Abstract
High-density single-nucleotide polymorphism (SNP) genotyping technology enables extensive genotyping as well as the detection of increasingly smaller chromosomal aberrations. In this study, we assess molecular karyotyping as first-round analysis of patients with mental retardation and/or multiple congenital abnormalities (MR/MCA). We used different commercially available SNP array platforms, the Affymetrix GeneChip 262K NspI, the Genechip 238K StyI, the Illumina HumanHap 300 and HumanCNV 370 BeadChip, to detect copy number variants (CNVs) in 318 patients with unexplained MR/MCA. We found abnormalities in 22.6% of the patients, including six CNVs that overlap known microdeletion/duplication syndromes, eight CNVs that overlap recently described syndromes, 63 potentially pathogenic CNVs (in 52 patients), four large segments of homozygosity and two mosaic trisomies for an entire chromosome. This study shows that high-density SNP array analysis reveals a much higher diagnostic yield as that of conventional karyotyping. SNP arrays have the potential to detect CNVs, mosaics, uniparental disomies and loss of heterozygosity in one experiment. We, therefore, propose a novel diagnostic approach to all MR/MCA patients by first analyzing every patient with an SNP array instead of conventional karyotyping.
Keywords: SNP array, mental retardation, copy number variants, diagnostic workflow
Introduction
Mental retardation (MR) is a life-long disability with a major impact on the lives of the patients and their families. The prevalence of MR is 2–3%, and the underlying cause remains unknown in 65–80% of patients.1, 2, 3 Diagnosing is a challenge because of the broad spectrum of potentially underlying disorders and the wide range of available tests. Knowing the cause is necessary for assessing recurrence risk, short- and long-term prognosis and to decide on treatment options.
Changes in genetic dosage of one or more genes are common causes of MR.3 Routine microscopic analysis of chromosomes isolated from peripheral blood lymphocytes has been used successfully to identify such genetic imbalances over the past 50 years. This conventional karyotyping has the advantage of surveying the entire genome for chromosome abnormalities in a single experiment, but it cannot detect imbalances smaller than approximately 5 Mb. Smaller chromosomal aberrations can be identified with fluorescent in situ hybridization (FISH) and multiplex ligation-dependent probe amplification (MLPA) analysis. These techniques are used either to confirm a clinical suspicion by screening for well-known microdeletion syndromes associated with MR or for the analysis of all subtelomeric regions of the genome. The subtelomeric regions are known to be frequently affected in MR.1 The use of FISH and MLPA analysis is limited because only a few genomic regions can be screened in a single experiment and it can therefore not be applied genome wide.
Patients with unexplained MR with or without multiple congenital abnormalities (MR/MCA), who are referred to genetic laboratories, are initially screened with conventional karyotyping and, if required, with targeted FISH or MLPA analysis. The combined diagnostic yield of these analyses is approximately 5–10%.4 Consequently, a clinical diagnosis is lacking in the majority of these patients, which impedes the development of treatment strategies and adequate genetic counseling. Therefore, new high-resolution whole-genome technologies facilitating an increased detection rate of subtle chromosome imbalances are needed to improve the diagnosis of MR/MCA patients.
Recent developments in array technology allow whole-genome analysis for copy number variants (CNVs) at a resolution 10–10 000 times higher than that of conventional karyotyping. Comparative genome hybridization (CGH) studies using arrays with large insert clones (usually bacterial artificial clones (BACs)) have shown the potential of array technology to identify diagnostic CNVs in generally 16.7% of the unexplained MR/MCA patients.4, 5, 6, 7, 8, 9, 10, 11 The pathogenic CNVs detected in CGH studies range in size from 0.25 to 15 Mb.12 Resolution is limited by the size of the probes and the distance between the clones, that is 100 kb to 1 Mb. Therefore, the ideal technique would identify abnormalities with an even higher resolution. The single-nucleotide polymorphism (SNP) arrays have been widely used for genotyping and can identify submicroscopic CNVs as well as low-level chromosomal mosaicisms and uniparental disomies (UPDs).2, 13, 14, 15
We performed SNP array analysis on DNA from 318 patients with unexplained MR/MCA and an apparently balanced karyotype to search for potentially pathogenic submicroscopic CNVs with two different commercially available SNP array platforms. In this study, we show the importance of implementing the SNP array analysis in a diagnostic setting and advocate a whole-genome copy number screening using an SNP array as a new diagnostic tool for every MR/MCA patient rather than conventional karyotyping.
Materials and methods
Patients
A total of 318 patients referred for MR/MCA were recruited without further selection. Previously performed conventional karyotyping, targeted FISH or molecular tests revealed no etiological diagnosis. Detailed phenotypic information on all patients found to have a pathogenic or potentially pathogenic CNV is provided in Supplementary Table 1. DNA was extracted from whole blood using a Gentra Puregene DNA Purification Kit (Gentra Systems, Minneapolis, MN, USA), following the manufacturer's instructions. The study was approved by the Leiden University Medical Center Clinical Research Ethics Board, conforming to Dutch law and the World Medical Association Declaration of Helsinki.
SNP arrays
The Affymetrix GeneChip Human Mapping 262K NspI and 238K StyI arrays (Affymetrix, Santa Clara, CA, USA) contain 262 262 and 238 304 25-mer oligonucleotides, respectively, with an average spacing of approximately 12 kb per array. An amount of 250 ng DNA was processed according to the manufacturer's instructions. SNP copy number was assessed using the software program CNAG version 2.0.16
The Illumina HumanHap300 BeadChip (Illumina Inc., San Diego, CA, USA) contains 317 000 TagSNPs, with an average spacing of approximately 9 kb. The Illumina HumanCNV370 BeadChip (Illumina) contains 317 000 TagSNPs and 52 000 non-polymorphic markers for specifically targetting nearly 14 000 known CNVs. This array has an average spacing of approximately 7.7 kb. A total of 750 ng DNA was processed according to the manufacturer's instructions. SNP copy number (log R ratio) and B-allele frequency were assessed using the software programs BeadStudio version 3.2 (Illumina) and Partek Genomics Suite version 6.3 (Partek Inc., St Louis, MO, USA).
Evaluation of CNVs
Deletions of at least five adjacent SNPs or of a minimum region of 150 kb and duplications of at least seven adjacent SNPs or of a minimum region of 200 kb were analyzed.17 This approach was adopted to minimize the number of false-positive findings. The detected CNVs were classified into three different groups: I, known pathogenic CNVs (known microdeletion or microduplication syndrome); II, potentially pathogenic CNVs, not described in the Database of Genomic Variants (DGV; The Centre for Applied Genomics, The Hospital for Sick Children, Toronto, Canada, http://projects.tcag.ca/variation/); and III, known polymorphic CNVs described in the DGV or observed in our in-house reference set (60 controls), whereby at least three individuals must be reported with the same rearrangement. All type III CNVs were further excluded from this study.
All type II CNVs were assessed with Ensembl (Wellcome Trust Genome Campus, Hinxton, Cambridge, UK, http://www.ensembl.org: Ensembl release 52 – December 2008) and DECIPHER (Wellcome Trust Genome Campus, Hinxton, Cambridge, UK) for gene content and similar cases, respectively. All patients with a type II CNV were added to DECIPHER when consent was obtained.
Validation of CNVs
The known and potentially pathogenic CNVs were confirmed with MLPA, FISH or another type of SNP array on a second independent sample. If parents were available, segregation analysis was performed by MLPA, FISH or SNP array.
MLPA experiments were performed as described.18 At least two synthetic MLPA the probes were designed within the CNV and probes were commercially obtained from Biolegio (Malden, The Netherlands). Amplification products were identified and quantified by capillary electrophoresis on an ABI 3130 genetic analyzer (Applied Biosystems, Nieuwerkerk aan de IJssel, The Netherlands). Fragment analysis was performed with the GeneMarker Software V1.51 (SoftGenetics, State College, PA, USA). Thresholds for deletions and duplications were set at 0.75 and 1.25, respectively.
FISH analysis was carried out by standard procedures as described.19 BAC clones mapping to the CNVs were selected on the basis of their physical location within the affected region (http://www.ensembl.org: Ensembl release 49 – March 2008).
Results
A total of 318 patients were screened for submicroscopic CNVs. All patients had an apparently normal balanced karyotype, and targeted FISH or molecular tests, if performed, revealed no rearrangements. The Affymetrix GeneChip was applied to 132 patients and the Illumina BeadChip platform was applied to 186 patients. Eight (5.71%) Affymetrix and two (1.06%) Illumina experiments failed. On average, two CNVs per patient were obtained (Affymetrix 3 and Illumina 1.7). All polymorphic CNVs were excluded from further research.
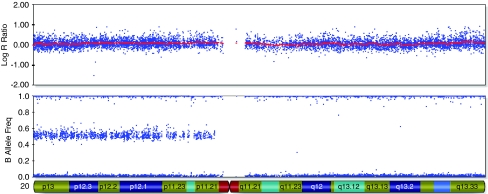
Supplementary Table 1 shows a summary of all detected CNVs. Six patients showed a CNV that has a clear clinical significance as it overlaps a known microdeletion/duplication syndrome. In eight patients, we detected a CNV that was recently described as a new microdeletion/duplication syndrome.20, 21, 22, 23, 24, 25, 26 63 Potentially pathogenic CNVs were observed in 52 patients (16.4%). Four patients showed striking regions of loss of heterozygosity (LOH) (Table 1 and Figure 1). Regions of homozygosity, ranging in size from 200 kb to 15 Mb, are common in healthy individuals.27 Here, four patients showed regions of LOH extending more than 15 Mb. Two patients showed a single segment of LOH (BC227 and BC318) and one patient a single segment, however in mosaic form (BC302), and one patient, two segments (BC308). The parents of the patients were not related.
Table 1. Regions of LOH detected in patients with no consanguine parents.
| ID | Chromosome | Starting SNP | Ending SNP | Size in Mb |
|---|---|---|---|---|
| BC227 | 12q15q23.2 | rs3905279 | rs7312593 | 33.78 |
| BC302a | 16q11.2qter | rs166161 | rs7498985 | 43.31 |
| BC311 | 13q14.2q22.1 | rs12875203 | rs9318228 | 27.16 |
| 20p11.21qter | rs6138531 | rs10485816 | 37.26 | |
| BC318 | 17q21.32q23.2 | rs6503455 | rs758466 | 15.17 |
LOH was found in mosaic.
Figure 1.
37.26 Mb region of LOH on chromosome 20q in case BC311 detected with the Illumina 317K BeadChip. Beadstudio log R ratio estimate for each individual SNP in the first plot and genotype call for every SNP in the second plot. The x axis shows the position on the chromosome.
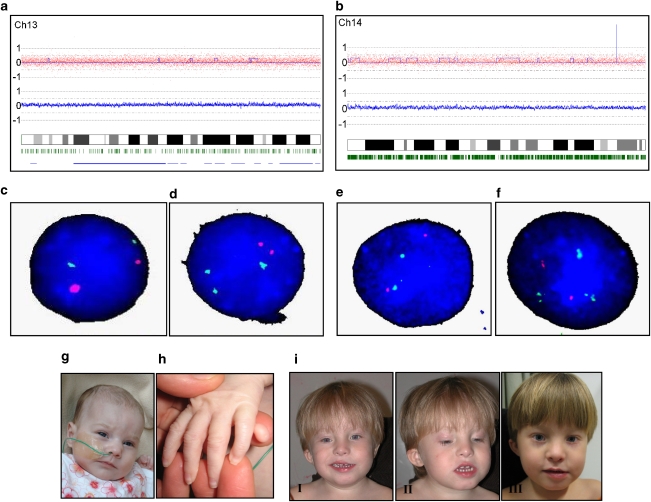
Two patients showed a low-level chromosomal mosaicism. Patient CR355 was a girl diagnosed with microcephaly, ventricular septum defect, diaphragmatic hernia, umbilical hernia and postaxial polydactyly of the left hand (Figure 2g and h). Pregnancy was conceived by in vitro fertilization and the girl was born at a gestational age of 36 5/7 weeks, with a birthweight of 2475 g. Her psychomotor development was delayed and she failed to thrive. She developed severe respiratory insufficiency and died at the age of 7 months. Initial conventional karyotyping of five metaphases did not show rearrangements. SNP array analysis showed a subtle increase in copy number for chromosome 13, suggesting an extra copy of chromosome 13 in 14% of the cells (Figure 2a). FISH experiments confirmed the presence of trisomy 13 in 18% of cultured lymphocytes (Figure 2c and d) and supplementary karyotyping detected in 7 of the 50 (13%) metaphases an extra chromosome 13.
Figure 2.
(a) CNAG copy number analysis for patient CR355 using the Affymetrix 262K GeneChip. Log R ratio estimate for each individual SNP in the first plot and for an average of 10 SNPs in the second plot. Both plots show a slight increase in log R ratio for whole chromosome 13. Blue line in first plot: copy number estimate calculated with the Hidden Markov Model. The x axis shows the position on the chromosome. Green stripes: heterozygous SNP calls. (b) CNAG copy number analysis output for patient CR377 using the Affymetrix 262K GeneChip. Both plots show a slight increase in log R ratio for chromosome 14. (c) FISH experiment (probes LSI13 (green) and LSI21 (red); Vysis, Abbott Laboratories, Abbott Park, IL, USA) showing a normal cell. (d) FISH experiment showing the presence of a mosaic trisomy 13 in 18% of the 200 cells analyzed. (e) FISH experiment (probes LSI CCNDI, 11q13 (red) and LSI IGH, 14q32 (green); Vysis) showing a normal cell. (f) FISH experiment showing the presence of a mosaic trisomy 14 in 9% of the 200 cells analyzed. (g) Facial picture of patient CR355. Facial dysmorphisms included upslant of palpebral fissures, a broad nasal bridge and uplifted earlobes. (h) Picture of postaxial polydactyly of the left hand of CR355. (i) Facial pictures of case CR377, 3 years and 7 months (I and II), and 4 years and 8 months (III). Note marked asymmetry when smiling, asymmetric upslanted palpebral fissures, left-sided epicanthus, hypertelorism, low-set and small right ear.
Patient CR377 was a boy referred at the age of 2 years and 9 months because of short stature, speech delay and motor delay (Figure 2i). Pregnancy had been uneventful and the boy was born at a gestational age of 40 5/7 weeks after vacuum extraction, with a birthweight of 3610 g. In early childhood, he suffered from recurrent respiratory infections and recurrent otitis media. At referral, his height was 84 cm (−3.4 SD). He had a broad thorax, pectus excavatum, a right-sided simian crease and short second phalanges of both digiti V. On follow-up at the age of 3 years and 7 months, his height was even more compromised (−4.2 SD). At the age of 4 years and 8 months, a marked discrepancy in leg length was noted, the right being shorter. At that time, the skin around both wrists and ankles showed an apparent reticular pattern of hypo- and hyperpigmentation. The body asymmetry combined with an abnormal skin pigmentation pointed in the direction of a mosaic condition. Conventional karyotyping on 31 metaphases had shown one cell with trisomy 14, which, confirming to professional guidelines, was interpreted as an artifact. SNP array results displayed a subtle increase in copy number for chromosome 14, suggesting an extra copy of chromosome 14 in 19% of the cells, and mosaicism was confirmed with FISH experiments on cultured lymphocytes (9%) (Figure 2b,e and f). UPD of chromosome 14 for the normal cells was excluded (results not shown).
Discussion
In this study, SNP arrays were used to search for pathogenic CNVs in patients with unexplained MR/MCA. The detected CNVs can be divided into the following groups: clearly pathogenic CNVs that overlap known microdeletion/duplication syndromes, CNVs that overlap recently described syndromes, potentially pathogenic CNVs and polymorphic CNVs (Supplementary Table 1). In total, we detected known syndromes in six patients, recently described CNVs in eight patients and 63 potentially pathogenic CNVs in 52 patients (in total, 20.7%). The polymorphic CNVs were excluded from further research.
Six CNVs were considered pathogenic as they are associated with well-established microdeletion syndromes. These syndromes were recognized afterwards by a clinical geneticist, which underlines the difficulty of establishing a diagnosis by clinical observation. Eight patients showed CNVs that were recently identified in other studies. For these new syndromes, no obvious phenotype has been established yet, and more patients with the same abnormalities are needed to unravel the associated phenotype. The discovery of these ‘known' CNVs highlights the advantage of the whole-genome screening methods to detect a known deletion or duplication syndrome in one single experiment.
Unraveling the clinical relevance for the potentially pathogenic CNVs is a new challenge. Regions containing coding genes can be present in variable copy number without obvious clinical manifestations, which makes it very hard to determine whether a subtle CNV has a clinical significance. Recent papers have already presented flow schemes for the interpretation of these CNVs.28, 29 In this study, first, all polymorphic CNVs were excluded by comparing against the DGV and our in-house reference set. Second, for all CNVs containing coding genes annotated by Ensembl (release 52, December 2008), the inheritance was determined by checking both parents (if available).
For 27 potentially pathogenic CNVs, we could establish that the rearrangement was inherited from one of the unaffected parents. Several studies have shown that some CNVs are indeed polymorphisms contributing to common variations in healthy individuals.30, 31 A large number of small rearrangements, detected in patients with MR and inherited from phenotypically normal parents, have been reported, whereby it was speculated that some of these imbalances may indeed be benign variations and others are likely to represent susceptibility loci for disease.32, 33 A particularly intriguing example is the submicroscopic 1q21 deletion, characteristic for thrombocytopenia absent radius syndrome, which is found in all patients with the syndrome, but is inherited from a phenotypically normal parent in a subset of cases.34 It is becoming increasingly clear that many CNVs come with a highly variable phenotype, including what is considered as ‘normal.' Among the many examples are the 22q11 deletion and duplication,21 the 16p11.2 deletion,23, 24, 25 and the Xp deletions involving the neuroligin and VCX genes.35 Mechanisms that can explain why some inherited CNVs occasionally result in abnormal development have been postulated.32, 36 These mechanisms include: a mutation in the same region on the other chromosome; a mutation in one or more unlinked modifying genes; imprinting; mosaicism in one of the parents; or any other unidentified genetic, epigenetic or environmental factor.21, 32 Furthermore, it is frequently assumed that parents are phenotypically normal, although a closer inspection by a clinical geneticist might reveal subtle anomalies.32
Twenty-two de novo potentially pathogenic CNVs are detected and are likely to be relevant for the phenotype of the patient. For 14 potentially pathogenic CNVs, the inheritance could not be determined. Interpretation of these CNVs is even more difficult. Attempts should be made to receive DNA from the parents or, alternatively, other relatives. However, for all potentially pathogenic CNVs, phenotypically concordant patients with the same abnormality need to be found to be sure of their pathogenicity. Therefore, databases such as DECIPHER (https://decipher.sanger.ac.uk/) have been created to compile molecular cytogenetic data from clinical studies all over the world to provide the basis for understanding the role of different CNVs in genetic diseases. For the 63 potentially pathogenic CNVs detected in this study, no complete overlapping cases were described in DECIPHER. More array data on MR patients and healthy controls will be needed to determine the clinical relevance of these CNVs.
In 9 of the 26 de novo CNVs (pathogenic and potentially pathogenic), DNA from the parents was tested on SNP array, enabling us to determine the parental origin. Seven CNVs occurred in the paternally derived chromosome. Only two CNVs occurred in maternally derived chromosomes, giving a paternal–maternal ratio of 6:2. Parental origins of microdeletions and duplications have been investigated in several genomic disorders. Deletions in Williams and DiGeorge syndrome were equally of paternal and maternal origin equally.37 Deletions in neurofibromatosis type 1 and in 1p36 syndrome were predominant on the maternally derived chromosome.38, 39 By contrast, duplications in Charcot-Marie-Tooth disease type I and deletions in Wolf–Hirschhorn and Cri Du Chat syndromes occur more frequently in the paternally derived chromosome.40, 41, 42 Much more parent-of-origin data are needed to document the possible existence of regional parental bias.
ArrayCGH (aCGH) screenings performed on mentally retarded patients are a powerful tool for the detection of CNVs.4, 5, 6, 7, 8, 9, 10, 11 These arrays consist of large-insert clones, and the smallest pathogenic CNVs detected are approximately 0.25 Mb. The high-density whole-genome SNP arrays, which were initially developed for genotyping, are now widely used to search for smaller CNVs.2, 13, 14, 15 In approximately 25% of patients with unexplained MR/MCA, CNVs are detected by aCGH and SNP array studies. The array technology is the most effective method resulting in the most clinical diagnoses compared with conventional karyotyping, FISH analysis and mutation screening. Although it was suspected that array analysis would not be able to detect mosaicisms, the aCGH and SNP array techniques actually appear to be more sensitive in detecting low-level mosaicism than conventional karyotyping.43, 44 If mosaicism is not suspected, the number of cells counted with conventional karyotyping may not be sufficient to detect the aberrant subset of cells, and a single abnormal cell might be interpreted as an artifact of cell culture.45 Two such cases of low-level mosaicism were reported in this study. Our patients with mosaic trisomy 13 and 14 have phenotypical characteristics that resemble the reported phenotypes of mosaic trisomy 13 and 14 (Figures 2).46, 47, 48, 49, 50, 51
A major advantage of SNP array analysis is the extra SNP genotyping information, which enables the detection of copy number neutral chromosomal aberrations such as UPD and LOH.52 UPD, which arises when an individual inherits two copies of a chromosome pair from one parent and no copy of the other parent, can result in rare recessive disorders, or developmental problems because of the effects of imprinting.53 Examples of genetic diseases linked to UPD include the Prader–Willi syndrome (MIM 176270), Angelman syndrome (MIM 105830), Beckwith–Wiedemann syndrome (MIM 130650) and Silver–Russell syndrome (MIM 180860). SNP array analysis is able to detect uniparental isodisomy and uniparental heterodisomy (when both parents are included in the experiment), but the interpretation of new UPD regions is difficult and further research is required to confirm the clinical consequences. Recessive and normally non-penetrant alleles in isodisomic form (two copies of the same parental chromosome) may cause recessive diseases. Gene defects underlying autosomal recessive disorders can be localized and identified by homozygosity mapping. Furthermore, patients with consanguineous parents display many regions of homozygosity (LOH) that might result in a recessive disorder. In this study, we identified an extended segment of LOH in four patients, with no consanguineous parents. To identify the responsible gene or genes is a challenge now, but it may become a realistic possibility with next-generation high-throughput DNA-sequencing technology. Finally, the information on the SNP genotype could be used to verify biological parentage and cases of suspected incest.
Conversely, a disadvantage of using arrays instead of conventional karyotyping is the inability to detect balanced rearrangements. Around 6% of antenatal cases with balanced reciprocal translocations and inversions are associated with abnormal phenotypes.54 In these cases, the breakpoints of the rearrangement probably disrupt a gene, or small duplications or deletions beyond microscopic resolution are present. The SNP array analysis will (depending on the resolution) detect the small abnormalities, but the disruption of genes will remain unknown. A retrospective Dutch study showed that only approximately 0.78% of potentially pathogenic balanced rearrangements of all referrals will be undetectable by array analysis without conventional karyotyping.55
The absence of an aberration or the presence of only polymorphic CNVs after SNP array analysis does not exclude a syndrome caused by a mutation at the gene level. Therefore, we emphasize that MR/MCA patients with normal array results should always be referred to a clinical geneticist to exclude such known syndromes. Furthermore, genomic data obtained from the SNP array analysis can be used in future research for association between genetic markers and specific phenotypes to hopefully diagnose even more patients.
In 2006, Rauch et al3 compared the diagnostic yield of various techniques in MR/MCA patients. These authors suggested targeted analysis in patients with a clear diagnosis and conventional karyotyping and molecular screening in the remaining patients.3 Kriek et al56 proposed another diagnostic approach to MR/MCA patients, suggesting a screening with MLPA first and based on the outcome of additional aCGH or karyotyping. However, more recent studies have already mentioned the partial replacement of conventional karyotyping by molecular karyotyping.53, 57 In addition, Koolen et al29 described a workflow for the clinical interpretation of CNVs in individuals with MR. Our results show that high-density SNP arrays can be successfully used as a tool for the detection of CNVs, low-level mosaicism and copy number neutral abnormalities. Their high resolution and commercial availability make them attractive to implement in a routine diagnostic setting.
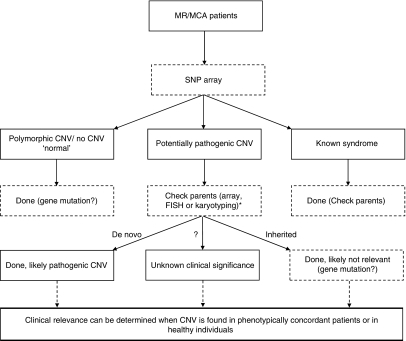
Here, we combine the flowcharts designed by Kriek et al56 and Koolen et al29 in a novel approach to the patient with MR/MCA (Figure 3). We recommend testing every patient first with an SNP array instead of conventional karyotyping. The results will be classified in patients with polymorphic CNVs or no CNV (‘normal'), patients with CNVs that overlap known syndromes and patients with potentially pathogenic CNVs. The ‘normal' patients could be screened for gene mutation in targeted genes or in the future with whole-genome next-generation high-throughput DNA-sequencing technology. The patients with CNVs overlapping known syndromes are diagnosed and family members could be checked for inheritance and recurrence risk. The inheritance of the potentially pathogenic CNVs should be tested and the patients should be reported in a database such as DECIPHER. The clinical relevance of these CNVs can be determined when the specific CNV is reported in adequate numbers of healthy individuals or phenotypically concordant patients.
Figure 3.
Flow chart for the new diagnostic approach to patients with mental retardation. *If the CNV exceeds 200 kb, we recommend additional FISH analysis to confirm the CNV in the patient and screen for balanced translocations or insertions in the parents. If the CNV is smaller than 200 kb, we recommend a second array analysis on the patient to confirm the CNV and on the parents to test heritability.
This new approach will diagnose a larger proportion of CNVs in the first round; however, the interpretation of the CNVs will be the major challenge. Eventually, more families will be informed about the cause of the disease of their family member. This will improve medical care and genetic counseling. Furthermore, as the SNP array approach will make targeted FISH and MLPA analysis redundant, less laboratory tests will be needed, which leads to a substantial reduction of cost.
Acknowledgments
We are grateful to the patients and their parents, and to the clinicians of the Clinical Genetics Department of the Leiden University Medical Centre for referring the patients. Further, we would thank the technicians of the Laboratory of Diagnostic Genome Analysis, Leiden University Medical Centre, for performing FISH experiments. In addition, we also thank Drs Nicole de Leeuw and Rolph Pfundt from the Department of Human Genetics, Radboud University Nijmegen Medical Centre, for sharing their knowledge about the implementation of arrays in their diagnostic setting. This work was supported by EuroGentest, an EU-FP6-supported NoE contract number 512148.
Footnotes
Supplementary Information accompanies the paper on European Journal of Human Genetics website (http://www.nature.com/ejhg)
Electronic database information
Affymetrix: http://www.affymetrix.com
CNAG: http://www.genome.umin.jp/
Database of Genomic Variants: http://projects.tcag.ca/variation/
DECIPHER: https://decipher.sanger.ac.uk/
ECARUCA: http://ecaruca.net/
ENSEMBL: http://www.ensembl.org/index.html
Illumina: http://www.illumina.com/
MRC Holland: http://www.mlpa.com/pages/indexpag.html
Supplementary Material
References
- Flint J, Knight S. The use of telomere probes to investigate submicroscopic rearrangements associated with mental retardation. Curr Opin Genet Dev. 2003;13:310–316. doi: 10.1016/s0959-437x(03)00049-2. [DOI] [PubMed] [Google Scholar]
- Hoyer J, Dreweke A, Becker C, et al. Molecular karyotyping in patients with mental retardation using 100K single-nucleotide polymorphism arrays. J Med Genet. 2007;44:629–636. doi: 10.1136/jmg.2007.050914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rauch A, Hoyer J, Guth S, et al. Diagnostic yield of various genetic approaches in patients with unexplained developmental delay or mental retardation. Am J Med Genet A. 2006;140:2063–2074. doi: 10.1002/ajmg.a.31416. [DOI] [PubMed] [Google Scholar]
- de Vries BB, Pfundt R, Leisink M, et al. Diagnostic genome profiling in mental retardation. Am J Hum Genet. 2005;77:606–616. doi: 10.1086/491719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menten B, Maas N, Thienpont B, et al. Emerging patterns of cryptic chromosomal imbalance in patients with idiopathic mental retardation and multiple congenital anomalies: a new series of 140 patients and review of published reports. J Med Genet. 2006;43:625–633. doi: 10.1136/jmg.2005.039453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg C, Knijnenburg J, Bakker E, et al. Array-CGH detection of micro rearrangements in mentally retarded individuals: clinical significance of imbalances present both in affected children and normal parents. J Med Genet. 2006;43:180–186. doi: 10.1136/jmg.2005.032268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoumans J, Ruivenkamp C, Holmberg E, Kyllerman M, Anderlid BM, Nordenskjold M. Detection of chromosomal imbalances in children with idiopathic mental retardation by array based comparative genomic hybridisation (array-CGH) J Med Genet. 2005;42:699–705. doi: 10.1136/jmg.2004.029637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw-Smith C, Redon R, Rickman L, et al. Microarray based comparative genomic hybridisation (array-CGH) detects submicroscopic chromosomal deletions and duplications in patients with learning disability/mental retardation and dysmorphic features. J Med Genet. 2004;41:241–248. doi: 10.1136/jmg.2003.017731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyson C, Harvard C, Locker R, et al. Submicroscopic deletions and duplications in individuals with intellectual disability detected by array-CGH. Am J Med Genet A. 2005;139:173–185. doi: 10.1002/ajmg.a.31015. [DOI] [PubMed] [Google Scholar]
- Vissers LE, de Vries BB, Osoegawa K, et al. Array-based comparative genomic hybridization for the genomewide detection of submicroscopic chromosomal abnormalities. Am J Hum Genet. 2003;73:1261–1270. doi: 10.1086/379977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan YS, Jayakar P, Zhu H, et al. Detection of pathogenic gene copy number variations in patients with mental retardation by genomewide oligonucleotide array comparative genomic hybridization. Hum Mutat. 2007;28:1124–1132. doi: 10.1002/humu.20581. [DOI] [PubMed] [Google Scholar]
- Kirov G, Gumus D, Chen W, et al. Comparative genome hybridization suggests a role for NRXN1 and APBA2 in schizophrenia. Hum Mol Genet. 2008;17:458–465. doi: 10.1093/hmg/ddm323. [DOI] [PubMed] [Google Scholar]
- Altug-Teber O, Dufke A, Poths S, et al. A rapid microarray based whole genome analysis for detection of uniparental disomy. Hum Mutat. 2005;26:153–159. doi: 10.1002/humu.20198. [DOI] [PubMed] [Google Scholar]
- Friedman JM, Baross A, Delaney AD, et al. Oligonucleotide microarray analysis of genomic imbalance in children with mental retardation. Am J Hum Genet. 2006;79:500–513. doi: 10.1086/507471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruno DL, Ganesamoorthy D, Schoumans J, et al. Detection of cryptic pathogenic copy number variations and constitutional loss of heterozygosity using high resolution SNP microarray Analysis in 117 patients referred for cytogenetic analysis and impact on clinical practice. J Med Genet. 2009;46:123–131. doi: 10.1136/jmg.2008.062604. [DOI] [PubMed] [Google Scholar]
- Nannya Y, Sanada M, Nakazaki K, et al. A robust algorithm for copy number detection using high-density oligonucleotide single nucleotide polymorphism genotyping arrays. Cancer Res. 2005;65:6071–6079. doi: 10.1158/0008-5472.CAN-05-0465. [DOI] [PubMed] [Google Scholar]
- Hehir-Kwa JY, Egmont-Petersen M, Janssen IM, Smeets D, van Kessel AG, Veltman JA. Genome-wide copy number profiling on high-density bacterial artificial chromosomes, single-nucleotide polymorphisms, and oligonucleotide microarrays: a platform comparison based on statistical power analysis. DNA Res. 2007;14:1–11. doi: 10.1093/dnares/dsm002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White SJ, Vink GR, Kriek M, et al. Two-color multiplex ligation-dependent probe amplification: detecting genomic rearrangements in hereditary multiple exostoses. Hum Mutat. 2004;24:86–92. doi: 10.1002/humu.20054. [DOI] [PubMed] [Google Scholar]
- Dauwerse JG, Kievits T, Beverstock GC, et al. Rapid detection of chromosome 16 inversion in acute nonlymphocytic leukemia, subtype M4: regional localization of the breakpoint in 16p. Cytogenet Cell Genet. 1990;53:126–128. doi: 10.1159/000132911. [DOI] [PubMed] [Google Scholar]
- Rauch A, Zink S, Zweier C, et al. Systematic assessment of atypical deletions reveals genotype–phenotype correlation in 22q11.2. J Med Genet. 2005;42:871–876. doi: 10.1136/jmg.2004.030619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courtens W, Schramme I, Laridon A. Microduplication 22q11.2: a benign polymorphism or a syndrome with a very large clinical variability and reduced penetrance? – Report of two families. Am J Med Genet A. 2008;146A:758–763. doi: 10.1002/ajmg.a.31910. [DOI] [PubMed] [Google Scholar]
- Hannes FD, Sharp AJ, Mefford HC, et al. Recurrent reciprocal deletions and duplications of 16p13.11: the deletion is a risk factor for MR/MCA while the duplication may be a rare benign variant. J Med Genet. 2009;46:223–232. doi: 10.1136/jmg.2007.055202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar RA, KaraMohamed S, Sudi J, et al. Recurrent 16p11.2 microdeletions in autism. Hum Mol Genet. 2008;17:628–638. doi: 10.1093/hmg/ddm376. [DOI] [PubMed] [Google Scholar]
- Weiss LA, Shen Y, Korn JM, et al. Association between microdeletion and microduplication at 16p11.2 and autism. N Engl J Med. 2008;358:667–675. doi: 10.1056/NEJMoa075974. [DOI] [PubMed] [Google Scholar]
- Bijlsma EK, Gijsbers AC, Schuurs-Hoeijmakers JH, et al. Extending the phenotype of recurrent rearrangements of 16p11.2: deletions in mentally retarded patients without autism and in normal individuals Eur J Med Genet 2009(in press). [DOI] [PubMed]
- Ballif BC, Theisen A, Coppinger J, et al. Expanding the clinical phenotype of the 3q29 microdeletion syndrome and characterization of the reciprocal microduplication. Mol Cytogenet. 2008;1:8. doi: 10.1186/1755-8166-1-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lencz T, Lambert C, DeRosse P, et al. Runs of homozygosity reveal highly penetrant recessive loci in schizophrenia. Proc Natl Acad Sci USA. 2007;104:19942–19947. doi: 10.1073/pnas.0710021104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C, Iafrate AJ, Brothman AR. Copy number variations and clinical cytogenetic diagnosis of constitutional disorders. Nat Genet. 2007;39:S48–S54. doi: 10.1038/ng2092. [DOI] [PubMed] [Google Scholar]
- Koolen DA, Pfundt R, de Leeuw N, et al. Genomic microarrays in mental retardation: a practical workflow for diagnostic applications. Hum Mutat. 2009;30:283–292. doi: 10.1002/humu.20883. [DOI] [PubMed] [Google Scholar]
- Iafrate AJ, Feuk L, Rivera MN, et al. Detection of large-scale variation in the human genome. Nat Genet. 2004;36:949–951. doi: 10.1038/ng1416. [DOI] [PubMed] [Google Scholar]
- Sebat J, Lakshmi B, Troge J, et al. Large-scale copy number polymorphism in the human genome. Science. 2004;305:525–528. doi: 10.1126/science.1098918. [DOI] [PubMed] [Google Scholar]
- Barber JC. Directly transmitted unbalanced chromosome abnormalities and euchromatic variants. J Med Genet. 2005;42:609–629. doi: 10.1136/jmg.2004.026955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Ravel TJ, Balikova I, Thienpont B, et al. Molecular karyotyping of patients with MCA/MR: the blurred boundary between normal and pathogenic variation. Cytogenet Genome Res. 2006;115:225–230. doi: 10.1159/000095918. [DOI] [PubMed] [Google Scholar]
- Klopocki E, Schulze H, Strauss G, et al. Complex inheritance pattern resembling autosomal recessive inheritance involving a microdeletion in thrombocytopenia-absent radius syndrome. Am J Hum Genet. 2007;80:232–240. doi: 10.1086/510919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mochel F, Missirian C, Reynaud R, Moncla A. Normal intelligence and social interactions in a male patient despite the deletion of NLGN4X and the VCX genes. Eur J Med Genet. 2008;51:68–73. doi: 10.1016/j.ejmg.2007.11.002. [DOI] [PubMed] [Google Scholar]
- Lesnik Oberstein SA, Kriek M, White SJ, et al. Peters Plus syndrome is caused by mutations in B3GALTL, a putative glycosyltransferase. Am J Hum Genet. 2006;79:562–566. doi: 10.1086/507567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumer A, Dutly F, Balmer D, et al. High level of unequal meiotic crossovers at the origin of the 22q11. 2 and 7q11.23 deletions. Hum Mol Genet. 1998;7:887–894. doi: 10.1093/hmg/7.5.887. [DOI] [PubMed] [Google Scholar]
- Lazaro C, Gaona A, Ainsworth P, et al. Sex differences in mutational rate and mutational mechanism in the NF1 gene in neurofibromatosis type 1 patients. Hum Genet. 1996;98:696–699. doi: 10.1007/s004390050287. [DOI] [PubMed] [Google Scholar]
- Wu YQ, Heilstedt HA, Bedell JA, et al. Molecular refinement of the 1p36 deletion syndrome reveals size diversity and a preponderance of maternally derived deletions. Hum Mol Genet. 1999;8:313–321. doi: 10.1093/hmg/8.2.313. [DOI] [PubMed] [Google Scholar]
- Lopes J, Vandenberghe A, Tardieu S, et al. Sex-dependent rearrangements resulting in CMT1A and HNPP. Nat Genet. 1997;17:136–137. doi: 10.1038/ng1097-136. [DOI] [PubMed] [Google Scholar]
- Wieczorek D, Krause M, Majewski F, et al. Effect of the size of the deletion and clinical manifestation in Wolf–Hirschhorn syndrome: analysis of 13 patients with a de novo deletion. Eur J Hum Genet. 2000;8:519–526. doi: 10.1038/sj.ejhg.5200498. [DOI] [PubMed] [Google Scholar]
- Mainardi PC, Perfumo C, Cali A, et al. Clinical and molecular characterisation of 80 patients with 5p deletion: genotype–phenotype correlation. J Med Genet. 2001;38:151–158. doi: 10.1136/jmg.38.3.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballif BC, Rorem EA, Sundin K, et al. Detection of low-level mosaicism by array CGH in routine diagnostic specimens. Am J Med Genet A. 2006;140:2757–2767. doi: 10.1002/ajmg.a.31539. [DOI] [PubMed] [Google Scholar]
- Powis Z, Kang SH, Cooper ML, et al. Mosaic tetrasomy 12p with triplication of 12p detected by array-based comparative genomic hybridization of peripheral blood DNA. Am J Med Genet A. 2007;143A:2910–2915. doi: 10.1002/ajmg.a.31959. [DOI] [PubMed] [Google Scholar]
- Cheung SW, Shaw CA, Scott DA, et al. Microarray-based CGH detects chromosomal mosaicism not revealed by conventional cytogenetics. Am J Med Genet A. 2007;143A:1679–1686. doi: 10.1002/ajmg.a.31740. [DOI] [PubMed] [Google Scholar]
- Delatycki M, Gardner RJ. Three cases of trisomy 13 mosaicism and a review of the literature. Clin Genet. 1997;51:403–407. doi: 10.1111/j.1399-0004.1997.tb02499.x. [DOI] [PubMed] [Google Scholar]
- Delatycki MB, Pertile MD, Gardner RJ. Trisomy 13 mosaicism at prenatal diagnosis: dilemmas in interpretation. Prenat Diagn. 1998;18:45–50. doi: 10.1002/(sici)1097-0223(199801)18:1<45::aid-pd216>3.0.co;2-c. [DOI] [PubMed] [Google Scholar]
- Toledo SP, Wajntal A. 47XX,+13/46,XX mosaicism: a case report. Acta Genet Med Gemellol (Roma) 1977;26:71–79. doi: 10.1017/s0001566000010217. [DOI] [PubMed] [Google Scholar]
- Shinawi M, Shao L, Jeng LJ, et al. Low-level mosaicism of trisomy 14: phenotypic and molecular characterization. Am J Med Genet A. 2008;146A:1395–1405. doi: 10.1002/ajmg.a.32287. [DOI] [PubMed] [Google Scholar]
- Cheung SW, Kolacki PL, Watson MS, Crane JP. Prenatal diagnosis, fetal pathology, and cytogenetic analysis of mosaic trisomy 14. Prenat Diagn. 1988;8:677–682. doi: 10.1002/pd.1970080908. [DOI] [PubMed] [Google Scholar]
- Johnson VP, Aceto T, Jr, Likness C. Trisomy 14 mosaicism: case report and review. Am J Med Genet. 1979;3:331–339. doi: 10.1002/ajmg.1320030404. [DOI] [PubMed] [Google Scholar]
- Zhang ZF, Ruivenkamp C, Staaf J, et al. Detection of submicroscopic constitutional chromosome aberrations in clinical diagnostics: a validation of the practical performance of different array platforms. Eur J Hum Genet. 2008;16:786–792. doi: 10.1038/ejhg.2008.14. [DOI] [PubMed] [Google Scholar]
- Cassidy SB, Dykens E, Williams CA. Prader–Willi and Angelman syndromes: sister imprinted disorders. Am J Med Genet. 2000;97:136–146. doi: 10.1002/1096-8628(200022)97:2<136::aid-ajmg5>3.0.co;2-v. [DOI] [PubMed] [Google Scholar]
- Warburton D. De novo balanced chromosome rearrangements and extra marker chromosomes identified at prenatal diagnosis: clinical significance and distribution of breakpoints. Am J Hum Genet. 1991;49:995–1013. [PMC free article] [PubMed] [Google Scholar]
- Hochstenbach R, van Binsbergen E, Engelen J, et al. Array analysis and karyotyping: workflow consequences based on a retrospective study of 36 325 patients with idiopathic developmental delay in the Netherlands Eur J Med Genet 2009(in press). [DOI] [PubMed]
- Kriek M, Knijnenburg J, White SJ, et al. Diagnosis of genetic abnormalities in developmentally delayed patients: a new strategy combining MLPA and array-CGH. Am J Med Genet A. 2007;143:610–614. doi: 10.1002/ajmg.a.31593. [DOI] [PubMed] [Google Scholar]
- Vermeesch JR, Fiegler H, de Leeuw N, et al. Guidelines for molecular karyotyping in constitutional genetic diagnosis. Eur J Hum Genet. 2007;15:1105–1114. doi: 10.1038/sj.ejhg.5201896. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.