On page 1067 in this issue of PNAS, Hannaert et al. (1) present evidence for the idea that trypanosomes once possessed a chloroplast that they lost some time in their distant evolutionary past. It would probably be going overboard to call trypanosomes algae in disguise, but for a protozoan parasite without any trace of chloroplast remnants, the trypanosome is now shown to possess a remarkable number of enzymes of plant-related origin. This finding advances our understanding of trypanosome biology and could even help efforts to discover drugs to treat trypanosomatid-caused diseases.
Typanosomatids are unicellular eukaryotes belonging to the order of Kinetoplastidae; they are among the most versatile parasites in nature, infecting mammals, fish, and plants, and are usually transmitted by insect vectors. Major human diseases caused by trypanosomatids are leishmaniases (Leishmania species), sleeping sickness (Trypanosoma brucei variants), and Chagas' disease (Trypanosoma cruzi). Drugs for treating these diseases are few and toxic, and resistance is on the rise (2, 3). Because these diseases predominantly affect poor people in (sub)tropical developing countries, development of new drugs is not a high priority of the pharmaceutical industry. Yet academic labs are hunting for new drug targets, so the discovery of plant-like enzymes in T. brucei is good news indeed.
The evidence for secondary loss of plastids in trypanosomes stems from the genome sequencing programs that are now well underway for Leishmania, T. brucei, and T. cruzi.§ Hints for the presence of plant- and plastid-derived genes in trypanosomatids came from analyses of two enzymes involved in the hexose phosphate shunt (4) and of trypanosomatid peroxidases (5). Hannaert et al. (1) now report a substantial number of plant-like trypanosomatid genes, most of which are involved in core carbohydrate metabolism. For many of these genes, the sequence comparisons reveal a specific evolutionary affinity between the trypanosome gene and a higher plant, algal, or cyanobacterial homolog, indicating that at some time in the evolutionary past, some photosynthetic cell donated quite a few genes into the trypanosomatid lineage. One of the most conclusive of the green trypanosomatid genes is that encoding sedoheptulose-1,7-bisphosphatase (SPBase), an enzyme specific to the Calvin cycle in plant chloroplasts. In terms of both sequence similarities and its enzymatic properties, this enzyme is specific to plants, because cyanobacteria use a side activity of their highly distinct fructose-1,6-bisphosphatase for the corresponding reaction and lack a specific SPBase altogether.
It would probably be going overboard to call trypanosomes algae in disguise.
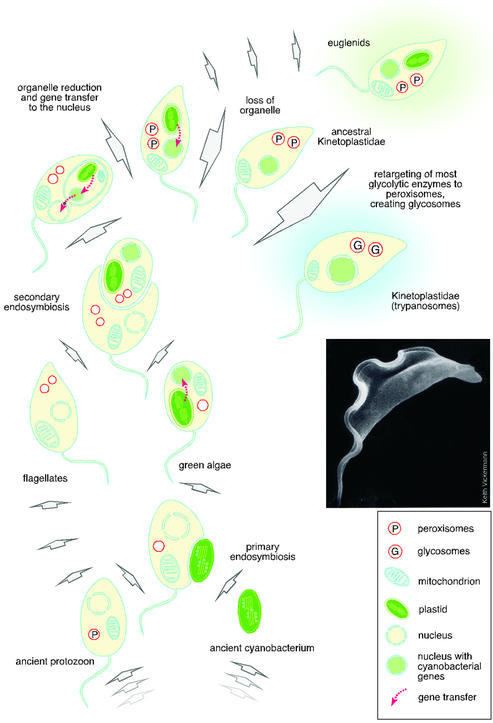
These new findings raise an obvious evolutionary question: What photosynthetic lineage was the source of these plant-like traits and genes? Hannaert et al. point to the euglenids, which contain numerous photosynthetic members including Euglena gracilis. On the basis of comparative cytology and morphology, taxonomists have traditionally placed the trypanosomatids and the euglenids in a well-circumscribed common group, the Euglenozoa. They share many unusual and defining molecular traits as well, including the unusual base “J” (β-d-glucosyl-hydroxymethyluracil) in their nuclear DNA (6). The plastids of the euglenids are surrounded by three membranes, which is the sure sign of secondary endosymbiosis, that is, Euglena acquired its plastid through engulfment of a eukaryotic green alga, rather than through engulfment of a cyanobacterium (primary symbiosis) (7). Hannaert et al. suggest, as the most reasonable and simplest of all possible explanations, that trypanosomatids and euglenids possessed one and the same green secondary plastid, which was retained in the euglenids but lost through reduction in the trypanosomatid lineage.
The new findings by Hannaert et al. are reminiscent of the malaria parasite story. Two major apicomplexan parasites, Plasmodium falciparum, the infectious agent of malaria, and Toxoplasma gondii, which causes toxoplasmosis, were long known to contain an enigmatic organelle in their cytosol, called the hohlzylinder. In 1984, a 35-kb circular DNA molecule was found in Toxoplasma (8), and Ian Wilson and colleagues later showed that this DNA is a highly reduced chloroplast genome (9), whereas Geoff McFadden and colleagues discovered that the hohlzylinder itself is in fact a highly reduced plastid (10). These insights led to the discovery that some compounds that inhibit plant-specific pathways, much the way that herbicides do, also kill apicomplexan parasites, opening up new opportunities for the development of treatments for apicomplexan-caused maladies (11).
Yet in contrast to the apicomplexans, the trypanosomatids have retained no detectable trace of a membrane-bounded organelle that might represent a plastid relict: the plastid compartment has been reduced to the point that it has disappeared entirely (Fig. 1). All that remains are a number of genes in the nucleus that were transferred from the symbiont's genome, the unmistakable imprint of endosymbiosis. How many algal genes might the trypanosomatid lineage have acquired during its prior life as an alga? In Arabidopsis, as much as 18% of all nuclear genes seem to be acquisitions from the genome of their primary plastid (12). Complete genome data will be forthcoming for Leishmania and possibly for other trypanosomatids in the course of 2003 (3). This finding will permit a more detailed analysis of the numbers and kinds of genes that these organisms acquired from their plastid.
Figure 1.
Schematic diagram indicating how trypanosomes may have come to acquire their plant-like genes. The common ancestry indicated here for the euglenid and trypanosome plastid is the simplest of several possibilities to account for the current findings. Electron micrograph courtesy of Keith Vickermann.
A flood of recent papers indicates that the acquisition of genes from organelles has been a major force in the evolution of unicellular eukaryotes (13). Wholesale incorporation of genes and genomes acquired via endosymbiosis was at least as important, if not more so, to genome evolution in these organisms than the laborious route of gene duplication and genetic drift among endogenously generated surplus copies. “You are what you eat,” wrote Doolittle (14), when it comes to gene donations from organelles. When organelles lyse, a whole organelle-genome's worth of DNA is suddenly in the cytosol, just waiting to get into the nuclear chromosomes via illegitimate recombination. When host cells lyse, on the other hand, the result is not transfer to the organelle, but cell death. This one-way street will inexorably drive the integration of organelle genes into the chromosomes of their host (14). For functionally equivalent genes, the intruding genes may have an advantage for fixation as long as a copy remains in the organelle, because if one integration does not lead to fixation, the next one can, or the next, and so forth.
Newer findings suggest that the main mechanism of organelle-to-nucleus transfer is in fact the direct integration of bulk DNA from organelles, probably lysed ones (15, 16). In Arabidopsis, for example, a copy of the entire 367-kb mitochondrial genome, 99% identical to the organelle copy, can be found on chromosome 2 (17, 18), and in rice, a contiguous 33-kb fragment of the chloroplast genome, 99.7% identical to the organelle copy, can be found on nuclear chromosome 10 (19). Such transfer events are likely to have been as prevalent in the past as they are today, and the encoded proteins can be targeted to organelles other than the ones from which the genes were donated (12), in line with the newer findings from trypanosomes (1).
When a host cell acquires an endosymbiont, the latter must have a genome complete enough to support a free-living lifestyle, but contemporary organelle genomes are the most highly reduced genomes known. Only a few percent of the genes that were once present in the genomes of ancestral chloroplasts and mitochondria are still retained in these organelles today (20). The remainder was either discarded or put to new use in the host. As Palenik (21) put it in a recent commentary in these pages: Hosts keep the baby (the plastid) and the bath water (additional genes donated by the endosymbiont that need not be encoded in the organelle). The trypanosomatids have gone one step further: They have kept the bath water (the acquired genes) but have tossed out the baby (the plastid).
The trypanosomatids are by no means the only organisms to have lost a plastid. In fact, plastid loss is turning out to be a surprisingly common theme in cell evolution (22), particularly among parasites. A striking example concerns the oomycetes, a group of fungus-like protists that have cell walls of cellulose (rather than chitin, as is typical of fungi) and that contains many important agricultural pathogens of crop plants, among them the agent of potato blight, Phytophthora infestans. Molecular studies have revealed that the oomycetes are not fungi at all, but rather are derived from algae that became secondarily nonphotosynthetic through loss of their plastids (22, 23).
Moreover, secondary loss of organelles is not restricted to plastids, either. Mitochondria can be lost, or “lost and found” as well, as newer findings have shown. A prime example here is Entamoeba histolytica, an important intestinal pathogen of humans, which was long thought to be without mitochondria (amitochondriate) and which obtains all of its ATP through a cytosolic pathway called extended glycolysis (24). However, a small highly reduced relict mitochondrion was recently discovered in Entamoeba, and this apparently has no function in core ATP production (25, 26). One suspicion is that such relict mitochondria might be retained because they are needed for the assembly of iron–sulfur clusters, an essential function present in mitochondria (27) and in hydrogenosomes (28), anaerobic H2-producing forms of mitochondria found among many protists that inhabit niches too poor in oxygen to support typical respiration (24).
Another striking example of secondary loss involves the microsporidia, which cause secondary infection in AIDS patients. These protists were long thought to be amitochondriate and furthermore to be one of the earliest-branching eukaryotic lineages based on their position in the rRNA tree. Recent genome-based work has, however, shown that they are in fact highly modified fungi (29) and, perhaps even more remarkably, that they have not completely lost their mitochondrion after all (30). Furthermore, there is the case of Giardia intestinalis. This pathogen of the human intestinal tract has yet to reveal cytological evidence for a mitochondrion-derived organelle, but it possesses mitochondrion-derived genes in its genome, indicating the past presence of a mitochondrion that was secondarily lost (31, 32), much like the case of the trypanosomatid plastid.
In one respect, the trypanosomatids differ from all other parasites that have lost an endosymbiotically derived organelle: They have invented a new organelle not present in any other eukaryotic group, the glycosome. Glycosomes are specialized microbodies that contain most of the enzymes of the glycolytic pathway (33, 34). Interestingly, some of these glycolytic enzymes now turn out to have a plastid-related origin, which does not mean that the glycosome is a withered evolutionary remnant of a plastid (1). The glycosome is an undisputed member of the peroxisome family; it is bounded by a single membrane that contains proteins that are clearly homologous to their counterparts in peroxisomes of other eukaryotes. Furthermore, import of proteins into glycosomes requires the same targeting signals as in peroxisomes. Thus, it seems that some enzymes that were “abandoned” by their disappearing chloroplast have found a new home inside the glycosome (1).
The evolutionary Odyssey of genes encoding the enzymes reported by Hannaert et al. is sketched in Fig. 1. Several of these genes entered into the eukaryotic cell through the (primary) cyanobacterial ancestor of the chloroplast and were transferred to the nucleus during the evolution of the green algae lineage. When a green alga was engulfed to give rise to the secondary plastid of the euglenids and trypanosomatids, genes were transferred once again, this time from the nucleus of the eukaryotic endosymbiont to the nucleus of the eukaryotic host. This model predicts that trypanosomes and euglenids possessed one and the same plastid, but other more complicated scenarios cannot be excluded. For example, the trypanosome lineage might have already possessed a primary plastid (35) before the euglenid lineage acquired a secondary plastid. The analysis of homologues from the Euglena genome should clarify such questions.
For parasitologists, the disappearing plastid opens up new avenues for understanding trypanosomatids and combating the diseases that they cause. For evolutionary biologists, these findings are a reminder that, fortunately or unfortunately, things are rarely as simple as they seem. With genome projects of other protozoa now ongoing, data analysts should be on the lookout for further unexpected genes of cyanobacterial or algal origin and the tell-tale trace of organelles long gone.
Footnotes
See companion article on page 1067.
For T. brucei, see www.sanger.ac.uk/Projects/T_brucei/ and www.tigr.org/tdb/mdb/tbdb/index.shtml; for L. major, see www.sanger.ac.uk/Projects/L_major/; for T. cruzi, see www.tigr.org/tdb/e2k1/tca1.
References
- 1.Hannaert V, Saavedra E, Duffieux F, Szikora J-P, Rigden D J, Michels P A M, Opperdoes F R. Proc Natl Acad Sci USA. 2003;100:1067–1071. doi: 10.1073/pnas.0335769100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gelb M H, Hol W G J. Science. 2002;297:343–344. doi: 10.1126/science.1073126. [DOI] [PubMed] [Google Scholar]
- 3.Urbina J A. Curr Opin Infect Dis. 2001;14:733–741. doi: 10.1097/00001432-200112000-00012. [DOI] [PubMed] [Google Scholar]
- 4.Krepinsky K, Plaumann M, Martin W, Schnarrenberger C. Eur J Biochem. 2001;268:2678–2686. doi: 10.1046/j.1432-1327.2001.02154.x. [DOI] [PubMed] [Google Scholar]
- 5.Wilkinson S R, Obado S O, Mauricio I L, Kelly J M. Proc Natl Acad Sci USA. 2002;99:13453–13458. doi: 10.1073/pnas.202422899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dooijes D, Chaves I, Kieft R, Martin W, Borst P. Nucleic Acids Res. 2000;28:3017–3021. doi: 10.1093/nar/28.16.3017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stoebe B, Maier U-G. Protoplasma. 2002;219:123–130. doi: 10.1007/s007090200013. [DOI] [PubMed] [Google Scholar]
- 8.Borst P, Overdulve J P, Weijer P J, Fase-Fowler F, van den Berg M. Biochim Biophys Acta. 1984;781:100–111. doi: 10.1016/0167-4781(84)90128-3. [DOI] [PubMed] [Google Scholar]
- 9.Williamson D H, Gardner M J, Preise P, Moore D J, Rangachari K, Wilson R J M. Mol Gen Genet. 1994;243:249–252. doi: 10.1007/BF00280323. [DOI] [PubMed] [Google Scholar]
- 10.McFadden G I, Reith M E, Munholland J, Lang-Unnasch N. Nature. 1996;381:482. doi: 10.1038/381482a0. [DOI] [PubMed] [Google Scholar]
- 11.Jomaa H, Wiesner J, Sanderbrand S, Altincicek B, Weidemeyer C, Hintz M, Turbachova I, Eberl M, Zeidler J, Lichtenthaler H K, et al. Science. 1999;285:1573–1576. doi: 10.1126/science.285.5433.1573. [DOI] [PubMed] [Google Scholar]
- 12.Martin W, Rujan T, Richly E, Hansen A, Cornelsen S, Lins T, Leister D, Stoebe B, Hasegawa M, Penny D. Proc Natl Acad Sci USA. 2002;99:12246–12251. doi: 10.1073/pnas.182432999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Katz L A. Int J Syst Evol Microbiol. 2002;52:1893–1900. doi: 10.1099/00207713-52-5-1893. [DOI] [PubMed] [Google Scholar]
- 14.Doolittle W F. Trends Genet. 1998;14:307–311. doi: 10.1016/s0168-9525(98)01494-2. [DOI] [PubMed] [Google Scholar]
- 15.Ricchetti M, Fairhead C, Dujon B. Nature. 1999;402:96–100. doi: 10.1038/47076. [DOI] [PubMed] [Google Scholar]
- 16.Henze K, Martin W. Trends Genet. 2001;17:383–387. doi: 10.1016/s0168-9525(01)02312-5. [DOI] [PubMed] [Google Scholar]
- 17.Lin X, Kaul S, Rounsley S, Shea T P, Benito M I, Town CD, Fujii C Y, Mason T, Bowman C L, Barnstead M, et al. Nature. 1999;402:761–768. doi: 10.1038/45471. [DOI] [PubMed] [Google Scholar]
- 18.Stupar R M, Lilly J W, Town C D, Cheng Z, Kaul S, Buell C R, Jiang J. Proc Natl Acad Sci USA. 2001;98:5099–5103. doi: 10.1073/pnas.091110398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yuan Q, Hill J, Hsiao J, Moffat K, Ouyang S, Cheng Z, Jiang J, Buell C R. Mol Genet Genom. 2002;267:713–720. doi: 10.1007/s00438-002-0706-1. [DOI] [PubMed] [Google Scholar]
- 20.Allen J F. Philos Trans R Soc London B. 2003;358:19–38. doi: 10.1098/rstb.2002.1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Palenik B. Proc Natl Acad Sci USA. 2002;99:11996–11997. doi: 10.1073/pnas.202486299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cavalier-Smith T. Trends Plant Sci. 2000;5:174–182. doi: 10.1016/s1360-1385(00)01598-3. [DOI] [PubMed] [Google Scholar]
- 23.van't Klooster J W, van den Berg-Velthuis G, van West P, Govers F. Gene. 2000;249:145–151. doi: 10.1016/s0378-1119(00)00151-7. [DOI] [PubMed] [Google Scholar]
- 24.Müller M. In: Evolutionary Relationships Among Protozoa. Coombs G H, Vickermann K, Sleigh M A, Warren A, editors. Dordrecht, The Netherlands: Kluwer; 1998. pp. 109–131. [Google Scholar]
- 25.Tovar J, Fischer A, Clark C G. Mol Microbiol. 1999;32:1013–1021. doi: 10.1046/j.1365-2958.1999.01414.x. [DOI] [PubMed] [Google Scholar]
- 26.Mai Z, Ghosh S, Frisardi M, Rosenthal B, Rogers R, Samuelson J. Mol Cell Biol. 1999;19:2198–2205. doi: 10.1128/mcb.19.3.2198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kispal G, Csere P, Prohl C, Lill R. EMBO J. 1999;18:3981–3989. doi: 10.1093/emboj/18.14.3981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tachezy J, Sanchez L B, Müller M. Mol Biol Evol. 2001;18:1919–1928. doi: 10.1093/oxfordjournals.molbev.a003732. [DOI] [PubMed] [Google Scholar]
- 29.Keeling P J, Fast N M. Annu Rev Microbiol. 2002;56:93–116. doi: 10.1146/annurev.micro.56.012302.160854. [DOI] [PubMed] [Google Scholar]
- 30.Williams B A, Hirt R P, Lucocq J M, Embley T M. Nature. 2002;418:865–869. doi: 10.1038/nature00949. [DOI] [PubMed] [Google Scholar]
- 31.Horner D S, Embley T M. Mol Biol Evol. 2001;18:1970–1975. doi: 10.1093/oxfordjournals.molbev.a003737. [DOI] [PubMed] [Google Scholar]
- 32.Tielens A G M, Rotte C, van Hellemond J, Martin W. Trends Biochem Sci. 2002;27:564–572. doi: 10.1016/s0968-0004(02)02193-x. [DOI] [PubMed] [Google Scholar]
- 33.Opperdoes F R, Borst P. FEBS Lett. 1977;80:360–364. doi: 10.1016/0014-5793(77)80476-6. [DOI] [PubMed] [Google Scholar]
- 34.Michels P A M, Hannaert V, Bringaud F. Parasitol Today. 2000;16:482–489. doi: 10.1016/s0169-4758(00)01810-x. [DOI] [PubMed] [Google Scholar]
- 35.Andersson J O, Roger A J. Curr Biol. 2002;12:115–119. doi: 10.1016/s0960-9822(01)00649-2. [DOI] [PubMed] [Google Scholar]