Abstract
Populations of northeastern Europe and the Uralic mountain range are found in close geographic proximity, but they have been subject to different demographic histories. The current study attempts to better understand the genetic paternal relationships of ethnic groups residing in these regions. We have performed high-resolution haplotyping of 236 Y-chromosomes from populations in northwestern Russia and the Uralic mountains, and compared them to relevant previously published data. Haplotype variation and age estimation analyses using 15 Y-STR loci were conducted for samples within the N1b, N1c1 and R1a1 single-nucleotide polymorphism backgrounds. Our results suggest that although most genetic relationships throughout Eurasia are dependent on geographic proximity, members of the Uralic and Slavic linguistic families and subfamilies, yield significant correlations at both levels of comparison making it difficult to denote either linguistics or geographic proximity as the basis for their genetic substrata. Expansion times for haplogroup R1a1 date approximately to 18 000 YBP, and age estimates along with Network topology of populations found at opposite poles of its range (Eastern Europe and South Asia) indicate that two separate haplotypic foci exist within this haplogroup. Data based on haplogroup N1b challenge earlier findings and suggest that the mutation may have occurred in the Uralic range rather than in Siberia and much earlier than has been proposed (12.9±4.1 instead of 5.2±2.7 kya). In addition, age and variance estimates for haplogroup N1c1 suggest that populations from the western Urals may have been genetically influenced by a dispersal from northeastern Europe (eg, eastern Slavs) rather than the converse.
Keywords: Y-chromosome, Y-STRs, northeastern Europe, phylogenetics
Introduction
Relatively recent archaeological evidence indicates that northeastern Europe was initially occupied by modern humans during the transition from the Middle to Upper Paleolithic periods (approximately 35–45 000 YBP).1 However, the last glacial maximum (LGM) forced the contraction of the entire European populace to a number of refugia in the Iberian Peninsula, present day Ukraine and the northern Balkans.2 The region was impacted again 12 200–13 000 years ago, by an expansion from southwestern Europe during the final stage of the LGM, an event still imprinted in the mtDNA landscape of the area.3 The next group of migrants to arrive in the locality is theorized to have been the Comb Ware people (predecessors of Finno-Ugric-speaking tribes, a branch of the Uralic language family) from the Uralic mountains about 6900 YBP.4
Populations within the Urals are characterized by high levels of genetic heterogeneity and various degrees of admixture between Europeans and Asians.5 It has been reported that these groups possess some Asian maternal DNA components.6, 7 Additional investigations utilizing the autosomal VNTR markers, D1S80 and 3′ApoB,8, 9, 10 TP53 single-nucleotide polymorphism (SNP) haplotypes11 along with Y-chromosomal analyses12 signal both Asian and European genetic constituents. For example, Y-chromosomal haplogroup N (specifically sub-haplogroups N1c and N1b), believed to be of Asian ancestry,13, 14, 15, 16 is found at high frequencies within the Urals; and its pronounced presence in the Baltic countries (Lithuania, Latvia, and Estonia), as well as in the Nordic Peninsula (Finland) and in the Saami of Sweden, argue for an Uralic genetic signature throughout northeastern Europe.17
Despite the marked genetic similarities between Finno-Ugric speakers (Finns, Estonians, the Saami, and groups found in the slopes of the Urals) and Latvians and Lithuanians, peoples from the latter two Baltic countries speak languages belonging to the Balto-Slavic branch of the Indo-European language family. The Indo-European languages are believed to have been initially spread by the Kurgan horse culture about 10 000 YBP.12, 13, 18 In spite of this, a lack of consensus on the roots of this civilization is reflected in the existence of varying theories claiming the Ukraine,13 the Central Asian steppes,12 and northern India18 all as plausible cradles for Proto-Indo-Europeans. Proto-Baltic ancestors, in turn, are speculated to have arrived from Central and southeastern Europe 5000–4000 YBP,19 triggering the contraction of the already present Finno-Ugric tribes to the north. Early genetic analyses based on blood groups and serum protein marker distributions indicate that the contemporary Balts constitute a composite of the Finno-Ugrians and Slavic groups.20 More recent work, utilizing Y-chromosomal short tandem repeats (STRs), suggests that the Baltic populations of Latvia and Lithuania are phylogenetically closer to each other than either is to their Finno-Ugric Estonian neighbors.21
The eastern Slavic populations (the present-day Russians, Byelorussians, and Ukrainians) are speculated to have descended from Proto-Slavic-speaking groups that extended into northeastern Europe from Central Europe during the early middle ages,22 yet the origins of these migrant tribes is widely debated.23 Two theories have been proposed on the origins of eastern Slavs: the hybridization and transformation hypotheses. According to the former, these groups arose as a result of fusion between the invading Slavic tribes and populations inhabiting Eastern Europe. Alternatively, the transformation model proposes that eastern Slavic groups gradually evolved in situ from ancient groups autochthonous to the area. Mitochondrial DNA,24, 25 Y-STR haplotypes,26, 27 and autosomal STR diversity distributions8, 28 endorse the hybridization theory supporting the Central European Slavic infusion into tribes previously residing in Eastern Europe.
A recent Y-chromosomal study addressing the intra-ethnic variation in Russian populations revealed that central and southern Slavic Russian groups cluster closely together, whereas northern groups exhibit genetic and phylogenetic affinities to Finno-Ugric peoples, suggesting an assimilation of the Uralic substrata throughout the area,23 a phenomenon previously observed using other marker systems, such as mtDNA,24, 25 Y-STR haplotypes,26, 27 and autosomal STR loci.8, 28 These and other publications14, 29 also claim that geographic partitioning rather than ethnolinguistic boundaries constitutes the main genetic barriers throughout Europe. Nevertheless, the complexity of the region (especially of northeastern groups) and the fusion of a plethora of people make the scenarios portrayed by this claim simplistic in nature.
To date, several studies have been performed to genetically characterize populations both within northeastern Europe and northwestern Asia; yet, the data are fragmentary and uneven in geographic scope, heterogeneous in the marker systems used, and at times contradictory. In addition, limited work has been conducted to integrate the existing information comprehensively in order to delineate migratory patterns and phylogenetic relationships. In the current study, high-resolution Y-chromosome binary markers were used to shed light onto the paternal genetic histories of populations from the aforementioned regions and their relationships to previously published collections. Furthermore, 15 Y-STR loci were assayed for individuals from the SNP backgrounds, R1a1, N1c1, and N1b, to ascertain population expansion times and elucidate possible migratory scenarios.
Materials and methods
Sample collection and DNA isolation
Blood samples were collected in Vacutainer tubes from a total of 236 unrelated male individuals residing in the East European region of Russia (Arkhangelski (n=28), Kursk (n=40), Tver (n=38), Izhemski Komi (n=54), and Priluzski Komi (n=49)) and Siberia (Khanty (n=27)). Genealogical information was recorded for at least three generations to establish regional ancestry. Table 1 lists the sampling sizes, geographic locations, linguistic affiliations, and references of the previously published, geographically targeted populations under study.
Table 1. Populations examined in Y-SNP analyses.
| Geographic region and populations | N | Abbreviation | Linguistic affiliation | References |
|---|---|---|---|---|
| North Caucasus | ||||
| Kabardinians | 59 | KAB | North Caucasian, Northwest Caucasian | 30 |
| Lezgi (Dagestan) | 25 | LEZ | North Caucasian, East Caucasian | 31 |
| Ossetians (Ardon) | 28 | OSA | Indo-European, Indo-Iranian | 31 |
| Ossetians (Digora) | 31 | OSD | Indo-European, Indo-Iranian | 31 |
| South Caucasus | ||||
| Armenia | 100 | ARM | Indo-European, Armenian | 30 |
| Azerbaijan | 72 | AZE | Altaic, Turkic | 30 |
| Georgia | 77 | GEO | Kartvelian, Georgian | 30 |
| Levant and Anatolia | ||||
| Iraq | 139 | IRQ | Afro-Asiatic, Semitic | 32 |
| Lebanon | 31 | LEB | Afro-Asiatic, Semitic | 13 |
| Syria | 20 | SYR | Afro-Asiatic, Semitic | 13 |
| Turkey | 523 | TUR | Altaic, Turkic | 33 |
| Middle East | ||||
| North Iran | 33 | NIR | Indo-European, Indo-Iranian | 34 |
| South Iran | 117 | SIR | Indo-European, Indo-Iranian | 34 |
| North Pakistan | 176 | NPA | Indo-European, Indo-Iranian | 18 |
| South Pakistan | 176 | SPA | Indo-European, Indo-Iranian | 18 |
| Central Asia | ||||
| Kazakhstan | 30 | KAZ | Altaic, Turkic | 35 |
| Turkmenistan | 30 | TUK | Altaic, Turkic | 12 |
| Uzbekistan | 54 | UZB | Altaic, Turkic | 35 |
| Northeastern Europe | ||||
| Belarus | 41 | BEL | Indo-European, Slavic | 29 |
| Estonia | 207 | EST | Uralic, Finnic | 29 |
| Finland | 57 | FIN | Uralic, Finnic | 29 |
| Latvia | 34 | LAT | Indo-European, Baltic | 29 |
| Lithuania | 38 | LIT | Indo-European, Baltic | 29 |
| Poland | 112 | POL | Indo-European, Slavic | 29 |
| Southeastern Europe | ||||
| Romania | 45 | ROM | Indo-European, Italic | 29 |
| Slovakia | 70 | SLO | Indo-European, Slavic | 29 |
| Ukraine | 50 | UKR | Indo-European, Slavic | 13 |
| Russia (European) | ||||
| Arkhangelsk | 28 | ARK | Indo-European, Slavic | Present study |
| Belgorod | 143 | BEG | Indo-European, Slavic | 23 |
| Krasnoborsk | 91 | KRA | Indo-European, Slavic | 23 |
| Kursk | 40 | KUR | Indo-European, Slavic | Present study |
| Livni | 110 | LIV | Indo-European, Slavic | 23 |
| Mezen | 54 | MEZ | Indo-European, Slavic | 23 |
| Ostrov | 75 | OST | Indo-European, Slavic | 23 |
| Pinega | 114 | PIN | Indo-European, Slavic | 23 |
| Roslavl | 107 | ROS | Indo-European, Slavic | 23 |
| Tver | 38 | TVE | Indo-European, Slavic | Present study |
| Unzha | 52 | UNZ | Indo-European, Slavic | 23 |
| Vologda | 121 | VOL | Indo-European, Slavic | 23 |
| Russia (Uralic Mountains) | ||||
| Komi Izhemski | 54 | KOI | Uralic, Finno-Ugric | Present study |
| Komi Priluzski | 49 | KOP | Uralic, Finno-Ugric | Present study |
| Mari | 48 | MAR | Uralic, Finno-Ugric | 29 |
| Russia (Siberia) | ||||
| Evenks | 50 | EVE | Altaic, Manchu-Tungus | 36 |
| Khakassians | 53 | KAK | Altaic, Turkic | 36 |
| Khanty | 27 | KHA | Uralic, Finno-Ugric | Present study |
| Tuvinians | 113 | TUV | Altaic, Turkic | 36 |
Total nucleic acid was isolated by standard phenol–chloroform extraction, as described by Antunez-de-Mayolo and collaborators.37 DNA was ethanol-precipitated and stored in 0.010 M Tris-EDTA (pH 8.0) at −80°C as stock solutions. The samples were procured with informed consent following all ethical guidelines as stipulated by all research institutions involved in the project.
Y-chromosome haplotyping
A total of 105 binary markers were hierarchically genotyped by PCR-RFLP, allele-specific PCR,38, 39 and amplicon size detection of the YAP polymorphic Alu insertion.40 Detailed information on the locations, allelic states, primer sequences, and references for each marker can be found at the Y-chromosome consortium web page (http://ycc.biosci.arizona.edu/nomenclature_system/index.html) and in subsequent publications.18, 41, 33
Y-STR genotyping
A total of 17 Y-STR markers (DYS19, DYS385 a/b, DYS389 I/II, DYS390, DYS391, DYS392, DYS393, DYS437, DYS438, DYS439, DYS448, DYS456, DYS458, DYS635, and Y-GATA H4) were PCR-amplified using the AmpF/STR Y Filer Kit (Applied Biosystems) according to the manufacturer's specifications for samples under the SNP backgrounds R1a1 (M198), N1c1 (M178) and N1b (P43). Fragment separation was conducted with an ABI Prism Genetic Analyzer (Applied Biosystems). The electropherogram profiles were then analyzed using the Genescan 3.7 and Genotype 3.7 NT softwares.
Time estimations
DYS385a and DYS385b were not included in time estimation or variance calculations, given their duplicative nature. Variance estimations were ascertained using the Vp function as shown by Kayser et al42 and were based on seven Y-STR loci (DYS19, DYS389 I, DYS389 II, DYS390, DYS391, DYS392, and DYS393) for R1a1 and six loci (DYS389 I, DYS389 II, DYS390, DYS391, DYS392, and DYS393) for N1c1- and N1b-derived samples given the limited number of loci reported for the previously published reference populations (Supplementary Table 1). Haplotype expansion times were defined using the programs NETWORK 4.2.00 and BATWING, assuming an average Y-STR mutation rate of 6.9 × 10−4,43 an intergeneration time of 25 and 32 years,44 and exponential population growth from a constant size ancestral population.45 Assumptions for the BATWING analysis were followed, as previously described by Cinnioğlu et al,33 with the exemption of the population growth rate (α) using γ (1.01,1) instead.46 Median joining networks were constructed, also excluding DYS385 a/b, with the aid of the NETWORK 4.2.0045 software package (SNP-STR references and number of individuals are provided in Supplementary Table 1).
Unfortunately, BATWING did not generate credible 95% CIs (confidence intervals) for most comparisons, and as such most of the values generated grossly disagree with coalescence time estimates performed by other authors.15, 18 As such, unless otherwise stated, age estimates used throughout the Results and Discussion sections will be the NETWORK estimations using a 25-year intergeneration time for comparison purposes, as most reports base their calculations on this time frame. BATWING estimates will only be referred to when credible 95% CIs are attained. Nevertheless, given that BATWING estimates did not generate credible 95% CIs in most instances, NETWORK calculations should be taken with caution, as there are likely to be violations of the dating method's assumptions.
Phylogenetic and statistical analyses
A correspondence analysis (CA) based on the frequencies of the binary markers defining major haplogroups (A–R) was generated to gauge genetic similarities among the populations using the NTSYSpc 2.02i software.47 CAs based on Y-STR haplotype frequencies were also conducted. Analyses of Molecular Variance (AMOVAs) and Fst distances were calculated using the Arlequin software package (version 3.11).48, 49 Significance was ascertained at α=0.05.
Results
Haplogroup phylogeography
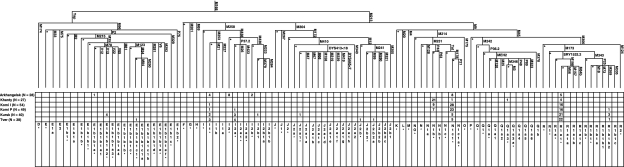
Of 105 binary markers typed, 48 were found to be polymorphic (Arkhangelski (18), Khanty (13), Izhemski Komi (13), Priluzski Komi (16), Kursk (29), and Tver (26)) in the 236 individuals who were examined (Figure 1). Sub-haplogroup N1c1 (M178) is shared across all the European and Uralic populations at varying frequencies, with the highest level detected in the Izhemski Komi collection (52%) and the lowest in the Siberian Khanty (4%), which exhibits a considerable proportion of haplogroup N1b (78%) (Figure 1). These findings parallel the result from other northeastern European populations (eg, Finland, Estonia, Lithuania and Latvia), which contain comparable frequencies of N1c; however, M178 was not typed in a previously published report.39 Haplogroup N1b is also found at appreciable quantities in the Izhemski and Priluzski Komi groups (17% and 14%, respectively).
Figure 1.
Haplogroup phylogeography throughout Northwestern Russia. Y-Chromosome markers typed hierarchically along with current haplogroup denominations and numbers of individuals belonging to each haplogroup.
R1a1 (defined by mutation M198) is shared across all the populations genotyped in this study, with frequencies ranging from 0.15 in the Khanty collection to 53 and 58% in Kursk and Tver, respectively (Figure 1). Haplogroup I derivatives, specifically I1 and I2a (defined by M253 and P37, respectively), are found at substantial proportions in the Slavic populations of Kursk and Tver (Figure 1), adding up to 13 and 18% of each population's paternal gene pool, respectively. The Arkhangelski group displays similar levels of I1 (14%) and is completely lacking I2a, exhibiting high frequencies of I2* (absent in both Tver and Kursk). The haplogroup distribution within Central Eurasia based on the six genotyped populations in this study and the reference collections are illustrated in Figure 2.
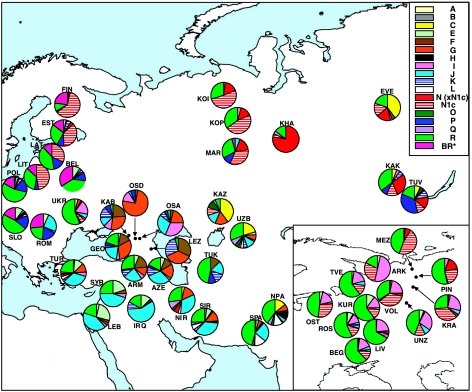
Figure 2.
Haplogroup distributions throughout Central Eurasia. Population names and abbreviations: KAB (Kabardinians), LEZ (Lezgi), OSA (Ossetians Ardon), OSD (Ossetians Digora), ARM (Armenia), AZE (Azerbaijan), GEO (Georgia), IRQ (Iraq), LEB (Lebanon), SYR (Syria), TUR (Turkey), NIR (North Iran), SIR (South Iran), NPA (North Pakistan), SPA (South Pakistan), KAZ (Kazakhstan), TUK (Turkmenistan), UZB (Uzbekistan), BEL (Belarus), EST (Estonia), FIN (Finland), LAT (Latvia), LIT (Lithuania), POL (Poland), ROM (Romania), SLO (Slovakia), UKR (Ukraine), ARK (Arkhangelski), BEG (Belgorod), KRA (Krasnoborsk), KUR (Kursk), LIV (Livni), MEZ (Mezen), OST (Ostrov), PIN (Pinega), ROS (Roslavl), TVE (Tver), UNZ (Unzha), VOL (Vologda), KOI (Komi Izhemski), KOP (Komi Priluzski), MAR (Mari), EVE (Evenks), KAK (Khakassians), KHA (Khanty), and TUV (Tuvinians).
Population relationships
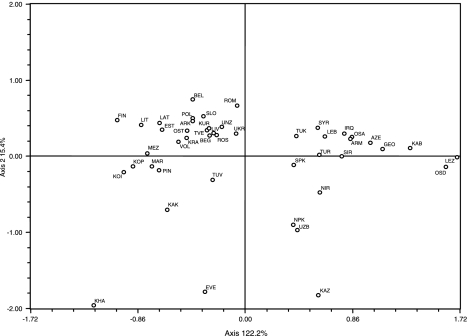
Genetic similarities between Finno-Ugric and Balto-Slavic populations are illustrated in Figure 3. The Slavic populations cluster tightly in the upper-left quadrant, with the Finno-Ugric Estonians and Finnish partitioning loosely to the left of the aforementioned grouping along with their geographical neighbors Lithuania and Latvia. The Uralic populations segregate to the bottom-left quadrant midway between the Slavic cluster and a poorly defined Siberian grouping. The Khanty collection strays away from any pairings and lays to the extreme lower corner of the same portion of the graph. To the right half of the projection, Caucasian, Middle Eastern, and Central Asian populations follow an almost geographical cline from the North Caucasus toward the Middle East and then into Central Asia from the extreme right midway between the upper and lower quadrants to the center of the lower-right portion of the graph.
Figure 3.
Correspondence Analysis (CA) based on major Y-chromosome haplogroup bifurcations (A–R).
When a continentally based AMOVA was conducted, variance components suggest a greater affinity for geographical influences rather than for linguistic ties (Table 2), supporting earlier findings.15, 29, 50 However, when only Balto-Slavic and Uralic groups are evaluated, both linguistic and geographic components yield similar variance component percentages, making it difficult to ascertain whether linguistics or geographical connections influence genetic relationships (Table 2).
Table 2. Analysis of molecular variance.
| % Variation attributable to | |||||||
|---|---|---|---|---|---|---|---|
| Among groups | Among populations within groups | Within groups | |||||
| Grouping | Fst | % Variation | P-value | % Variation | P-value | % Variation | P-value |
| Geographical (8 groups) | 0.16676 | 8.81 | <0.00001 | 7.87 | <0.00001 | 83.32 | <0.00001 |
| Linguistic (5 groups) | 0.15835 | 6.51 | <0.00001 | 10.10 | <0.00001 | 83.39 | <0.00001 |
| Geographical (4 groups) | 0.16676 | 6.62 | <0.00001 | 7.11 | <0.00001 | 86.28 | <0.00001 |
| Linguistic (2 groups) | 0.15117 | 6.92 | <0.00001 | 8.19 | <0.00001 | 84.99 | <0.00001 |
Geographical partitioning (8 groups): 1, Caucasus (Kabardinians, Lezgi, Ossetians Ardon, Ossetians Digora, Armenia, Azerbaijan, Georgia); 2, Levant and Anatolia (Lebanon, Syria, Iraq, Turkey); 3, Middle East (North Iran, South Iran, North Pakistan, South Pakistan); 4, Central Asia (Uzbekistan, Kazhakstan, Turkmenistan); 5, northeastern Europe (Lithuania, Latvia, Belarus, Finland, Estonia, Kursk, Tver, Arkhangelsk, Poland, Belgorod, Livni, Roslavl, Ostrov, Unzha, Vologda, Krashoborsk, Pinega, Mezeh); 6, southeastern Europe (Slovakia, Ukraine, Romania); 7, Uralic (Komi Priluzski, Komi Izhemski, Mari); 8, Siberia (Khanty, Khakassians, Evenks, Tuva).
Geographical partitioning (4 groups): 1, Slavic Russians (Kursk, Tver, Arkhangelsk, Belgorod, Livni, Roslavl, Ostrov, Unzha, Vologda, Krashoborsk, Pinega, Mezeh); 2, Europe (Slovakia, Ukraine, Romania, Poland, Lithuania, Latvia, Belarus, Finland, Estonia); 3, Uralic (Komi Priluzski, Komi Izhemski, Mari); 4, Siberia (Khanty).
Linguistic partitioning (5 groups): 1, Caucasian (Kabardinians, Lezgi, Georgia); 2, Indo-European (Ossetians Ardon, Ossetians Digora, Armenians, North Iran, South Iran, North Pakistan, South Pakistan, Lithuania, Latvia, Belarus, Kursk, Tver, Arkhangelsk, Poland, Slovakia, Ukraine, Romania, Belgorod, Livni, Roslavl, Ostrov, Unzha, Vologda, Krashoborsk, Pinega, Mezeh); 3, Afro-Asiatic (Lebanon, Syria, Iraq); 4, Altaic-Turkic (Turkey, Azerbaijan, Uzbekistan, Kazhakstan, Turkmenistan, Khakassians, Evenks, Tuva); 5, Uralic (Finland, Estonia, Mari, Komi Priluzski, Komi Izhemski, Khanty).
Linguistic partitioning (2 groups): 1, Balto-Slavic (Lithuania, Latvia, Belarus, Kursk, Tver, Arkhangelsk, Poland, Slovakia, Ukraine, Romania, Belgorod, Livni, Roslavl, Ostrov, Unzha, Vologda, Krashoborsk, Pinega, Mezeh); 2, Uralic (Finland, Estonia, Mari, Komi Priluzski, Komi Izhemski, Khanty).
Pairwise Fst distances are presented in Supplementary Table 2. Values of pairwise comparisons reported in red represent statistically nonsignificant distances at α=0.05, whereas estimates displayed in blue correspond to distances found nonsignificant after applying the Bonferroni correction for Type 1 errors at α=0.05/1050=0.000048. The northeastern European populations of Latvia and Lithuania were found to be more similar genetically to the Finno-Ugric populations than to their Slavic neighbors. Populations from Central Asia and Siberia (excluding Khanty) exhibit comparable average distance values (0.09540 and 0.090254, respectively) when compared among themselves (all generating significant Fst values), whereas populations from the Caucasus, which are found in much closer geographical proximity to each other than the aforementioned groups, display an average value of 0.09759, including several significant pairwise distance values (Supplementary Table 2).
Y-STR variance, age estimates, and network projections
Distribution and Age Estimates of Haplogroup R1a1
The oldest age estimates dating back to Mesolithic times (approximately 18 000 YBP) for Haplogroup R1a1 have been detected in West India (16.7±4.3), South India (18.2±5.5), South Pakistan (18.7±4.7), and Serbia (17.3±5.9) (Table 3). However, STR variance is highest in southern India with a value of 0.505 (Table 3).
Table 3. Haplotype variance and age estimations for haplogroup R1a1 (M198).
| Network time estimations | Network time estimations | BATWING expansion times using 25-year generation time (kya) | BATWING expansion times using 32-year generation time (kya) | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Population | N | Haplotype variance based on 7 Y-STR loci | Network shape | with 25-year generation time (kya)a | with 32-year generation time (kya)a | Mean | Median | 2.50% | 97.50% | Mean | Median | 2.50% | 97.50 |
| Arkhangelsk | 5 | 0.271 | Star-like | 10.4±3.9 | 13.3±5.0 | 39.7 | 10.9 | 0.0 | 751.4 | 50.9 | 13.9 | 961.8 | |
| Arkhangelsk (15 loci) | Star-like | 10.4±2.9 | 10.1±2.9 | 36.4 | 9.7 | 0.0 | 707.5 | 46.6 | 12.4 | 0.0 | 905.7 | ||
| Komi Izhemski | 16 | 0.228 | Star-like | 5.8±2.6 | 7.5±3.3 | 9.1 | 1.4 | 0.0 | 343.1 | 11.6 | 1.8 | 0.0 | 439.2 |
| Komi Izhemski (15 loci) | Star-like | 10.1±2.4 | 12.9±3.1 | 5.9 | 1.0 | 0.0 | 222.4 | 7.5 | 1.2 | 0.0 | 284.7 | ||
| Komi Priluzski | 16 | 0.154 | Non-Star | 7.8±3.2 | 9.9±4.1 | 9.8 | 1.7 | 0.0 | 356.8 | 12.5 | 2.1 | 0.0 | 456.7 |
| Komi Priluzski (15 loci) | Star-like | 11.3±2.6 | 14.5±3.3 | 14.4 | 1.0 | 0.0 | 542.7 | 18.4 | 1.3 | 0.0 | 694.7 | ||
| Kursk | 20 | 0.191 | Star-like | 6.5±2.2 | 8.3±2.8 | 7.6 | 4.0 | 0.3 | 59.7 | 9.7 | 5.1 | 0.4 | 76.5 |
| Kursk (15 loci) | Star-like | 9.3±2.0 | 11.9±2.6 | 14.5 | 3.8 | 0.1 | 413.7 | 18.6 | 4.9 | 0.2 | 529.5 | ||
| Tver | 20 | 0.280 | Non-Star | 9.8±2.8 | 12.6±3.6 | 34.2 | 14.4 | 0.2 | 366.6 | 43.8 | 18.5 | 0.2 | 469.2 |
| Tver (15 loci) | Star-like | 11.8±2.2 | 15.2±2.8 | 24.6 | 11.2 | 0.2 | 362.0 | 31.5 | 14.4 | 0.3 | 463.3 | ||
| Finland | 38 | 0.353 | Star-like | 12.1±2.7 | 15.5±3.4 | 8.8 | 7.0 | 1.8 | 40.1 | 11.3 | 9.0 | 2.3 | 51.4 |
| Herzegovina | 17 | 0.222 | Star-like | 9.1±3.4 | 11.7±4.3 | 13.7 | 9.8 | 0.5 | 100.1 | 17.6 | 12.5 | 0.7 | 128.2 |
| India (North) | 31 | 0.346 | Star-like | 14.0±3.7 | 18.0±4.7 | 13.9 | 11.2 | 2.1 | 58.7 | 17.8 | 14.4 | 2.7 | 75.1 |
| India (East) | 18 | 0.250 | Non-Star | 12.9±4.1 | 16.6±5.3 | 13.0 | 3.4 | 0.1 | 411.0 | 16.7 | 4.4 | 0.1 | 526.1 |
| India (West) | 17 | 0.426 | Non-Star | 16.7±4.6 | 21.4±5.9 | 21.3 | 5.7 | 0.0 | 683.3 | 27.3 | 7.3 | 0.0 | 874.6 |
| India (South) | 37 | 0.505 | Star-like | 18.2±5.5 | 23.3±7.0 | 10.2 | 7.3 | 1.6 | 104.8 | 13.0 | 9.4 | 2.0 | 134.1 |
| Khakassians | 18 | 0.180 | Non-Star | 7.2±3.1 | 9.2±3.9 | 9.5 | 1.3 | 0.0 | 340.2 | 12.2 | 1.7 | 0.0 | 435.5 |
| Pakistan (North) | 14 | 0.243 | Non-Star | 9.6±2.7 | 12.3±3.5 | 24.4 | 1.4 | 0.0 | 773.6 | 31.3 | 1.8 | 0.0 | 990.2 |
| Pakistan (South) | 29 | 0.475 | Non-Star | 18.7±4.7 | 24.0±6.1 | 8.4 | 5.6 | 0.4 | 52.3 | 10.7 | 7.2 | 0.5 | 67.0 |
| Serbia | 18 | 0.295 | Non-Star | 17.3±5.4 | 22.1±6.9 | 22.5 | 4.9 | 0.1 | 644.2 | 28.8 | 6.3 | 0.1 | 824.6 |
| Turkey | 36 | 0.298 | Star-like | 12.3±3.3 | 15.8±4.2 | 7.9 | 5.9 | 1.1 | 40.4 | 10.1 | 7.6 | 1.4 | 51.7 |
| Tuva | 15 | 0.184 | Star-like | 8.3±3.1 | 10.6±3.9 | 19.1 | 5.3 | 0.0 | 387.1 | 24.5 | 6.8 | 0.0 | 495.5 |
| Hungary | 8 | 0.247 | Star-like | 7.1±3.0 | 9.1±3.8 | 27.5 | 6.6 | 0.0 | 468.1 | 35.2 | 8.4 | 0.0 | 599.1 |
| Latvia | 4 | 0.131 | Non-Star | 9.1±5.3 | 11.6±6.8 | 17.5 | 3.2 | 0.0 | 505.6 | 22.4 | 4.1 | 0.0 | 647.1 |
| Lithuania | 11 | 0.169 | Non-Star | 7.5±3.3 | 9.6±4.2 | 13.0 | 4.4 | 0.1 | 298.1 | 16.7 | 5.7 | 0.1 | 381.5 |
| Poland | 52 | 0.252 | Non-Star | 9.1±2.4 | 11.6±3.0 | 20.2 | 9.8 | 0.6 | 206.3 | 25.8 | 12.5 | 0.8 | 264.1 |
| Ukraine | 10 | 0.281 | Star-like | 6.7±2.1 | 8.6±2.7 | 19.4 | 10.4 | 0.3 | 194.7 | 24.8 | 13.3 | 0.4 | 249.2 |
A mutation rate of 0.00069 mutations per locus per generation was used to estimate generation times.
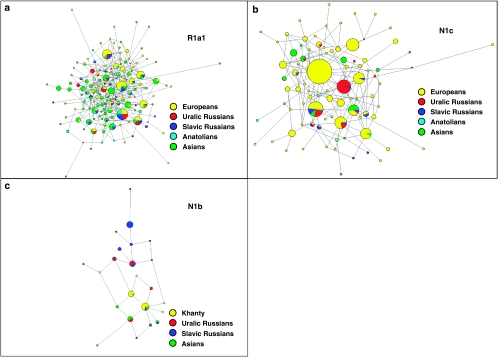
A NETWORK projection based on the Y-STR profiles of all R1a1 individuals is presented in Figure 4a. It is readily observed that the diversity of Asian haplotypes is far greater than that found in European populations. There are several specific clades exclusive to Asian groups; however, the same is not true for Europeans. The microsatellite distributions are especially interesting in Turkey (the only Anatolian group included), given the plethora of haplotypes present in the population. Supplementary Figure 1 displays the genetic relationships among R1a1 individuals of Russian Slavic descent and Uralic groups across 15 Y-STR loci in a Network projection. The distribution does not reflect population-specific partitioning or ancestral–descendant relationships, but rather all the collections appear to contain a widespread distribution of haplotypes suggesting multiple founders. A CA plot based on the Y-STR profiles of individuals belonging to haplogroup R1a1 is presented in Supplementary Figure 1a.
Figure 4.
NETWORK Projections for all populations analyzed. (a) R1a1 using 7 Y-STR loci; (b) N1c using 6 Y-STR loci; and (c) N1b using 6 Y-STR loci.
Haplogroups N1c (Tat) and N1c1 (M178)
Y-STR variance estimates for N1c1 reach levels as low as 0.079 in the Izhemski Komi group and as high as 0.226 in the Arkhangelski group (Table 4). Age estimates for haplogroup N1c1 (based on six STR loci) range from 7.2±3.4 in Tver to 9.7±5.8 in the Priluzski Komi population. Similar age estimates for other northeastern European populations were attained (Table 4); however, not all the reference populations were typed for M178 (samples typed for M178 are designated as N1c1-derived individuals in Table 4). However when using 15 STR loci, these values range from as low as 8.2±2.5 kya in the Arkhangelski collection to as high as 13.0±4.2 kya in the Komi from Priluzski. Yet, both Komi populations (Izhemski and Priluzski) exhibit N1c1 Network topologies consisting of two subclusters (Supplementary Figures 2b and c), each subcluster generating considerably lower ages; the values are 5.6±2.0 and 2.4±1.7 kya for the Izhemski Komi, and 5.5±2.0 and 2.1±0.8 kya for the Priluzski Komi (Table 4). Two distinct independent clusters are also observed when the two Komi populations are pooled together for age determinations (Supplementary Figure 2d).
Table 4. Haplotype variance and age estimations for haplogroups N1c (Tat), N1c1 (M178) and N1b (P43).
| Haplotype variance | Network time estimations with 25-year | Network time estimations with 32-year generation | BATWING expansion times using 25-year generation time (kya) | BATWING expansion times using 32-year generation time (kya) | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Population | Haplotype | N | based on 6 Y-STR loci | Network Shape | generation time (kya) (6 loci)a | time (kya) (6 loci)a | Mean | Median | 2.50% | 97.50% | Mean | Median | 2.50% | 97.50% |
| Arkhangelsk | N1c1 | 8 | 0.226 | Non-Star | 9.1±3.7 | 11.6±4.7 | 11.5 | 3.5 | 0.0 | 251.1 | 14.7 | 4.5 | 0.0 | 321.5 |
| Arkhangelsk (15 loci) | N1c1 | Star-like | 8.2±2.5 | 10.4±4.3 | 16.8 | 2.0 | 0.0 | 540.8 | 21.6 | 2.5 | 0.0 | 692.3 | ||
| Komi Izhemski | N1c1 | 28 | 0.079 | Star-like | 9.3±5.1 | 11.9±6.5 | 5.0 | 1.2 | 0.0 | 110.6 | 6.4 | 1.5 | 0.0 | 141.6 |
| Komi Izhemski (15 loci) | N1c1 | Star-like | 12.4±3.6 (5.6±2.0 and 2.4±1.7)b | 15.9±4.6 (7.2±2.6 and 3.1±2.2)b | 5.2 | 0.7 | 0.0 | 208.6 | 6.6 | 0.9 | 0.0 | 267.0 | ||
| Komi Priluzski | N1c1 | 23 | 0.121 | Star-like | 9.7±5.8 | 12.4±7.4 | 6.4 | 2.2 | 0.0 | 103.1 | 8.2 | 2.8 | 0.0 | 131.9 |
| Komi Priluzski (15 loci) | N1c1 | Star-like | 13.0±4.2 (5.5±2.0 and 2.1±0.8)b | 16.7±5.3 (7.1±2.5 and 2.7±1.0)b | 3.8 | 1.8 | 0.0 | 33.7 | 4.8 | 2.3 | 0.0 | 43.2 | ||
| Kursk | N1c1 | 5 | 0.167 | Non-Star | 8.5±4.0 | 10.8±5.1 | 33.8 | 10.4 | 0.0 | 574.3 | 43.3 | 13.3 | 0.0 | 735.1 |
| Kursk (15 loci) | N1c1 | Star-like | 9.7±2.6 | 12.4±3.4 | 39.6 | 11.7 | 0.1 | 681.3 | 50.6 | 14.9 | 0.1 | 872.0 | ||
| Tver | N1c1 | 5 | 0.183 | Non-Star | 7.2±3.4 | 9.3±4.4 | 28.4 | 8.1 | 0.0 | 569.2 | 36.3 | 10.4 | 0.0 | 728.5 |
| Tver (15 loci) | N1c1 | Star-like | 9.2±2.4 | 11.7±8.1 | 35.1 | 12.6 | 0.2 | 499.3 | 44.9 | 16.1 | 0.2 | 639.1 | ||
| China | N1c | 5 | 0.300 | Non-Star | 10.9±5.5 | 13.9±7.1 | 20.0 | 3.8 | 0.0 | 591.8 | 25.6 | 4.9 | 0.0 | 757.5 |
| Slovakia | N1c | 4 | 0.181 | Non-Star | 7.5±4.0 | 9.7±5.1 | 36.3 | 9.8 | 0.0 | 612.2 | 46.5 | 12.6 | 0.1 | 783.6 |
| Estonia | N1c | 6 | 0.206 | Non-Star | 10.1±4.7 | 12.9±6.0 | 35.8 | 10.3 | 0.0 | 634.5 | 45.8 | 13.2 | 0.0 | 812.1 |
| Finland | N1c | 312 | 0.223 | Star-like | 7.6±2.2 | 9.7±2.8 | 4.7 | 3.8 | 1.6 | 12.4 | 6.0 | 4.9 | 2.1 | 15.9 |
| Turkey | N1c | 5 | 0.450 | Non-Star | 12.1±4.8 | 15.5±6.2 | 44.4 | 10.9 | 0.1 | 1000.4 | 56.9 | 13.9 | 0.1 | 1280.5 |
| Tuva | N1c | 4 | 0.139 | Non-Star | 6.0±3.7 | 7.7±4.7 | 28.7 | 8.0 | 0.0 | 498.4 | 36.8 | 10.2 | 0.0 | 637.9 |
| Yakuts | N1c | 16 | 0.082 | Non-Star | 4.5±3.1 | 5.8±3.9 | 5.6 | 1.1 | 0.0 | 185.0 | 7.1 | 1.4 | 0.0 | 236.8 |
| Inner Mongolia | N1c1 | 6 | 0.056 | Star-like | 2.0±1.4 | 2.6±1.8 | 10.2 | 3.1 | 0.0 | 172.0 | 13.0 | 4.0 | 0.0 | 220.4 |
| Outer Mongolia | N1c1 | 5 | 0.083 | Non-Star | 3.6±2.7 | 4.6±3.5 | 14.7 | 4.6 | 0.0 | 255.6 | 18.8 | 5.9 | 0.0 | 327.2 |
| Other Siberian and Mongolian groupsc | N1c1 | 8 | 0.429 | Star-like | 10.6±4.0 | 13.5±5.1 | 28.0 | 5.8 | 0.0 | 710.8 | 35.9 | 7.5 | 0.0 | 909.9 |
| Khanty | N1b | 21 | 0.098 | Star-like | 4.0±2.6 | 5.1±3.4 | 5.8 | 3.0 | 0.0 | 93.4 | 7.4 | 3.8 | 0.0 | 119.5 |
| Khanty (15 loci) | N1b | Star-like | 2.8±1.2 | 3.5±1.5 | 3.1 | 0.8 | 0.0 | 101.4 | 3.9 | 1.0 | 0.0 | 129.9 | ||
| Komi Izhemski | N1b | 9 | 0.181 | Star-like | 6.7±4.2 | 8.6±5.4 | 10.7 | 1.2 | 0.0 | 343.8 | 13.7 | 1.5 | 0.0 | 440.1 |
| Komi Izhemski (15 loci) | N1b | Star-like | 5.6±2.0 | 7.2±2.5 | 6.7 | 2.2 | 0.0 | 216.7 | 8.6 | 2.8 | 0.0 | 277.4 | ||
| Komi Priluzski | N1b | 7 | 0.611 | Star-like | 12.9±4.1 | 16.6±5.3 | 30.6 | 3.7 | 0.0 | 1010.3 | 24.3 | 7.6 | 0.0 | 426.1 |
| Komi Priluzski (15 loci) | N1b | Star-like | 13.1±2.9 | 16.8±3.7 | 23.6 | 1.6 | 0.0 | 981.4 | 30.2 | 2.0 | 0.0 | 1256.2 | ||
| Mezen | N1b | 4 | 0.083 | Star-like | 3.0±2.1 | 3.9±2.7 | 17.2 | 5.1 | 0.0 | 299.4 | 22.0 | 6.5 | 0.0 | 383.3 |
| Mezen (15 loci) | N1b | Star-like | 5.4±2.0 | 7.0±2.6 | 22.3 | 6.5 | 0.0 | 415.5 | 28.5 | 8.3 | 0.0 | 531.9 | ||
| Pinega | N1b | 15 | 0.163 | Star-like | 5.6±2.9 | 7.2±3.7 | 7.2 | 1.5 | 0.0 | 210.8 | 9.2 | 2.0 | 0.0 | 269.9 |
| Pinega (15 loci) | N1b | Star-like | 4.2±1.6 | 5.4±2.0 | 3.3 | 1.3 | 0.0 | 63.4 | 4.3 | 1.6 | 0.0 | 81.1 | ||
| Other Slavsd | N1b | 9 | 0.653 | Star-like | 18.1±6.4 (6.0±3.7 and 6.0±3.2)b | 23.2±8.2 (7.7±4.7 and 7.7±4.1)b | 13.9 | 5.6 | 0.0 | 194.7 | 17.8 | 7.2 | 0.0 | 249.2 |
| Other Slavs (15 loci)d | N1b | Star-like | 12.1±3.3 (5.7±2.0 and 5.4±2.0)b | 15.5±4.2 (7.4±2.6 and 7.0±3.2)b | 10.6 | 3.2 | 0.0 | 233.3 | 13.6 | 4.1 | 0.0 | 298.6 | ||
| Hezhens | N1b | 8 | 0.077 | Star-like | 3.0±1.8 | 3.9±2.4 | 15.8 | 4.4 | 0.0 | 255.2 | 20.2 | 5.6 | 0.0 | 326.7 |
| Other Siberian and Mongolian groupse | N1b | 7 | 0.159 | Star-like | 5.2±2.7 | 6.6±3.5 | 19.0 | 6.0 | 0.0 | 332.9 | 24.3 | 7.6 | 0.0 | 426.1 |
A mutation rate of 0.00069 mutations per locus per generation was used to estimate generation times.
Subclustering of haplotypes led to separate time estimates for each.
Uygur Yili, Xibe, Han Harbin, Daur.
Krasnoborsk, Vologda, Belgorod, Cossacs, Livni, Porhov.
Uygur, Oroqen, Outer Mongolian.
A Network projection based on N1c haplotype distributions exhibits a segregation between Asian and European groups despite some haplotypic sharing between these two (Figure 4b). Close relationships are observed among the Slavic and Uralic Russians, as most haplotypes present in one subset of populations are also present in the other. They both share clusters with European groups as well. Supplementary Figure 2e displays a Network Analysis of N1c1-derived individuals based on the 15 Y-STR loci. The Slavic populations (Arkhangelski, Kursk, and Tver) are found to the right of the projection along with some Komi haplotypes (shown in red and green); however, a split to the left of the NETWORK, shared only by the Priluzski and Izhemski Komi populations, suggests a different source for N1c1 (M178) in these Uralic groups or, alternatively, the expansion into the Komi territory of Slavic individuals. Supplementary Figure 2a illustrates a CA graph based on the Y-STR profile (six loci) of individuals possessing the N1c1 haplogroup.
Haplogroup N1b (P43)
N1b is the predominant haplogroup in the Khanty population; however, Y-STR variance values are much higher for both the Izhemski and Priluzski Komi groups (0.098 versus 0.181 and 0.611, respectively) (Table 4). Similarly, time estimates for the Khants reveal a rather recent entrance of the haplogroup into the population (4.0±2.6), whereas much later dates are obtained for the Izhemski (6.7±4.2) and Priluzski (12.9±4.1) Komi populations (Table 4). Variance calculations for the Pinega and Mezen populations yield Vp values of 0.163 and 0.083, respectively. Conversely, the other Slavs group attains a variance value of 0.653 and age estimate of 18.1±6.4; however, a bipartite structure is observed with separate clusters that attain ages of 6.0 ± 3.7 and 6.0±3.2. A Network Analysis, including Khanty, and the Uralic and Slavic Russian groups at a resolution of 15 Y-STR loci, displays a clear partition between the Slavic groups and the Khanty collection (Supplementary Figure 3b). Interestingly, the Izhemski Komi partitions to the portion of the projection encompassing the Slavic groups while the Komi from Priluzski shares haplotypes with both clusters.
A Network projection based on Y-STR distributions of haplogroup N1b is presented in Figure 4c. Haplotype distributions in Uralic groups are widespread throughout the projection sharing clusters with both Asian and European Slavic populations. The Siberian Khanty collection segregates into one portion of the graph composed of Asian haplotypes, but shows some affinities to Uralic groups as well. A CA based on the Y-STR haplotype frequencies of these populations is presented in Supplementary Figure 3a.
Haplogroup N
Grouped age estimates based on the major bifurcations of haplogroup N were performed to achieve a consensus on the antiquity of each of its sub-haplogroups on a regional basis (Europe and Asia) and in specific ethnic groups (Russian Slavic and Russian Uralic) (Table 5). Estimates for M231 (N*) are highest among Mongolian/Siberian groups reaching 23.7±5.4 kya compared with an overall value for all populations of 19.1±4.2 kya. BATWING expansion times for the same comparison yield an overall age of 3.8 kya for M231. The age for haplogroup N1a (M128) is more recent (6.9±3.9 kya) than that of its sister clades N1b (15.8±5.4 kya), which exhibits a bipartite substructure leading to separate age estimates of 7.8±3.7 and 5.0±2.2 kya (BATWING yields an estimate of 1.1 kya for this haplogroup), and N1c (10.7±3.4 kya). BATWING estimates for N1c achieve an age of 3.0 kya. Calculations using all clades within the haplogroup (N*, N1a, N1b, and N1c) yield an average age of 13.4±4.0 kya, meanwhile BATWING calculations provide a value of 3.4 kya. It should be noted that these BATWING age estimates do exhibit a credible 95% CI; however, they dispute previous findings by Rootsi et al15 and provide recent haplogroup ages. NETWORK projections, on the other hand, provide estimates that go hand in hand with previous findings.15
Table 5. Grouped age estimations.
| Network time estimations with 25-year | Network time estimations with 32-year | BATWING expansion time using 25-year generation time (kya) | BATWING expansion time using 32-year generation time (kya) | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Haplogroup and population group | N | Network shape | generation time (kya)a | generation time (kya)a | Mean | Median | 2.50% | 97.50% | Mean | Median | 2.50% | 97.50% |
| N (M231) | ||||||||||||
| Asianb | 39 | Star | 19.5±4.2 | 25.0±5.2 | 13.9 | 11.3 | 3.0 | 55.9 | 17.8 | 14.5 | 3.9 | 71.6 |
| Chinesec | 14 | Non-Star | 12.1±3.7 | 15.5±4.7 | 14.9 | 10.3 | 0.5 | 122.8 | 19.1 | 13.2 | 0.7 | 157.2 |
| Mongolian and Siberiand | 23 | Non-Star | 23.7±5.4 | 30.3±7.0 | 27.1 | 22.0 | 7.0 | 100.1 | 34.7 | 28.1 | 9.0 | 128.1 |
| Europeane | 40 | Star-like | 10.9±4.0 (9.1±3.3 and 6.5±2.6)a | 13.9±5.2 (11.6±4.2 and 8.3±3.3)a | 6.9 | 5.1 | 1.4 | 29.2 | 8.8 | 6.5 | 1.8 | 37.3 |
| All | 79 | Star-like | 19.1±4.2 | 24.5±5.4 | 4.7 | 3.8 | 1.4 | 16.2 | 6.0 | 4.9 | 1.8 | 20.8 |
| N1a (M128) | ||||||||||||
| Asianf | 6 | Star-like | 6.9±3.9 | 8.8±4.9 | 17.0 | 3.4 | 0.0 | 405.7 | 21.8 | 4.3 | 0.0 | 519.3 |
| N1b (P43) | ||||||||||||
| Asiang | 15 | Non-Star | 7.2±3.6 | 9.3±4.6 | 14.2 | 5.2 | 0.1 | 283.8 | 18.1 | 6.6 | 0.1 | 363.2 |
| Russian Slavich | 28 | Star-like | 12.1±4.7 (9.1±4.9 and 6.0±3.7)a | 15.5±6.0 (11.6±6.3 and 7.7±4.7)a | 6.4 | 1.6 | 0.0 | 118.2 | 8.1 | 2.1 | 0.0 | 151.2 |
| Russian Slavic (15 loci)h | Star-like | 11.6±3.1 (8.7±2.3 and 3.3±1.0)a | 14.9±4.0 (11.1±2.9 and 4.3±1.3)a | 2.9 | 1.7 | 0.1 | 30.6 | 3.7 | 2.1 | 0.1 | 39.2 | |
| Russian Uralici | 16 | Star-like | 12.8±5.0 | 16.4±6.4 | 10.9 | 1.9 | 0.0 | 384.4 | 13.9 | 2.4 | 0.0 | 492.1 |
| Russian Uralic (15 loci)i | 14.6±3.5 (10.5±3.2 and 4.1±1.3)a | 18.7±4.4 (13.4±4.1 and 5.3±1.6)a | 8.7 | 2.4 | 0.0 | 219.6 | 11.1 | 3.1 | 0.0 | 281.1 | ||
| Russian Uralic and Khantsj | 37 | Star-like | 9.8±4.2 (6.6±4.1 and 3.4±2.1)a | 12.5±5.4 (8.4±5.3 and 4.5±2.6)a | 7.9 | 1.5 | 0.0 | 292.7 | 10.2 | 1.9 | 0.0 | 374.6 |
| Russian Uralic and Khants (15 loci)j | 8.7±2.2 (5.3±1.9 and 4.2±1.2)a | 11.2±2.8 (6.7±2.4 and 5.4±1.8)a | 9.3 | 1.6 | 0.0 | 235.1 | 11.9 | 2.0 | 0.1 | 300.9 | ||
| All | 80 | Star-like | 15.8±5.4 (7.8±3.7 and 5.0±2.2)a | 20.3±7.0 (9.9±4.7 and 6.4±2.8)a | 2.0 | 1.1 | 0.2 | 21.7 | 2.5 | 1.4 | 0.2 | 27.8 |
| N1c (Tat) | ||||||||||||
| Russian Slavick | 18 | Star-like | 8.1±2.5 | 10.3±3.2 | 15.9 | 5.1 | 0.2 | 235.1 | 20.3 | 6.5 | 0.2 | 300.9 |
| Russian Slavic (15 loci)k | 8.3±1.6 | 10.7±2.1 | 18.4 | 9.4 | 0.4 | 231.9 | 23.6 | 12.1 | 0.5 | 296.9 | ||
| Russian Uralicl | 51 | Star-like | 9.5±5.3 (4.5±2.2 and 0.6±0.3)a | 12.1±6.8 (5.8±2.8 and 0.7±0.4)a | 3.2 | 2.4 | 0.7 | 16.4 | 4.1 | 3.1 | 0.9 | 21.0 |
| Russian Uralic (15 loci)l | 11.2±2.8 (5.4 ±1.5 and 2.8±1.3)a | 14.3±3.6 (7.0±1.8 and 3.1±1.6)a | 11.2 | 2.3 | 0.2 | 195.4 | 14.4 | 2.9 | 0.2 | 250.1 | ||
| Russian Slavic and Russian Uralicm | 69 | Star-like | 8.3±3.4 | 10.6±4.8 | 2.0 | 1.6 | 0.1 | 11.3 | 2.6 | 2.1 | 0.1 | 14.4 |
| Russian Slavic and Russian Uralic (15 loci)n | 26.5±5.5 (15.0±2.8 and 2.9±1.3)a | 33.9±7.0 (19.2±3.6 and 3.7±1.6)a | 1.9 | 1.4 | 0.3 | 9.5 | 2.4 | 1.7 | 0.3 | 12.2 | ||
| Asiann | 44 | Star-like | 15.9±4.9 | 20.4±6.2 | 20.0 | 0.9 | 0.0 | 898.7 | 25.5 | 1.2 | 0.0 | 1150.3 |
| Europeano | 399 | Star-like | 9.4±2.6 | 12.1±3.3 | 4.4 | 4.0 | 1.8 | 10.3 | 5.6 | 5.2 | 2.3 | 13.1 |
| All | 443 | Star-like | 10.7±3.4 | 13.6±4.3 | 3.4 | 3.0 | 1.3 | 8.4 | 4.4 | 3.8 | 1.7 | 10.8 |
| N (overall N-N1c) | ||||||||||||
| All | 580 | Star-like | 13.4±4.0 | 17.2±5.1 | 4.0 | 3.4 | 1.5 | 10.5 | 5.2 | 4.4 | 1.9 | 13.5 |
Subclustering of haplotypes led to separate time estimates for each.
Tibet (3), Hani (4), Han (5), Manchu (2), Uygur (2), Xibe (4), Japan (2), Korea (1), China (14), Philippines (1), Cambodia (1).
Tibet (3), Hani (4), Han (5), Manchu (2), Uygur (2), Xibe (4), Japan (2), Korea (1).
Finland (19); Hungary, Sweden (5); Czech Republic (2); England, Poland (2); Latvia, Russia (2); Lithuania (2); Czechoslovakia, Belarus, Germany, Ireland, Norway.
Manchu (2), Buyi (2), Xibe, Kazah.
Manchu, Uygur (2); Oroqen (2); Outer Mongolia (2); Hezhen (8).
Pinega (15); Mezen (4); Krasnoborsk (3); Vologda (2); Belgorod, Cossacs, Livni, Porhov.
Komi Izhemski (9), Komi Priluzski (7).
Komi Izhemski (9), Komi Priluzski (7), Khanty (21).
Arkhangelsk (8), Kursk (5), Tver (5).
Komi Izhemski (28), Komi Priluzski (23).
Arkhangelsk (8), Kursk (5), Tver (5), Komi Izhemski (28), Komi Priluzski (23).
Daur (4); Mongola, Oroqen (2); Xibe (3); Yakut (16); Inner Mongolia (6), Outer Mongolia (5); Han, Uygur (2); Tuva (4).
Arkhangelsk (8), Estonia (6), Finland (312), Komi Izhemski (28), Komi Priluzski (23), Kursk (5), Slovakia (4), Turkey (5), Tver (5), Khanty (3).
Discussion
Population relationships
The CA (Figure 3) based on major Y-SNP haplogroups reveals several distinct groupings reflecting both geographic and linguistic affiliation. With the exception of the Khanty, a clear cluster is formed among the Uralic-speaking populations where Finland segregates at a distance from the rest. This partitioning may be related to Finland's low-effective population size for long periods of time and local isolation of small groups, possibly causing major bottlenecks, which are significantly limiting the current diversity of the population, allowing for genetic drift.4
Lithuania and Latvia, both Indo-European-speaking groups, are also found within the Uralic assemblage. This phylogenetic connection between the Baltic- and Uralic-speaking collections is also reflected in Fst distances (Supplementary Table 2), where both Lithuania and Latvia exhibit nonsignificant genetic distances with all Uralic speakers, excluding Khanty (Supplementary Table 2). When distances within the group are averaged, Fst values are lower when Lithuania and Latvia are included in the calculations (Fstavg=0.03622) than when they are removed (Fstavg=0.05322). On the other hand, analyzing these two populations with Slavic-speaking groups leads to an increase from 0.09078 to 0.09222 in Fst distances. These data lend support to previous findings by Kasperaviciute et al,51 who propose a close relationship between Lithuanians and Latvians with their Finno-Ugric-speaking neighbors (Estonia and Finland). The Y-haplogroup distributions of Latvia and Lithuania also exhibit greater affinity with those of Uralic populations than with the other Indo-European-speaking groups. For example, both Baltic populations display considerable frequencies of haplogroup N1c (33 and 47% in Latvia and Lithuania, respectively), whereas in other geographically proximal Indo-European-speaking groups (ie, Belarus, Slovakia, and Poland), this frequency is only 2–5% (Figure 2). These data corroborate results by Laitinen et al,52 indicating that males from these Baltic and Uralic populations exhibit common genetic patrimonies and suggests that the Uralic dominion encompassed a greater area than has been previously reported.
It has been reported that haplogroup distributions from western (Poles, Slovakians, Czechs, and Lusatians), southern (Slovenes, Croats, Bosnians, Montenegrins, Serbs, Macedonians, and Bulgarians), and eastern Slavs (Belarusians and Ukrainians) differ considerably from those of Russian Slavs, specifically northwestern Russians (also considered part of the eastern Slavs). For example, Slovakians, Ukrainians, Poles and Belarusians exhibit very low frequencies of N1c, whereas the haplogroup attains levels of 13, 13, and 29% in the Russian Slavic groups of Kursk, Tver, and Arkhangelski, respectively, despite the close geographical proximity of these groups. These differences are also observed between Russian groups, with southeastern Russians exhibiting frequencies of N1c as low as 5% in a collection from the Livni province and northeastern Russians possessing levels as high as 46% in the Mezen locality.23 N1c is particularly high in populations of Uralic descent and may signal genetic input from the autochthonous (former) groups of northeastern Europe. The Slavic Russian populations (Kursk, Tver, and Arkhangelski) also possess frequencies of haplogroup I of 15, 18, and 50%, respectively, found at 18% in Ukraine, where it may have arisen during the LGM;13, 53 similar frequency distributions of haplogroup I have been reported for other Russian groups.23 The distributions and clinal frequency gradients of N1c support the hybridization hypothesis for Slavic Russians and argue for considerably more genetic signals from Uralic tribes in northwestern Russian groups than in the rest of the eastern Slavic domain.
It should be noted that although statistically significant correlations are observed between linguistics as well as geography and genetics in the AMOVA, a closer relationship between geography and genetics (8.81% in the Among Groups comparison versus 7.87% in the Among Populations Within-Groups comparison) than between linguistics and genetics (6.51% variance attributable to the Among Groups comparison versus 10.10% to the Among Populations Within-Groups estimate) as has been stated previously,15, 28, 29, 50 is seen when populations throughout Eurasia are compared at the transcontinental level. When only members of the Balto-Slavic linguistic branch of the Indo-European language family and Uralic groups are compared, neither linguistic nor geographic ties appear to define the genetic structure of the populations in question, suggesting that other factors besides geographical proximity and linguistic affiliations have been involved in shaping the current genetic and phylogenetic relationships of members of these two linguistic families (Table 2).
A discontinuity is apparent between populations from North Caucasia and Baltic/Slavic/Uralic groups to the north in the distributions of haplogroups G and N (Figure 2). Haplogroup G is confined to the Caucasus and the Middle East and not detected in the northern groups (Slavic and other Eastern European populations) despite the lack of major geographical barrier between the northern Caucasus and the aforementioned areas. Conversely, haplogroup N is not observed within the Caucasus despite its high frequencies and widespread distribution throughout northeastern Europe, Siberia, and Central Asia (these apparent disconnections have also been reported by Fechner et al50). Phylogenetic relationships also illustrate a disconnection between northeastern European populations, which despite their proximal geographical locations map at opposite ends of the plot (Figure 3), suggesting linguistic, and/or ethnic obstacles to gene flow. Cultural barriers to genetic exchange have been previously observed in the Kalmyks, a group that after relocating to the area near the Caucasus from Mongolia has not received genetic inputs from North Caucasian groups.54 Populations from Caucasia, in turn, are described as traditional genetic isolates that have remained separate and independent from other groups for thousands of years.55
Haplogroup R1a1 is represented by complex diversity patterns
Haplogroup R1a1 (delineated by mutation M198) is believed to have originated in present day Ukraine13 following the LGM, and is thought to mark the expansion of the Kurgan horse culture.12 Kurgan migrations are believed to have occurred both into Europe and to the east, resulting in the dissemination of the Indo-European languages.56 Alternatively, Sengupta et al18 and Wells et al12 have proposed that the haplogroup originated in Northwestern India and in the Central Asian steppes, respectively, given the wide variety of R1a1 Y-STR haplotypes throughout these areas. Network age estimations from this study suggest that two separate groups exist within R1a1 with similar ages for populations found at the western (Serbia 17.3±5.4) and eastern (South Pakistan 18.7±4.7) poles of the expansion. These results along with time estimates for several other populations across Europe and Asia support the findings by Sengupta et al18 regarding the central Asian origins of the mutation. NETWORK projections also support an Asian origin to this haplogroup, given the plethora of STR haplotypes present in these groups versus those found in European populations (Figure 4a).
The R1a1 network projection in Supplementary Figure 1b based on 15 Y-STR loci lacks substructure along population lines. A central core of individuals and star-like topology is indicative of similar haplotypes from a common source for both the Slavic and Uralic Russians genotyped in this report. These results corroborate the comparable expansion time estimates based on 7 and 15 STR loci (Table 3).
Microevolutionary processes
The separation between the geographically proximal collections of the Priluzski Komi and Izhemski Komi in Supplementary Figure 2a is noteworthy. Similarly, North and South Pakistan partition distantly in the plot. In the case of South and North Pakistan, one possible explanation is the distinctive involvement of South Pakistan as a migratory corridor between the Middle East and Asia in the original migration of modern humans out of Africa followed by bidirectional dispersals.38 North Pakistan, on the other hand, located at the southwestern end of the Himalayan range, a known genetic as well as topo-geographical barrier,39 has more likely experienced limited dispersals allowing for the observed patterns.
The differences between the two Komi groups may reflect events regarding people with a common origin being differentially influenced genetically by unrelated migrations and/or genetically distinct populations adopting similar cultures and languages. It is possible that the observed genetic differences may reflect cultural and socioeconomical separations between the two groups, who despite inhabiting a close geographical area exhibit differing subsistence styles (the Komi from Priluzski are cattle breeders and farmers, whereas the Komi from Izhemski have adapted reindeer herding from neighboring Nenets).10 In support of this scenario, it is known that the Priluzski Komi belong to a group of populations that appear to have arisen much earlier historically than the Izhemski Komi, which, in turn, exhibit some peculiar linguistic traits not observed in other Komi populations.57 Yet, the profound differences in the Y-STR profiles and the separation from each other in the Network Analysis argue for populations with unique genetic backgrounds.
Possible origin and migration patterns of haplogroups N1c1 (M178) and N1b (P43)
Haplogroup N is found throughout North-Central Eurasia at varying frequencies with sub-haplogroup N1c being the most widespread.15 Proposed migratory routes based on Y-STR variance estimations have suggested that N1c carriers spread from northern China through Siberia to northeastern Europe.15 Sub-haplogroup N1c1 (defined by mutation M178), long believed to be restricted to Europe and to mark a recent Uralic migration into northern Europe,14 is now known to be widespread in northern China and Mongolia.16 However, Y-STR variance values from this study do not support a migratory route from the Urals to the northeastern Slavic domain, as Russian Slavic populations exhibit higher Y-STR diversity (as high as 0.226 in the Arkhangelski population) than those found in the Uralic groups (0.079 in the Izhemski Komi and 0.121 in the Priluzski Komi) (Table 4).
When Network projections are constructed for N1c1 using 15 Y-STR loci, topologies composed of two clusters observed for both Komi populations (Supplementary Figures 2a and b), leading to separate time estimates at the individual cluster level of 5.6±2.0 and 2.4±1.7 kya for the Komi from Izhemski, and 5.5±2.0 and 2.1±0.8 kya for the Komi from Priluzski. When the two populations are grouped (Supplementary Figure 3d), similar age estimates are attained for each subcluster (Table 5). On the other hand, the Network projections for the Russian Slavic populations do not show dual clustering, and their ages range from 8.2±2.5 kya in Arkhangelski to 9.7±2.6 kya in Kursk (Table 4), and 8.3±1.6 kya when the three Slavic populations are grouped (Table 6). The presence of dual clusters in these Komi groups may explain the high age estimates previously observed for this region, leading to the suggestion that an east to west dispersal of N1c1 was the most likely migratory route taken by the haplogroup's carriers.15 It is possible that the age values previously reported15 may be the result of subpopulation structure (known to lead to erroneously inflated accumulated ages) within the Uralic populations analyzed, probably resulting from the input from different source populations (eg, of Asian and European descent).
Similarly, haplotype variance calculations based on haplogroup N1c do not support an east to west dispersal, given that northeastern European populations, such as Finland (0.223), Estonia (0.206), Tver (0.183), Arkhangelski (0.226), and Kursk (0.167), possess higher variance levels than the Komi Izhemski and Priluzski collections (0.079 and 0.121, respectively). As such, these results suggest that, instead of the previously reported migratory scenario from the Urals to the west,14, 15 the flow of N1c may have occurred in the opposite direction. As older ages are observed when grouping All Asians versus All Europeans (Table 5) for N1c, the available data suggest that the mutation may have originated in northern China as previously reported,14, 15 but may have traversed through a different migratory route than has been postulated elsewhere,15 reaching northeastern European populations before the Urals. The presence of haplogroup I (of European descent) in both Komi populations (specifically I-M253), in turn, suggests that European groups have contributed to these populations' gene pools. The absence of I-M253 in the Khanty of West Siberia completes the demic decrease of this haplogroup (Europe–Urals–West Siberia), supporting the stipulated west to east Y-driven migration.
Haplogroup N1b has been reported to have separated into two clades of similar ages about 6.2 and 6.8 kya for Asia and Europe, respectively.15 Yet, time and variance estimations in this study indicate a much older origin for the haplogroup (12.9±4.1 kya) in the Priluzski Komi collection (Table 4). Comparable age estimates were obtained using two sets of 6 and 15 Y-STR markers; the battery of 6 loci is included in the group of 15. In the Network Analysis, the Khanty collection segregates into one portion of the bi-cluster topology observed (Supplementary Figure 3b) along with the Slavic populations identified as carrying Asian haplotypes,23 meanwhile the Komi from Izhemski and the Slavic Russian populations partition toward the other extreme of the projection. However, the Komi from Priluzski exhibit a bipartite distribution throughout the two star-like sub-clusters, providing an explanation for the population's high variance and old age estimates. These results make it possible to contemplate a scenario where the Komi from Priluzski have contributed differentially to populations within Asia and the Slavic domain. These findings should be further explored by examining other Uralic populations to elucidate whether the mutation did originate among the Priluzski Komi or whether other people within the region exhibit older age estimates and higher accumulated STR variance. Nevertheless, with the Khanty population, located eastward in northwest Siberia, exhibiting the most recent age and variance estimations, the data implicate migrations from the Urals into Siberia and Asia rather than the converse.
Acknowledgments
The work was partially supported by the programs of Presidium of Russian Academy of Sciences: ‘Molecular and Cell Biology' and ‘Fundamental Sciences for Medicine' (subprogram ‘Human Polymorphism'), and the Russian Foundation for Basic Research.
Footnotes
Supplementary Information accompanies the paper on European Journal of Human Genetics website (http://www.nature.com/ejhg)
Supplementary Material
References
- Pavlov P, Roebroeks W, Svedsen JI. The Pleistocene colonization of northeastern Europe: a report on recent research. J Hum Evol. 2004;47:3–17. doi: 10.1016/j.jhevol.2004.05.002. [DOI] [PubMed] [Google Scholar]
- Otte M.The northwestern European Plain around 18 000 BPIn Soffer O, Gamble C (eds): The World at 18 000 BP Unwin Hyman: London; 1990. Vol 1, pp54–68. [Google Scholar]
- Torroni A, Bandelt H, D'Urbano L, et al. mtDNA analysis reveals a major late Paleolithic population expansion from southwestern to northeastern Europe. Am J Hum Genet. 1998;62:1137–1152. doi: 10.1086/301822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lappalainen T, Koivumaki S, Salmela E, Huoponen K, Sistonen P, Savontaus ML, Lahermo P. Regional differences among the Finns: a Y-chromosomal perspective. Gene. 2006;376:207–215. doi: 10.1016/j.gene.2006.03.004. [DOI] [PubMed] [Google Scholar]
- Simchenko YuB.Early ethnogenesis of ethnic groups of the Ural language family from transpolar and circumpolar EurasiaIn Gurvich IS (ed): Ethnogenez narodov severa Moscow: Nauka; 198011–27. [Google Scholar]
- Sajantila A, Lahermo P, Anttinen T, et al. Genes and languages in Europe: an analysis of mitochondrial lineages. Genome Res. 1995;5:42–52. doi: 10.1101/gr.5.1.42. [DOI] [PubMed] [Google Scholar]
- Malyarchuk BA. Differentiation of the mitochondrial subhaplogroup U4 in the populations of Eastern Europe, Ural, and Western Siberia: implication to the genetic history of the Uralic populations. Genetika. 2004;40:1281–1287. [PubMed] [Google Scholar]
- Verbenko DA, Knjazev AN, Mikulich AI, Khusnutdinova EK, Bebyakova NA, Limborska SA. Variability of the 3′APOB minisatellite locus in eastern Slavonic populations. Hum Hered. 2005;60:10–18. doi: 10.1159/000087338. [DOI] [PubMed] [Google Scholar]
- Verbenko DA, Slominsky PA, Spitsyn VA, et al. Polymorphisms at locus D1S80 and other hypervariable regions in the analysis of Eastern European ethnic group relationships. Ann Hum Biol. 2006;33:570–584. doi: 10.1080/03014460601012077. [DOI] [PubMed] [Google Scholar]
- Khrunin A, Verbenko D, Nikitina K, Limborska S. Regional differences in the genetic variability of Finno-Ugric speaking Komi populations. Am J Hum Bio. 2007;19:741–750. doi: 10.1002/ajhb.20620. [DOI] [PubMed] [Google Scholar]
- Khrunin AV, Tarskaia LA, Spitsyn VA, Lylova OI, Bebyakova NA, Mikulich AI, Limborska SA. p53 polymorphisms in Russia and Belarus: correlation of the 2-1-1 haplotype frequency with longitude. Mol Genet Genomics. 2005;272:666–672. doi: 10.1007/s00438-004-1091-8. [DOI] [PubMed] [Google Scholar]
- Wells RS, Yuldasheva N, Ruzibakiev R, et al. The Eurasian heartland: a continental perspective on Y-chromosomal diversity. Proc Natl Acad USA. 2001;98:10244–10249. doi: 10.1073/pnas.171305098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semino O, Passarino G, Oefner PJ, et al. The genetic legacy of Paleolithic Homo sapiens sapiens in extant Europeans: A Y chromosome perspective. Science. 2000;290:1155–1159. doi: 10.1126/science.290.5494.1155. [DOI] [PubMed] [Google Scholar]
- Zerjal T, Dashnyam B, Pandya A, et al. Genetic relationships of Asians and Northern Europeans, revealed by Y-chromosomal DNA analysis. Am J Hum Genet. 1997;60:1174–1183. [PMC free article] [PubMed] [Google Scholar]
- Rootsi S, Zhivotovsky LA, Baldovic M, et al. A counter-clockwise northern route of the Y-chromosome haplogroup N from Southeast Asia towards Europe. Eur J Hum Genet. 2007;15:211–405. doi: 10.1038/sj.ejhg.5201748. [DOI] [PubMed] [Google Scholar]
- Xue Y, Zerjal T, Ban W, et al. Male demography in East Asia: a north–south contrast in human population expansion times. Genetics. 2006;172:2431–2439. doi: 10.1534/genetics.105.054270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guglielmino CR, Piazza A, Menozzi P, Cavalli-Sforza LL. Uralic genes in Europe. Am J Phys Anthropol. 1990;83:57–68. doi: 10.1002/ajpa.1330830107. [DOI] [PubMed] [Google Scholar]
- Sengupta S, Zhivotovsky LA, King R, et al. Polarity and temporality of high-resolution Y-chromosome distributions in India identify both indigenous and exogenous expansions and reveals minor genetic influence of Central Asian pastoralists. Am J Hum Genet. 2006;78:202–221. doi: 10.1086/499411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niitemaa V, Hovi K. Baltian Historia (Jyväskylä) Helsinki, Finland; 1991. pp. 272–276. [Google Scholar]
- Harvey RG, Tills D, Warlow A, et al. Genetic affinities of the Balts. A study of blood groups, serum proteins and enzymes of Lithuanians in the United Kingdom. Man (N. S.) 1983;18:535–552. [Google Scholar]
- Rudiger L, Edelmann J. Population data of Y-chromosomal STRs in Lithuanian, Latvian and Estonian males. Forensic Sci Int. 2001;120:223–225. doi: 10.1016/s0379-0738(01)00391-7. [DOI] [PubMed] [Google Scholar]
- Rebala K, Mikulich AI, Tsybovsky IS, et al. Y-STR variation among Slavs: evidence for the Slavic homeland in the Middle Dnieper Basin. J Hum Genet. 2007;52:406–414. doi: 10.1007/s10038-007-0125-6. [DOI] [PubMed] [Google Scholar]
- Balanovsky O, Rootsi S, Pshenichnov A, et al. Two sources of the Russian patrilineal heritage in their Eurasian context. Am J of Hum Genet. 2008;82:236–250. doi: 10.1016/j.ajhg.2007.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malyarchuk BA, Grzybowski T, Derenko MV, et al. Mitochondrial DNA variability in Poles and Russians. Ann Hum Genet. 2002;66:261–283. doi: 10.1017/S0003480002001161. [DOI] [PubMed] [Google Scholar]
- Belyaeva O, Bermisheva M, Khrunin A, et al. Mitochondrial DNA variations in Russian and Belorussian populations. Hum Biol. 2003;75:647–660. doi: 10.1353/hub.2003.0069. [DOI] [PubMed] [Google Scholar]
- Khar'kov VN, Stepanov VA, Borinskaya SA, et al. Gene pool structure of Eastern Ukrainians as inferred from the Y-chromosome haplogroups. Russ J Genet. 2004;40:326–331. [PubMed] [Google Scholar]
- Khrunin AV, Bebiakova NA, Ivanov VP, Solodilova MA, Limborskaia SA. Polymorphism of Y-chromosomal microsatellites in Russian populations from the northern and southern Russia as exemplified by the populations of Kursk and Arkhangelsk Oblast. Russ J Genet. 2005;41:922–927. [PubMed] [Google Scholar]
- Das K, Mastana SS. Genetic variation of three VNTR loci in three tribal populations of Orissa, India. Ann Hum Biol. 2003;30:237–249. doi: 10.1080/0301446031000064512. [DOI] [PubMed] [Google Scholar]
- Rosser ZH, Zerjal T, Hurles ME, Adojaan M, et al. Y-chromosomal diversity in Europe is clinal and influenced primarily by geography, rather than by language. Am J Hum Genet. 2000;67:1526–1543. doi: 10.1086/316890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasidze I, Sarkisian T, Kerimov A, Toneking M. Testing hypotheses of language replacement in the Caucasus: evidence from the Y chromosome. Hum Genet. 2003;112:255–261. doi: 10.1007/s00439-002-0874-4. [DOI] [PubMed] [Google Scholar]
- Nasidze I, Ling EYS, Quinque D, et al. Mitochondrial DNA and Y-chromosome variation in the Caucasus. Ann Hum Genet. 2004;68:205–222. doi: 10.1046/j.1529-8817.2004.00092.x. [DOI] [PubMed] [Google Scholar]
- Al-Zahery N, Semino O, Benuzzi G, et al. Y-chromosome and mtDNA polymorphisms in Iraq, a crossroad of the early human dispersal and of post-Neolithic migrations. Mol Phylogenet Evol. 2003;28:458–472. doi: 10.1016/s1055-7903(03)00039-3. [DOI] [PubMed] [Google Scholar]
- Cinnioğlu C, King R, Kivisild R, et al. Excavating Y-chromosome haplotype strata in Anatolia. Hum Genet. 2004;114:127–148. doi: 10.1007/s00439-003-1031-4. [DOI] [PubMed] [Google Scholar]
- Regueiro M, Cadenas AM, Gayden T, Underhill PA, Herrera RJ. Iran: tricontinental nexus for Y-chromosome driven migration. Hum Hered. 2006;61:132–143. doi: 10.1159/000093774. [DOI] [PubMed] [Google Scholar]
- Karafet T, Xu L, Du R, et al. Paternal population history of East Asia: sources, patterns, and microevolutionary processes. Am J Hum Genet. 2001;69:615–628. doi: 10.1086/323299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derenko M, Malyarhuk B, Denisova GA, et al. Contrasting patterns of Y-chromosome variation in South Siberian populations from Baikal and Altai-Sayan regions. Hum Genet. 2006;118:591–604. doi: 10.1007/s00439-005-0076-y. [DOI] [PubMed] [Google Scholar]
- Antunez-de-Mayolo G, Antunez-de-Mayolo A, Antunez-de-Mayolo P, et al. Phylogenetics of Worldwide populations as determined by polymorphic Alu insertions. Electrophoresis. 2002;23:3346–3356. doi: 10.1002/1522-2683(200210)23:19<3346::AID-ELPS3346>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- Cadenas AM, Zhivotovsky LA, Cavalli-Sforza LL, Underhill PA, Herrera RJ. Y chromosome diversity characterizes the Gulf of Oman. Eur J Hum Genet. 2008;16:374–386. doi: 10.1038/sj.ejhg.5201934. [DOI] [PubMed] [Google Scholar]
- Gayden T, Cadenas AM, Regueiro M, et al. The Himalayas as a directional barrier to gene flow. Am J Hum Genet. 2007;80:884–894. doi: 10.1086/516757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammer MF, Horai S. Y chromosomal DNA variation and the peopling of Japan. Am J Hum Genet. 1995;56:951–962. [PMC free article] [PubMed] [Google Scholar]
- Underhill PA, Passarino G, Lin AA, et al. The phylogeography of Y chromosome binary haplotypes and the origins of modern human populations. Ann Hum Genet. 2001;65:43–62. doi: 10.1046/j.1469-1809.2001.6510043.x. [DOI] [PubMed] [Google Scholar]
- Kayser M, Krawczak M, Excoffier L, et al. An extensive analysis of Y-chromosomal microsatellite haplotypes in globally dispersed human populations. Am J Hum Genet. 2001;68:990–1018. doi: 10.1086/319510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhivotovsky LA, Underhill PA, Cinnioğlu C, et al. The effective mutation rate at Y chromosome short tandem repeats, with application to human population divergence time. Am J Hum Genet. 2004;74:50–61. doi: 10.1086/380911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fenner JN. Cross-cultural estimation of the human generation interval for use in genetics-based population divergence studies. Am J Phys Ant. 2005;128:415–423. doi: 10.1002/ajpa.20188. [DOI] [PubMed] [Google Scholar]
- Röhl A.Network 2.0b. A program package for phylogenetic networks. Mathematisches Seminar, Universität HamburgAvailable at http://www.fluxus-engineering.com , 1997
- Luis JR, Rowold DJ, Regueiro M, et al. The Levant versus the Horn of Africa: evidence for bidirectional corridors of human migrations. Am J Hum Genet. 2004;74:532–544. doi: 10.1086/382286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohlf F. NTSYSpc. Setauket, NY: Exter Publishing; 2002. [Google Scholar]
- Guo S, Thompson E. Performing the exact test of Hardy–Weinberg proportion for multiple alleles. Biometrics. 1992;48:361–372. [PubMed] [Google Scholar]
- Schneider S, Kueffer JM, Roessli D, Excoffler L. Arlequin v. 2.000: A Software for Population Genetics Data Analysis. Geneva: Genetics and Biometry Laboratory, University of Geneva; 2000. [Google Scholar]
- Fechner A, Quinque D, Rychkov S, et al. Boundaries and clines in the West Eurasian Y-chromosome landscape: insights from the European part of Russia. Am J Phys Ant. 2008;137:41–47. doi: 10.1002/ajpa.20838. [DOI] [PubMed] [Google Scholar]
- Kasperaviciute D, Kucinskas V, Stoneking M. Y chromosome and mitochondrial DNA variation in Lithuanians. Ann Hum Genet. 2004;68:438–452. doi: 10.1046/j.1529-8817.2003.00119.x. [DOI] [PubMed] [Google Scholar]
- Laitinen V, Lahermo P, Sistonen P, Savontaus ML. Y-chromosomal diversity suggests that Baltic males share common Finno-Ugric-speaking forefathers. Hum Hered. 2002;53:68–78. doi: 10.1159/000057985. [DOI] [PubMed] [Google Scholar]
- Pericic M, Barac L, Klaric IM, et al. High-resolution phylogenetic analysis of Southeastern Europe traces major episodes of paternal gene flow among Slavic populations. Mol Biol Evol. 2005;22:1964–1975. doi: 10.1093/molbev/msi185. [DOI] [PubMed] [Google Scholar]
- Nasidze I, Quinque D, Dupanloup I, Cordaux R, Kokshunova L, Stoneking M. Genetic evidence for the Mongolian ancestry of Kalmyks. Am J Phys Anthropol. 2005;128:846–854. doi: 10.1002/ajpa.20159. [DOI] [PubMed] [Google Scholar]
- Bulayeva K, Jorde LB, Ostler C, Watkins S, Bulayev O, Harpending H. Genetics and population history of Caucasus populations. Hum Biol. 2003;75:837–853. doi: 10.1353/hub.2004.0003. [DOI] [PubMed] [Google Scholar]
- Gimbutas M.Proto-Indo-European culture: the Kurgan culture during the 5th to the 3rd millennia BCIn Cardona G, Koenigswald HM, Senn A (eds): Indo-European and Indo-Europeans Philadelphia: University of Pennsylvania Press; 1970155–198. [Google Scholar]
- Savel'eva EA.(ed): Atlas of the Komi Republic Moscow: Dizain. Moscow: Inter'er Kartographiya; 2001552 [Google Scholar]
- Kayser M, Brauer S, Cordaux R, et al. Melanesian and Asian origins of Polynesians: mtDNA and Y chromosome gradients across the Pacific. Mol Biol Evol. 2006;23:2234–2244. doi: 10.1093/molbev/msl093. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.