Abstract
Recently, genome-wide analyses revealed that variants on chromosome 9p21 are associated with myocardial infarction. We investigated whether this association was also present in a Belgian population of coronary artery disease (CAD) patients. As CAD and ischemic cerebrovascular disease (CVD) are thought to share some pathogenic pathways, we further examined the association of 9p21 with this disease. SNP rs10757278 on chromosome 9 was genotyped in 926 patients with CAD from the CAREGENE study, in 648 patients with CVD from the Leuven Stroke Genetics Study (LSGS) and the Belgian Stroke Study (BSS) and in 828 unrelated controls. A systematic review and meta-analysis were carried out in both vascular diseases. The frequency of the risk allele, rs10757278*G, was 55% in CAD cases versus 48% in controls, odds ratio (OR)=1.35 (1.18–1.54), P=1.3 × 10−5. No association was found with CVD, OR=1.03 (0.89–1.19), P=0.73. Meta-analysis revealed a consistent relationship between the risk variant and CAD. However, using a meta-analytic approach in CVD, only a marginal association was observed, which was no longer present after excluding patients with a history of CAD. The risk variant on chromosome 9, tagged by rs10757278, is associated with coronary heart disease in the Belgian population, but not with isolated CVD. These findings suggest different pathogenic mechanisms in CAD versus CVD.
Keywords: 9p21, stroke, coronary heart disease
Introduction
Stroke can be thought of as a complex disease in which several genetic variants act together with environmental factors. Genome-wide association studies are being carried out in complex disorders to identify the genes involved in these diseases with the hope of identifying new pathways that might lead to new pathophysiological insights.1 In the last few years, this approach has been successful.2 A locus on chromosome 9 has been consistently identified in coronary artery disease (CAD) in four independent genome-wide association studies.3, 4, 5, 6 Two studies tagged the variant on chromosome 9p by the C allele of rs1333049,5, 6 a third identified the G-allele of rs107572783 (r2=1 with rs1333049 in the HapMap CEU population), and a fourth study discovered an association with the G-allele of rs107572744 (r2=0.86 with rs10757278). In all but one subsequent publication, these same variants and/or other highly correlated ones (eg, the G allele of rs2891168, r2=0.88 with rs10757278 in the study by Broadbent et al7 and the G allele of rs2383206 r2>0.90 with the three tagging SNPs in the study by Zhou et al8) were replicated.7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18 Additional analyses revealed that the same variant increased the risk of abdominal aortic aneurysms (AAA) and intracranial aneurysms (IA).12
Results in ischemic cerebrovascular disease (CVD) have been less clear and have been limited by small sample sizes.11, 12, 19 We undertook a replication study in CAD patients from the Belgian population and, given the contradictory findings in CVD, we also investigated this variant in the latter. As it has been suggested that risk factors could be subtype-specific,20 we also determined whether associations were present for subgroups of ischemic stroke. In addition, we performed meta-analyses in both vascular diseases.
Materials and methods
Study population
Unrelated Caucasian patients, who were referred to the ambulatory cardiac rehabilitation programme of the University Hospitals of Leuven, were selected for inclusion in this study. Details of these patients from the CAREGENE (CArdiac REhabilitation and GENetics of Exercise performance) study have been published earlier.21 In short, the patients were referred after myocardial infarction, percutaneous transluminal coronary angioplasty, coronary artery bypass grafting, stable angina pectoris that did not limit exercise performance, or a combination of these. Various covariates were obtained from all patients: age, gender, height, weight, underlying heart disease, interventions and all types of medication, family history of CAD, history of hypertension or diabetes, earlier and current smoking habits, angina or dyspnea during daily life activities, systolic and diastolic blood pressure at rest, resting heart rate, and exercise-induced ST depression or arrhythmia.
We included Caucasian patients with cerebral ischemia (ischemic stroke and TIA), who were admitted to the Stroke Unit of the University Hospitals in Leuven and enrolled in the Leuven Stroke Genetics Study (LSGS), or who participated in the Belgian Stroke Study (BSS), which was described earlier.22 The World Health Organization criteria for determining a diagnosis of stroke and TIA were used.23 All CVD patients from the LSGS underwent brain imaging (MRI in >91% of patients, CT in the remainder) and a standardized protocol, including laboratory examination, carotid ultrasound or CT angiography, and cardiac examination (TEE or TTE and Holter). Control individuals were selected from the same population. In the LSGS, controls were selected from unrelated partners of patients with multiple sclerosis, amyotrophic lateral sclerosis, and stroke or TIA (n=271) in which stroke-free status was ensured in 60% using the Questionnaire for Verifying Stroke Free Status. In addition, Amyotrophic Lateral Sclerosis (ALS) patients were selected after stroke- and CAD-free status was verified by checking hospital records. Controls in the BSS gender- and ethnicity-matched subjects (>60 years, n=326), without a history of stroke or CAD, were recruited from the general population. Our study was approved by the local ethics committee and all cases and controls gave informed consent.
We recorded age at admission, sex, hypertension (defined as use of antihypertensive medication or physician-measured blood pressure >140/90 mmHg before stroke onset), diabetes mellitus (fasting glucose above 125 mg per 100 ml or use of antidiabetic medication), hypercholesterolemia (elevated fasting cholesterol levels or use of statins) and smoking status of the patients. Stroke subtypes were determined using the TOAST criteria by a single-stroke neurologist who was blinded to genotype.
Genotyping
DNA was extracted from venous blood samples using standard methods.
Genotyping for SNP rs10757278 was carried out using Taqman-based chemistry with assay C_11841860_10 run on a 7300 Real-time PCR System (Applied Biosystems).
Statistics
Marker performance (genotyping success rate, Hardy–Weinberg equilibrium) was evaluated using the Merlin (Pedstats) package and association analysis was carried out with the Unphased 3.0.12. package and with univariate logistic regression in SPSS 10.0. Reported probability values are two-sided and all levels of significance were set at P<0.05. Power calculation was carried out with a Genetic Power Calculator. For power calculation on the genotypic odds ratios (ORs), a multiplicative model was assumed. In the power calculation, a high-risk and marker allele frequency of 0.49 was used with D′=1 between them, and unselected controls were assumed. This study had >80% power to detect an OR of ≥1.22 in CAD and ≥1.23 in CVD.
Systematic review and meta-analysis
We identified all studies in PubMed investigating the 9p locus and CAD or CVD. The following search terms were used in the PubMed search: ‘9p21,' ‘vascular disease,' ‘coronary heart disease,' ‘myocardial infarction,' ‘stroke' and ‘genetics.' Publications were included till the 1st of October 2008. All studies for which ORs and 95% confidence intervals (CIs) were given, or could be calculated on the basis of genotype counts and/or allele frequencies, were included in the meta-analysis. Meta-analysis was carried out with the Mantel–Haenszel test. If heterogeneity was detected (I2>0%), a random effects model was applied. Statistics were calculated with Comprehensive meta-analysis 2 (Biostat Inc., Englewood, NJ, USA) and Fisher's combined probability test.
Results
Study population
Clinical characteristics of the 648 patients with TIA (n=99, 15.3%) or stroke (n=549, 84.7%) and of the 926 patients with CAD, who were included in the study, are shown in Tables 1 and 2. We recruited 568 healthy controls and 260 patients with ALS. Given that we did not observe a significant difference in allele or genotype frequency between the two control populations (healthy controls and ALS patients), we combined them as a single control group. Mean age was 62.7±13.5 for all controls and 63.4±13.1 for CVD patients. There were 239 females with CVD (36.9%) versus 388 female controls (46.9%). The mean age of CAD patients was 56.4±9.2 and the percentage of females was 9.2% (n=75).
Table 1. Baseline characteristics of CVD patients.
| LSGS and BSS Belgium (n=648) | |
|---|---|
| Age, years | 63.4±13.1 |
| Female sex, n (%) | 239 (36.9) |
| Hypertension, n (%) | 403 (62.2) |
| Current smoking, n (%) | 212 (32.7) |
| Hypercholesterolemia, n (%) | 353 (54.5) |
| Diabetes, n (%) | 113 (17.4) |
| Stroke subtype, n (%) | |
| Large vessel | 179 (27.6) |
| Lacunar | 154 (23.8) |
| Cardio-embolic | 125 (19.3) |
| Cryptogenic | 160 (24.7) |
| Other or multiple causes | 30 (4.6) |
| Coronary artery disease, n (%) | 111 (17.1) |
Table 2. Baseline characteristics of CAD patients.
| Caregene Belgium (n=926) | |
|---|---|
| Age, years | 56.4±9.2 |
| Female sex, n (%) | 75 (9.2) |
| Hypertension, n (%) | 260 (28.1) |
| Smoking history, n (%) | 676 (73.0) |
| Hypercholesterolemia, n (%) | 572 (61.8) |
| Diabetes, n (%) | 47 (5.1) |
| Reason for inclusion, n (%) | |
| Myocardial infarction | 400 (43.2) |
| Coronary artery bypass grafting | 338 (36.5) |
| Percutaneous transluminal coronary | 135 (14.6) |
| Angioplasty | |
| Other | 53 (5.7) |
The genotyping success rate was >97.7% in all groups (CAD cases, CVD cases and controls) and no deviation from the Hardy–Weinberg equilibrium was observed in any of these groups (P>0.21). No differences were observed among 19 samples typed in duplicate.
Association with rs10757278*G replicates in Belgian CAD patients
G-Allele frequencies were 55.5% in CAD cases versus 48.1% in controls, OR: 1.35 (95% CI 1.18–1.54); P=1.30 × 10−5. This association remained after correction for age and gender, OR: 1.29 (95% CI 1.11–1.51), P=1.12 × 10−3 (Table 3).
Table 3. Association of rs10757278 on 9p21 with CAD.
| rs10757278 | Case frequency (n) | Continued frequency (n) | OR (95% CI) | P-value | OR adjusted (95% CI) | P-value adjusted |
|---|---|---|---|---|---|---|
| A | 0.445 (813) | 0.519 (840) | 1 | |||
| G | 0.555 (1015) | 0.481 (778) | 1.35 (1.18–1.54) | 1.30 × 10−5 | 1.29 (1.11–1.51) | 1.12 × 10−3 |
| A/A | 0.193 (176) | 0.281 (227) | 1 | |||
| A/G | 0.504 (461) | 0.477 (386) | 1.54 (1.21–1.96) | 3.92 × 10−4 | 1.38 (1.05–1.81) | 0.022 |
| G/G | 0.303 (277) | 0.242 (196) | 1.82 (1.39–2.39) | 1.19 × 10−5 | 1.68 (1.23–2.28) | 1.03 × 10−3 |
Meta-analysis of association of risk allele with CAD
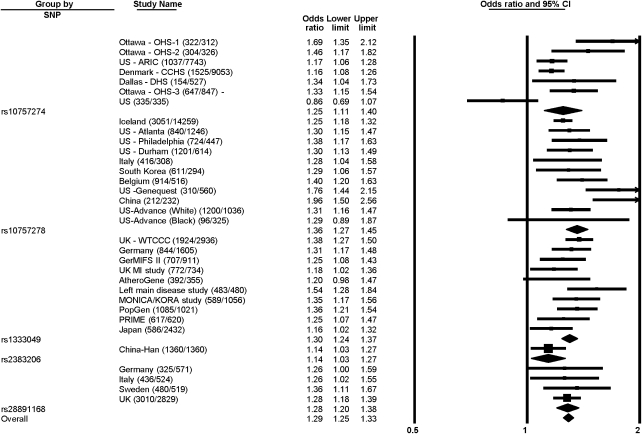
In addition, we carried out a meta-analysis of all studies published on the association of variants in this locus on chromosome 9p21 with CAD, reporting on four different SNPs. All showed an association with CAD (rs1333049*C, OR: 1.30 (95% CI 1.24–1.37); rs10757274*G, OR: 1.25 (95% CI 1.11–1.40); rs2891168*G, OR: 1.28 (95% CI 1.20–1.38); rs10757278*G, OR: 1.36 (95% CI 1.27–1.45); rs2383206*G, OR: 1.14 (95% CI 1.03–1.27)) with a combined P-value of 10−87 (Figure 1).
Figure 1.
Meta-analysis of case–control studies3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 14, 15, 16, 17, 18 with 5 SNPs, all tagging the 9p21 CAD risk variant. ORs were determined using a random effects model and are shown with 95% CIs. For the overall analysis, there is evidence of moderate heterogeneity (Cochran Q-value 72, I2=55%, P=0.00008). Heterogeneity is present among the populations that were studied with SNP rs10757274 (Cochran Q-value 25, I2=76%, P=0.0004) and rs10757278 (Cochran Q-value 21, I2=53%, P=0.02), but not between the other study populations (I2=0% for rs28891168, I2=26% for rs1333049). Only one study used the rs2383206 SNP. Ottawa – OHS-1, Ottawa – OHS-2, US – ARIC, Denmark – CCHS, Dallas – DHS, Ottawa – OHS-3: from McPherson et al4; US (335/335): from Zee and Ridker11; Iceland, US – Atlanta, US – Philadelphia, US – Durham: from Helgadotiir et al3; Italy (416/308), South Korea: from Shen et al9; US – Genequest: from Abdullah et al15; China (212/232): from Chen et al17; US – Advance (White), US – Advance (Black): Assimes et al16; UK – WTCCC, Germany (844/1605): from Samani et al5; GerMIFS II, UK MI study, AtheroGene, Left main disease study, MONICA/KORA study, PopGen, PRIME: from Schunkert et al14; Japan: from Hiura et al18; China – Han: from Zhou et al8; Germany (325/571), Italy (436/524), Sweden, UK: from Broadbent et al7.
No association for rs10757278 genotypes in CVD patients
G-Allele frequencies were not different between controls and cases, 48.1 versus 48.7% (OR: 1.03; 95% CI 0.89–1.19), nor after correction for age and gender (OR: 1.04; 95% CI 0.90–1.21). Genotype distribution did not show an association between rs10757278 and CVD either (Table 4). As Helgadottir et al12 reported an association for large artery and cardio-embolic stroke, we investigated whether certain subtypes of stroke might be associated with stroke in this population. However, rs10757278*G frequencies were not different for any of the subgroups compared with those for controls, nor after combining the large vessel and cardio-embolic subgroups. The G-allele frequencies were 52.0% for cardio-embolic stroke, 44.0% for cryptogenic stroke, 51.7% for large vessel disease, 49.3% for lacunar stroke, 39.7% for stroke due to other causes and 51.8% for large vessel and/or cardio-embolic combined. Moreover in the LSGS cohort, we analyzed all CVD patients with any evidence of atherosclerosis (defined as MI, symptomatic PAOD, high-grade atherosclerosis of carotid artery or vertebral artery; n=178) and found no association (OR: 1.09; 95% CI: 0.85–1.38). Furthermore, we observed a significant difference between the CAD and CVD patients with the G-allele being more frequent in CAD in comparison with all CVD patients (OR: 1.31; 95% CI: 1.14–1.52; P=2.01 × 10−4), an association that remained after correction for age and gender (OR: 1.27; 95% CI: 1.08–1.49; P=3.93 × 10−3).
Table 4. Allele and genotype distribution for rs10757278 in CVD.
| rs10757278 | Case frequency (n) | Continued frequency (n) | OR (95% CI) | P-value | OR adjusted (95% CI) | P-value adjusted |
|---|---|---|---|---|---|---|
| A | 0.513 (652) | 0.519 (840) | 1 | 1 | ||
| G | 0.487 (620) | 0.481 (778) | 1.03 (0.89–1.19) | 0.73 | 1.04 (0.90–1.21) | 0.60 |
| A/A | 0.267 (170) | 0.281 (227) | 1 | |||
| A/G | 0.491 (312) | 0.477 (386) | 1.08 (0.84–1.38) | 0.55 | 1.09 (0.85–1.40) | 0.49 |
| G/G | 0.242 (154) | 0.242 (196) | 1.05 (0.79–1.40) | 0.75 | 1.08 (0.81–1.45) | 0.61 |
Meta-analysis CVD
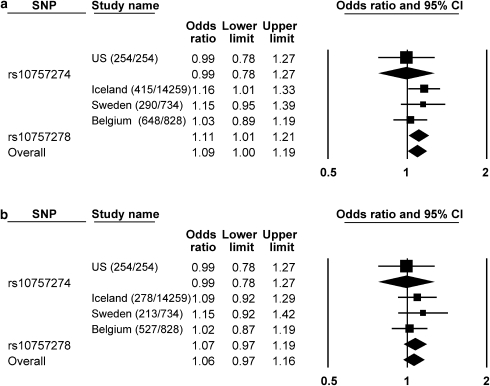
In a meta-analysis in CVD, combining all reported studies on this locus on chromosome 9p21 having typed rs10757278 and totaling 1353 cases and 15821 controls, a modest association was identified, OR: 1.11. (95% CI 1.01–1.21), P=0.03. Overall meta-analysis also, including one study reporting on rs10757274 totaling 1607 cases and 16075 controls, did not alter the relation, OR: 1.09 (95% CI 1.00–1.19), P=0.04 (Figure 2a). However, after excluding all patients with a history of CAD, the association was no longer present, OR: 1.06 (95% CI 0.97–1.16), P=0.22 as shown in Figure 2b.
Figure 2.
Meta-analysis of case–control studies 11, 12 with 2 SNPs tagging the 9p21 CAD risk variant in patients with CVD (a) and in CVD patients after excluding those who also had CAD (b). ORs were determined using a random effects model and are shown with 95% CIs. There is no evidence of heterogeneity (I2=0). US: from Zee and Ridker11; Iceland, Sweden: from Helgadottir et al12.
Discussion
Several groups have recently shown that a locus on chromosome 9p21 is associated with CAD.3, 4, 5, 7, 8, 9, 10, 13, 15, 16, 17, 18 We therefore investigated whether a relation would be present in a Belgian population with CAD. We clearly showed that the variant on chromosome 9p21 increases risk for this disease and that this relationship was present in almost all studied populations in CAD, as shown in our meta-analysis.
Variants in 9p21 have been studied in ischemic CVD as well,11, 12, 19 resulting in conflicting evidence. In the Framingham Study, the same region on 9p21 was associated with a cardiovascular phenotype that included, among three other diseases, stroke.13 In addition, the same sequence variant was investigated in four other arterial diseases and was found to be associated with IA and AAA but not consistently with ischemic stroke and peripheral arterial disease (PAD).12 Notably, the same genomic region has been associated with type 2 diabetes, but the effects are independent and conveyed by different variants.7
In our study population of 648 patients, we did not identify rs10757278*G to convey an increased risk for CVD. Similarly, no association was observed with large vessel disease, with any other subgroup of stroke or in CVD patients with evidence of atherosclerosis, although larger sample sizes might be necessary to exclude association in subgroups.
As the susceptibility in CAD cases with no clinical history of MI showed a trend toward stronger association, it was speculated that this variant might be implicated in plaque formation rather than in plaque rupture.7 This could partially explain the lack of association in CVD as atherothrombosis is particularly considered to be involved in a large portion of stroke patients. However, the same common sequence variants on 9p21 were earlier shown not to be associated with isolated PAD.12 Therefore, it might be concluded that variants in the described locus do not seem to be associated with atherosclerosis in general. In addition, this can be deduced from the fact that there is a clear relationship between the sequence variant on chromosome 9p and IA, in the pathogenesis of which atherosclerosis is not thought to play a major role.12 The risk allele, rs10757278*G, may be involved in a common mechanism like vascular remodeling and/or repair, although this has been shown to be involved in stroke24, 25 and PAD26 as well.
The variants on 9p21, which are most associated with CAD and intracranial and AAA, but not with stroke or PAD, are highly correlated in the European population and are present on two haplotypes that influence susceptibility in a yin-yang pattern.7 Whereas this allows tagging the haplotypes involved by genotyping a single SNP, prospects for further resolving the effect in the European population are therefore limited. Follow-up studies involving deep resequencing as well as functional experiments will be needed to unravel the exact mechanism by which this region of chromosome 9p21 is pathologically involved. The associated region lies upstream of the genes for two cyclin-dependent kinase inhibitors, CDKN2A and CDKN2B, known to play an important part in the cell cycle. The haplotype overlaps with exons of ANRIL, a large antisense non-coding RNA (ncRNA).7 ncRNAs are extensively being studied for their role as essential players in epigenetic mechanisms such as DNA methylation, histone modifications and chromatin conformational changes.27 Various studies have underscored the importance of epigenetics in atherosclerosis.28, 29, 30, 31 The understanding of these mechanisms has only begun to emerge32, 33 and the combined effect of epigenetics and microenvironment on an arterial phenotype may result in an atherosclerotic change only in particular vascular regions. This interplay might potentially explain why variants in ncRNAs might be implicated in some presentations of vascular diseases, but not in all. It is therefore appealing, but speculative, to hypothesize that the susceptibility of different regions of the vascular system to atherosclerosis can, at least to some extent, be determined by epigenetic pathways.
In conclusion, we replicated the association of a variant on chromosome 9p with CAD in a Belgian population, but were not able to observe a similar association with CVD. It is interesting that, in a meta-analysis of CVD, there was a marginal association that was no longer present after exclusion of patients with a history of CAD. These findings can be of importance for further genetic association studies that plan to investigate the potential role of recently identified SNPs in CAD in CVD. As patients presenting with stroke or TIA can have a higher incidence of past CAD, we would like to underline the importance of documenting the vascular medical history of patients in such studies. SNPs that are identified in these association studies may be risk markers of underlying CAD rather than be risk variants for CVD per se.
Acknowledgments
The BSS was supported by the Erasme Funds. AG is a Postdoctoral Fellow of the Research Fund Flanders (FWO-Vlaanderen). Vincent This is supported by FWO Research Foundation Flanders. WR is supported through the E von Behring Chair for Neuromuscular and Neurodegenerative Disorders, and by the Interuniversity Attraction Poles (IUAP) program P6/43 of the Belgian Federal Science Policy Office.
Footnotes
Conflicts of interest
The authors declare no conflict of interest.
References
- Topol EJ, Murray SS, Frazer KA. The genomics gold rush. JAMA. 2007;298:218–221. doi: 10.1001/jama.298.2.218. [DOI] [PubMed] [Google Scholar]
- Pennisi E. Breakthrough of the year. Human genetic variation. Science. 2007;318:1842–1843. doi: 10.1126/science.318.5858.1842. [DOI] [PubMed] [Google Scholar]
- Helgadottir A, Thorleifsson G, Manolescu A, et al. A common variant on chromosome 9p21 affects the risk of myocardial infarction. Science. 2007;316:1491–1493. doi: 10.1126/science.1142842. [DOI] [PubMed] [Google Scholar]
- McPherson R, Pertsemlidis A, Kavaslar N, et al. A common allele on chromosome 9 associated with coronary heart disease. Science. 2007;316:1488–1491. doi: 10.1126/science.1142447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samani NJ, Erdmann J, Hall AS, et al. Genomewide association analysis of coronary artery disease. N Engl J Med. 2007;357:443–453. doi: 10.1056/NEJMoa072366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wellcome Trust Case Control Consortium Genome-wide association study of 14 000 cases of seven common diseases and 3000 shared controls. Nature. 2007;447:661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broadbent HM, Peden JF, Lorkowski S, et al. Susceptibility to coronary artery disease and diabetes is encoded by distinct, tightly linked, SNPs in the ANRIL locus on chromosome 9p. Hum Mol Genet. 2008;17:806–814. doi: 10.1093/hmg/ddm352. [DOI] [PubMed] [Google Scholar]
- Zhou L, Zhang X, He M, et al. Associations between single nucleotide polymorphisms on chromosome 9p21 and risk of coronary heart disease in Chinese han population. Arterioscler Thromb Vasc Biol. 2008;28:2085–2089. doi: 10.1161/ATVBAHA.108.176065. [DOI] [PubMed] [Google Scholar]
- Shen GQ, Rao S, Martinelli N, et al. Association between four SNPs on chromosome 9p21 and myocardial infarction is replicated in an Italian population. J Hum Genet. 2008;53:144–150. doi: 10.1007/s10038-007-0230-6. [DOI] [PubMed] [Google Scholar]
- Shen GQ, Li L, Rao S, et al. Four SNPs on Chromosome 9p21 in a South Korean Population Implicate a Genetic Locus That Confers High Cross-Race Risk for Development of Coronary Artery Disease. Arterioscler Thromb Vasc Biol. 2008;28:360–365. doi: 10.1161/ATVBAHA.107.157248. [DOI] [PubMed] [Google Scholar]
- Zee RY, Ridker PM. Two common gene variants on chromosome 9 and risk of atherothrombosis. Stroke. 2007;38:e111. doi: 10.1161/STROKEAHA.107.497669. [DOI] [PubMed] [Google Scholar]
- Helgadottir A, Thorleifsson G, Magnusson KP, et al. The same sequence variant on 9p21 associates with myocardial infarction, abdominal aortic aneurysm and intracranial aneurysm. Nat Genet. 2008;40:217–224. doi: 10.1038/ng.72. [DOI] [PubMed] [Google Scholar]
- Larson MG, Atwood LD, Benjamin EJ, et al. Framingham Heart Study 100K project: genome-wide associations for cardiovascular disease outcomes. BMC Med Genet. 2007;8 Suppl 1:S5. doi: 10.1186/1471-2350-8-S1-S5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schunkert H, Gotz A, Braund P, et al. Repeated replication and a prospective meta-analysis of the association between chromosome 9p21.3 and coronary artery disease. Circulation. 2008;117:1675–1684. doi: 10.1161/CIRCULATIONAHA.107.730614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abdullah KG, Li L, Shen GQ, et al. Four SNPS on Chromosome 9p21 Confer Risk to Premature, Familial CAD and MI in an American Caucasian Population (GeneQuest) Ann Hum Genet. 2008;72 part 5:654–657. doi: 10.1111/j.1469-1809.2008.00454.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Assimes TL, Knowles JW, Basu A, et al. Susceptibility locus for clinical and subclinical coronary artery disease at chromosome 9p21 in the multi-ethnic ADVANCE study. Hum Mol Genet. 2008;17:2320–2328. doi: 10.1093/hmg/ddn132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z, Qian Q, Ma G, et al. A common variant on chromosome 9p21 affects the risk of early-onset coronary artery disease Mol Biol Rep 2008[E-pub ahead of print]. [DOI] [PubMed]
- Hiura Y, Fukushima Y, Yuno M, et al. Validation of the association of genetic variants on chromosome 9p21 and 1q41 with myocardial infarction in a Japanese population. Circ J. 2008;72:1213–1217. doi: 10.1253/circj.72.1213. [DOI] [PubMed] [Google Scholar]
- Matarin M, Brown WM, Singleton A, Hardy JA, Meschia JF. Whole genome analyses suggest ischemic stroke and heart disease share an association with polymorphisms on chromosome 9p21. Stroke. 2008;39:1586–1589. doi: 10.1161/STROKEAHA.107.502963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meschia JF. Ischaemic stroke: one or several complex genetic disorders. Lancet Neurol. 2003;2:459. doi: 10.1016/s1474-4422(03)00479-4. [DOI] [PubMed] [Google Scholar]
- Defoor J, Martens K, Zielinska D, et al. The CAREGENE study: polymorphisms of the beta1-adrenoceptor gene and aerobic power in coronary artery disease. Eur Heart J. 2006;27:808–816. doi: 10.1093/eurheartj/ehi737. [DOI] [PubMed] [Google Scholar]
- Abboud S, Karhunen PJ, Lutjohann D, et al. Proprotein Convertase Subtilisin/Kexin Type 9 (PCSK9) Gene Is a Risk Factor of Large-Vessel Atherosclerosis Stroke. PLoS ONE. 2007;2:e1043. doi: 10.1371/journal.pone.0001043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stroke--1989. Recommendations on stroke prevention, diagnosis, and therapy. Report of the WHO Task Force on Stroke and other Cerebrovascular Disorders. Stroke. 1989;20:1407–1431. doi: 10.1161/01.str.20.10.1407. [DOI] [PubMed] [Google Scholar]
- Ohab JJ, Fleming S, Blesch A, Carmichael ST. A neurovascular niche for neurogenesis after stroke. J Neurosci. 2006;26:13007–13016. doi: 10.1523/JNEUROSCI.4323-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Cui X, Zacharek A, et al. Niaspan increases angiogenesis and improves functional recovery after stroke. Ann Neurol. 2007;62:49–58. doi: 10.1002/ana.21160. [DOI] [PubMed] [Google Scholar]
- Palmer-Kazen U, Wahlberg E. Arteriogenesis in peripheral arterial disease. Endothelium. 2003;10:225–232. doi: 10.1080/10623320390246360. [DOI] [PubMed] [Google Scholar]
- Costa FF. Non-coding RNAs, epigenetics and complexity. Gene. 2008;410:9–17. doi: 10.1016/j.gene.2007.12.008. [DOI] [PubMed] [Google Scholar]
- Stenvinkel P, Karimi M, Johansson S, et al. Impact of inflammation on epigenetic DNA methylation – a novel risk factor for cardiovascular disease. J Intern Med. 2007;261:488–499. doi: 10.1111/j.1365-2796.2007.01777.x. [DOI] [PubMed] [Google Scholar]
- Chi JT, Chang HY, Haraldsen G, et al. Endothelial cell diversity revealed by global expression profiling. Proc Natl Acad Sci USA. 2003;100:10623–10628. doi: 10.1073/pnas.1434429100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Won D, Zhu SN, Chen M, et al. Relative reduction of endothelial nitric-oxide synthase expression and transcription in atherosclerosis-prone regions of the mouse aorta and in an in vitro model of disturbed flow. Am J Pathol. 2007;171:1691–1704. doi: 10.2353/ajpath.2007.060860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng DX, Tsalenko A, Vailaya A, et al. Differences in vascular bed disease susceptibility reflect differences in gene expression response to atherogenic stimuli. Circ Res. 2006;98:200–208. doi: 10.1161/01.RES.0000200738.50997.f2. [DOI] [PubMed] [Google Scholar]
- Lund G, Zaina S. Atherosclerosis, lipids, inflammation and epigenetics. Curr Opin Lipidol. 2007;18:699–701. doi: 10.1097/MOL.0b013e3282f21bb8. [DOI] [PubMed] [Google Scholar]
- Aird WC. Mechanisms of endothelial cell heterogeneity in health and disease. Circ Res. 2006;98:159–162. doi: 10.1161/01.RES.0000204553.32549.a7. [DOI] [PubMed] [Google Scholar]