10.1111/j.1439-0264.2008.00852.x. [DOI] [PubMed] [Google Scholar]
- Kirby ML. Cardiac development. Oxford: Oxford University Press; 2007. [Google Scholar]
- Stadtfeld M, Ye M, Graf T. Identification of interventricular septum precursor cells in the mouse embryo. Dev Biol. 2007;302:195–207. doi: 10.1016/j.ydbio.2006.09.025. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.