Complex I (NADH/quinone oxidoreductase) is the only enzyme of the membrane-bound respiratory chain to remain a “black box.” Atomic resolution structural models are available for three of the four energy-transducing respiratory complexes: the cytochrome bc1 complex, cytochrome oxidase, and the F1 domain of ATP synthase. Detailed mechanisms that integrate these structural models with the results of spectroscopic and functional studies are therefore being constructed (1). In comparison, due to its large size and subunit complexity, and because its cofactors are difficult to distinguish spectroscopically, complex I has proved intractable. The complex is L shaped (2) and, although the exact cofactor content and ligation remain unconfirmed, complex I is known to contain a noncovalently bound flavin mononucleotide at the active site for NADH oxidation and a number of iron–sulfur clusters (3). Complex I plays a pivotal role in energy transduction. It is the entry point for electrons into the respiratory chain, and it contributes to the proton-motive force (ΔμH+) across the inner-mitochondrial membrane, which is harnessed for the synthesis of ATP (1). In mitochondria, it is well established that complex I uses the free energy from transferring two electrons from NADH to ubiquinone to pump four protons across the inner-mitochondrial membrane (4, 5), and it has been widely assumed that all complexes I operate in the same way. In this issue of PNAS, Gemperli et al. (6) challenge this central tenet and present evidence that complex I from the enterobacterium Klebsiella pneumoniae pumps sodium ions rather than protons across the bacterial cytosolic membrane. In a reconstituted system, the sodium ion motive force (ΔμNa+) supports ATP synthesis by the sodium ion driven ATP synthase from Ilyobacter tartaricus. Complex I from Escherichia coli, closely related to the K. pneumoniae enzyme, has also been proposed to be a primary sodium ion pump (7).
Complex I from bovine heart mitochondria is composed of at least 45 different subunits (8), 14 of them being conserved throughout the complex I family. These 14 “core” subunits comprise the simpler bacterial enzymes (9) and therefore are deemed sufficient for catalysis and energy transduction. The core subunit conservation and homology suggest that all complexes I operate by a common mechanism. However, if bovine complex I pumps four protons across the membrane for each NADH oxidized but complex I from K. pneumoniae pumps two sodium ions (10), then this conclusion must be questioned. These different cation specificities and stoichiometries have fundamental implications for our understanding of the mechanism of complex I and its role in energy transduction and metabolism, and challenge the relevance of bacterial complexes I as models for the mammalian enzyme. It is therefore imperative that the sodium ion pumping activity of complex I from E. coli or K. pneumoniae be verified independently by other researchers. Furthermore, because only a limited number of systems have so far been characterized, equivalent analysis of the complexes I from other species is required before any significant generalization may be made. Two interesting possibilities are that the sodium ion specificity is more widespread than has been suspected hitherto, and that systematic differences in the electron/cation stoichiometry exist. It is conceivable that the more elaborate eukaryotic enzymes contain an extra “module” enabling them to pump two more protons than their bacterial counterparts. Alternatively, the stoichiometry may vary according to the free energy available: different quinones are used, and proton and sodium ion motive forces vary between species and in response to conditions.
Complex I from K. pneumoniae pumps sodium ions across the bacterial cytosolic membrane.
Determination of the cation specificity of a number of complexes I will allow meaningful sequence comparisons to be drawn, and these may help to identify the cation-binding sites. A similar approach was employed successfully for ATP synthase (11): most ATP synthases use a proton motive force, but closely related examples that use a sodium ion motive force have been identified in bacteria such as Propionigenium modestum. A family of sodium ion/proton antiporters identified in alkaliphilic bacteria contain stretches of sequence similarity with the membrane-bound ND2, 4, and 5 (NuoN, -M, and -L) subunits of complex I (12, 13). On this basis, these subunits have been proposed to bind cations and to constitute a conformationally driven ion pump (14, 15). However, the absence of any experimental support for these speculations demands that they be treated with caution.
Before discussing the mechanism of complex I, it is worth noting that the net redox reaction is the transfer of two electrons from NADH to quinone, with the concomitant uptake of two protons. Because the quinol product is released into the bilayer, these chemical protons cannot be substituted by sodium ions. The chemical reaction itself is nonelectrogenic (it does not contribute to the membrane potential), because the protons and electrons enter the bilayer from the same side; these protons are assumed to originate in the mitochondrial matrix to avoid dissipating the proton motive force. Only the vectorial protons, which cross the membrane completely, may be replaced by sodium ions.
The majority of proposals for the mechanism of complex I (16) can be classified as either Q cycle or redox pump mechanisms. Examples of these are now considered, under the assumption that the mechanism is conserved in both the sodium ion and proton-pumping enzymes.
Dutton et al. (17) proposed that two protons are translocated by complex I by a modified Q cycle based on that of the cytochrome bc1 complex, but energetically and vectorially complementary to it: complex I catalyzes the overall reduction of quinone instead of the oxidation of quinol. In support of this hypothesis, evidence for two bound semiquinones in complex I has been presented, and on the basis of their coupling to iron–sulfur cluster N2, they may be located at opposite sides of the membrane (18). However, the number and location of the quinone-binding sites in complex I are disputed, and there are two further arguments against this proposal. First, the Q cycle accounts for only two protons, and therefore it is insufficient to support the pumping of four protons by mitochondrial complex I. Second, during catalysis, all of the protons translocated across the membrane have been, or will be, bound to ubiquinol, which is free to diffuse in the membrane bilayer (19). These two protons cannot be replaced by sodium ions and therefore this mechanism, along with any mechanism based on a Q cycle, is precluded.
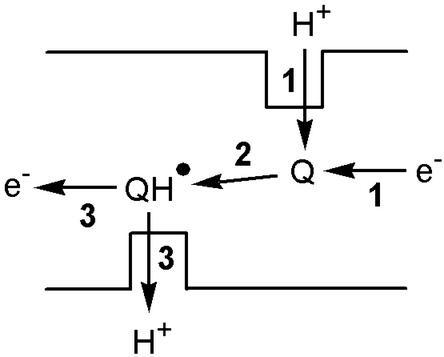
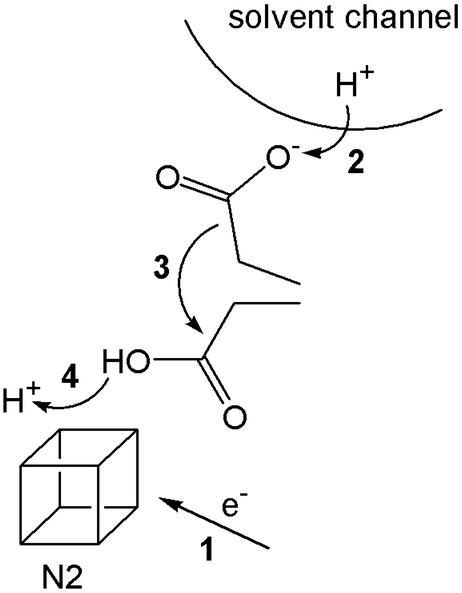
A redox pump uses the free energy from electron transfer to translocate protons across a hydrophobic barrier. The quinone/semiquinone proton pump suggested by Dutton et al. (17) to translocate the second pair of protons for each pair of electrons forms an illustrative example (see Fig. 1). The quinone gates a proton channel, which would otherwise permit unrestricted proton or ion movement in either direction. Reduction of the quinone causes a significant increase in its pK (electron and proton affinities are tightly coupled), and a proton is taken up from the matrix side. A conformational change to expose the protonated semiquinone to the intermembrane space is then required, before oxidation and the concomitant decrease in pK cause deprotonation and proton expulsion to the intermembrane space. The quinone must then return to its original conformation, ready for the next cycle, and so protons are shuttled across the hydrophobic barrier. A second potential redox pump in complex I involves iron–sulfur cluster N2, thought to be the electron donor to bound quinone (18). The reduction potential of cluster N2 is reported to be pH-dependent (20), suggesting that a proton binds at, or close to, the cluster, and the protonation state of a nearby carboxylate responds to the cluster oxidation state (21). This arrangement corresponds to the “proton-transfer module” found in a small iron–sulfur protein, ferredoxin I from Azotobacter vinelandii. In ferredoxin I, the reduction of a [3Fe-4S] cluster, buried below the solvent accessible surface, causes it to protonate. Proton transfer across the hydrophobic protein barrier is mediated by the swinging arm of an adjacent carboxylate residue (22). The pKs of both cluster and carboxylate depend on the cluster oxidation state (electron and proton affinities are tightly coupled), so that the proton moves only in response to movement of the electron. Similarly, in complex I, proton transfer may be gated by a carboxylate residue, carrying protons across a hydrophobic barrier in response to a change in the oxidation state of cluster N2 (see Fig. 2). A similar role has been suggested for a glutamate residue in cytochrome oxidase (23). To increase the stoichiometry above one proton per electron, pumping modules may be connected in series. However, both redox pumps described above depend on the covalent binding of a proton, and therefore, like the Q cycle, they cannot switch to pumping sodium ions.
Figure 1.
The quinone/semiquinone redox pump. (1) Reduction of the quinone causes protonation from the matrix side; (2) a conformational change exposes the semiquinone to the intermembrane space; and (3) oxidation of the semiquinone causes proton expulsion to the intermembrane space. The energy required to pump the proton across the membrane is provided by a decrease in the potential energy of the electron.
Figure 2.
Proton translocation across a hydrophobic barrier in response to the reduction of iron–sulfur cluster N2, mediated by a carboxylate residue. (1) Reduction of cluster N2; (2) the increased pK of the carboxylate causes it to collect a proton from the solvent channel; (3) the carboxylic acid group carries the proton across the hydrophobic barrier to the cluster; and (4) proton transfer from the carboxylic acid to the cluster (or a nearby group) that has a high pK in the reduced state.
Insights into how complex I transduces redox energy into a sodium ion motive force may be provided by the NADH/quinone oxidoreductase (Na+-NQR) from marine and pathogenic organisms such as Vibrio alginolyticus and Vibrio cholerae (24). Na+-NQR are primary sodium ion pumps, with a stoichiometry of two sodium ions pumped per NADH oxidized (25), but they display no sequence similarity to complex I and contain a different set of cofactors (26). Therefore, there is little reason to suppose that complex I and Na+-NQR operate by the same mechanism. Nevertheless, a common mechanism, analogous to the quinone/semiquinone proton pump described above, has been suggested (14). Because a sodium ion cannot be covalently bound by a semiquinone, the requirement for charge neutralization in a low dielectric is used to explain why semiquinone formation results in cation binding. However, it is difficult to appreciate how reduction and sodium ion binding can be coupled strongly enough, and how proton binding to the semiquinone is avoided. Furthermore, a low dielectric environment is difficult to reconcile with a hydrophilic channel that allows sodium ion access.
The results of Gemperli et al. (6) raise one further question: why should the complexes I from E. coli and K. pneumoniae be used to pump sodium ions instead of protons? Primary sodium ion pumps are widespread in prokaryotes. They include decarboxylases, such as the oxaloacetate decarboxylase of K. pneumoniae (27), the methyltransferases of methanogenic archaea (28), the Na+-NQR of marine and pathogenic organisms such as V. cholerae (24), and the V-type ATPase from Enterococcus hirae (29). Similarly, a sodium ion motive force can be used in various ways, most commonly for nutrient uptake (30) and as a buffer for pH homeostasis, in conjunction with sodium ion/proton antiporters (31). However, examples of a sodium ion motive force being used for ATP synthesis, for example in P. modestum (27), and to power the flagellar motor, for example in V. cholerae (32), are also known. Both the ATP synthase and the flagellar motor of E. coli are powered by the proton motive force, but E. coli does use a sodium ion motive force for the uptake of nutrients such as melibiose, glutamate, proline, and serine (30), and it contains a complement of sodium ion/proton antiporters for homeostasis. The factors that dictate a preference for sodium ions over protons are not clear, but primary sodium ion pumps are typical of organisms that live in environments of high external pH or high salinity, perhaps because of the increased importance of pH homeostasis, or to minimize the effect of an unfavorable contribution to ΔμH+ from ΔpH (31). Although E. coli is not halophilic or alkaliphilic, it may benefit from an increase in robustness and adaptability resulting from the use of two coupling cations instead of one. It is interesting to note that sodium ion translocating NADH/quinone oxidoreductases are common among pathogens (24). Metabolically, the identification of a sodium ion translocating complex I in E. coli is perhaps not so surprising.
In summary, complex I is an experimentally challenging system, due to its complexity and hydrophobicity, and because the flavin, iron–sulfur clusters, and quinones are difficult to study spectroscopically. As a result, and because no atomic resolution structural model is available, the formulation of a uniquely supported and convincing mechanism is not yet possible. The mechanistic hypotheses described above are those that appear most reasonable in the light of currently available experimental data, but they are strongly challenged by the assertion that complex I can pump either sodium ions or protons, and by apparent differences in stoichiometry between homologues. There remain many opportunities for rigorous and innovative experiments to establish a comprehensive understanding of complex I, which will provide a compelling challenge for the foreseeable future.
Footnotes
See companion article on page 839.
References
- 1.Saraste M. Science. 1999;283:1488–1493. doi: 10.1126/science.283.5407.1488. [DOI] [PubMed] [Google Scholar]
- 2.Grigorieff N. Curr Opin Struct Biol. 1999;9:476–483. doi: 10.1016/S0959-440X(99)80067-0. [DOI] [PubMed] [Google Scholar]
- 3.Walker J E. Q Rev Biophys. 1992;25:253–324. doi: 10.1017/s003358350000425x. [DOI] [PubMed] [Google Scholar]
- 4.Wikström M. FEBS Lett. 1984;169:300–304. doi: 10.1016/0014-5793(84)80338-5. [DOI] [PubMed] [Google Scholar]
- 5.Brown G C, Brand M D. Biochem J. 1988;252:473–479. doi: 10.1042/bj2520473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gemperli A C, Dimroth P, Steuber J. Proc Natl Acad Sci USA. 2003;100:839–844. doi: 10.1073/pnas.0237328100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Steuber J, Schmid C, Rufibach M, Dimroth P. Mol Microbiol. 2000;35:428–434. doi: 10.1046/j.1365-2958.2000.01712.x. [DOI] [PubMed] [Google Scholar]
- 8.Carroll J, Shannon R J, Fearnley I M, Walker J E, Hirst J. J Biol Chem. 2002;277:50311–50317. doi: 10.1074/jbc.M209166200. [DOI] [PubMed] [Google Scholar]
- 9.Yagi T, Yano T, Di Bernardo S, Matsuno-Yagi A. Biochim Biophys Acta. 1998;1364:125–133. doi: 10.1016/s0005-2728(98)00023-1. [DOI] [PubMed] [Google Scholar]
- 10.Gemperli A C, Dimroth P, Steuber J. J Biol Chem. 2002;277:33811–33817. doi: 10.1074/jbc.M204860200. [DOI] [PubMed] [Google Scholar]
- 11.Kaim G, Wehrle F, Gerike U, Dimroth P. Biochemistry. 1997;36:9185–9194. doi: 10.1021/bi970831q. [DOI] [PubMed] [Google Scholar]
- 12.Hamamoto T, Hashimoto M, Hino M, Kitada M, Seto Y, Kudo T, Horikoshi K. Mol Microbiol. 1994;14:939–946. doi: 10.1111/j.1365-2958.1994.tb01329.x. [DOI] [PubMed] [Google Scholar]
- 13.Mathiesen C, Hägerhäll C. Biochim Biophys Acta. 2002;1556:121–132. doi: 10.1016/s0005-2728(02)00343-2. [DOI] [PubMed] [Google Scholar]
- 14.Steuber J. Biochim Biophys Acta. 2001;1505:45–56. doi: 10.1016/s0005-2728(00)00276-0. [DOI] [PubMed] [Google Scholar]
- 15.Friedrich T. J Bioenerg Biomembr. 2001;33:169–177. doi: 10.1023/a:1010722717257. [DOI] [PubMed] [Google Scholar]
- 16.Brandt U. Biochim Biophys Acta. 1997;1318:79–91. doi: 10.1016/s0005-2728(96)00141-7. [DOI] [PubMed] [Google Scholar]
- 17.Dutton P L, Moser C C, Sled V D, Daldal F, Ohnishi T. Biochim Biophys Acta. 1998;1364:245–257. doi: 10.1016/s0005-2728(98)00031-0. [DOI] [PubMed] [Google Scholar]
- 18.Ohnishi T. Biochim Biophys Acta. 1998;1364:186–206. doi: 10.1016/s0005-2728(98)00027-9. [DOI] [PubMed] [Google Scholar]
- 19.Berry E A, Guergova-Kuras M, Huang L, Crofts A R. Annu Rev Biochem. 2000;69:1005–1075. doi: 10.1146/annurev.biochem.69.1.1005. [DOI] [PubMed] [Google Scholar]
- 20.Ingledew W J, Ohnishi T. Biochem J. 1980;186:111–117. doi: 10.1042/bj1860111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hellwig P, Scheide D, Bungert S, Mäntele W, Friedrich T. Biochemistry. 2000;39:10884–10891. doi: 10.1021/bi000842a. [DOI] [PubMed] [Google Scholar]
- 22.Chen K S, Hirst J, Camba R, Bonagura C A, Stout C D, Burgess B K, Armstrong F A. Nature. 2000;405:814–817. doi: 10.1038/35015610. [DOI] [PubMed] [Google Scholar]
- 23.Wikström M. Biochim Biophys Acta. 1998;1365:185–192. doi: 10.1016/s0005-2728(98)00095-4. [DOI] [PubMed] [Google Scholar]
- 24.Hayashi M, Nakayama Y, Unemoto T. Biochim Biophys Acta. 2001;1505:37–44. doi: 10.1016/s0005-2728(00)00275-9. [DOI] [PubMed] [Google Scholar]
- 25.Bogachev A V, Murtazina R A, Skulachev V P. FEBS Lett. 1997;409:475–477. doi: 10.1016/s0014-5793(97)00536-x. [DOI] [PubMed] [Google Scholar]
- 26.Barquera B, Zhou W, Morgan J E, Gennis R B. Proc Natl Acad Sci USA. 2002;99:10322–10324. doi: 10.1073/pnas.162361299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dimroth P. Biochim Biophys Acta. 1997;1318:11–51. doi: 10.1016/s0005-2728(96)00127-2. [DOI] [PubMed] [Google Scholar]
- 28.Gottschalk G, Thauer R K. Biochim Biophys Acta. 2001;1505:28–36. doi: 10.1016/s0005-2728(00)00274-7. [DOI] [PubMed] [Google Scholar]
- 29.Murata T, Kawano M, Igarashi K, Yamato I, Kakinuma Y. Biochim Biophys Acta. 2001;1505:75–81. doi: 10.1016/s0005-2728(00)00278-4. [DOI] [PubMed] [Google Scholar]
- 30.Wilson T H, Ding P Z. Biochim Biophys Acta. 2001;1505:121–130. doi: 10.1016/s0005-2728(00)00282-6. [DOI] [PubMed] [Google Scholar]
- 31.Krulwich T A, Ito M, Guffanti A. Biochim Biophys Acta. 2001;1505:158–168. doi: 10.1016/s0005-2728(00)00285-1. [DOI] [PubMed] [Google Scholar]
- 32.Yorimitsu T, Homma M. Biochim Biophys Acta. 2001;1505:82–93. doi: 10.1016/s0005-2728(00)00279-6. [DOI] [PubMed] [Google Scholar]