Abstract
The nuclear receptor PXR (pregnane X receptor) is a broad-specificity sensor that recognizes a wide variety of synthetic drugs and xenobiotic agents. On activation by these compounds, PXR coordinately induces a network of transporters, cytochrome P450 enzymes, and other genes that effectively clear xenobiotics from the liver and intestine. Like PXR, the majority of its target genes also possess a broad specificity for exogenous compounds. Thus, PXR is both a sensor and effector in a well integrated and generalized pathway for chemical immunity. Although it is clear that PXR responds to numerous foreign compounds, it is unclear whether it possesses an endogenous ligand. To address this issue, we noted that there is substantial overlap in the substrate specificities of PXR and its critical CYP3A target gene. This prompted us to ask whether endogenous CYP3A substrates also serve as PXR ligands. We demonstrate that 5β-cholestane-3α,7α,12α-triol (triol), a cholesterol-derived CYP3A substrate, is a potent PXR agonist that effectively induces cyp3a expression in mice. This defines a critical salvage pathway that can be autoinduced to minimize triol accumulation. In contrast, triol can accumulate to very high levels in humans, and unlike mice, these people develop the severe clinical manifestations of cerebrotendinous xanthomatosis. The reason for these dramatic species differences has remained unclear. We now demonstrate that triol fails to activate human PXR or induce the CYP3A-salvage pathway. This explains why humans are more susceptible to sterol accumulation and suggests that synthetic ligands for human PXR could be used to treat cerebrotendinous xanthomatosis and other disorders of cholesterol excess.
The nuclear receptor pregnane X receptor (PXR, NR1I2; refs. 1 and 2) is a promiscuous receptor that is activated by numerous drugs and foreign compounds (1, 2). On binding these ligands, PXR coordinately induces expression of a network of genes that control xenobiotic clearance from the liver and intestine. Among these target genes are heme-containing cytochrome P450 enzymes (CYP3A, CYP2C, and CYP2B) that promote oxidative (phase I) drug metabolism (3, 4); genes required for the synthesis of the common heme group and the P450 oxidoreductase subunit (aminolevulinic acid synthase, POR; ref. 4); phase II-conjugating enzymes that improve solubility of phase I metabolites (glutathione S-transferases, sulfotransferases, and UDP-glucoronosyltransferases; refs. 4–6); and xenobiotic transporters (MDR1, MRP2, MRP3, and OATP2; refs. 3 and 4) that mediate excretion of the above compounds. PXR can also autoinduce its own expression (4), thereby amplifying its initial responses. PXR is thus a master regulator of xenobiotic clearance, because it senses foreign compounds and induces their clearance via a host of complementary detoxification pathways.
The CYP3A enzymes (cyp3a11 in mice and CYP3A4 in humans) are among the most critical PXR targets because they metabolize ≈50% of all pharmaceutical agents (1, 2). Several of these CYP3A substrates are also known to activate PXR [e.g., taxol (3), ritonavir (7), clotrimazole (8), troglitazone (9), and others], suggesting an overlap in substrate recognition by these two proteins. This prompted us to explore the possibility that naturally occurring CYP3A substrates may represent endogenous PXR ligands.
Previous studies have demonstrated that the cholesterol metabolite 5β-cholestane-3α,7α,12α-triol (triol) is an endogenous CYP3A substrate (10, 11). Although CYP3A can metabolize triol, the predominant route of triol metabolism is catalyzed by CYP27 (sterol-27-hydroxylase). This metabolic conversion occurs exclusively in the liver and is an integral event in the conversion of cholesterol to bile acids (12). CYP27 catalyzes the addition of a hydroxyl group at position 27 of triol (see Fig. 2a), which is further oxidized to an intermediate destined for bile acid biosynthesis (5β-cholestanoic acid-3α,7α,12α-triol). Thus, animals with defects in CYP27 lack the enzymatic activity that catabolizes the bulk of this sterol.
Figure 2.
Triol is an endogenous activator of mPXR. (a) The metabolic pathways that underlie the production and elimination of triol are shown schematically. Triol is produced as an intermediate in the hepatic conversion of cholesterol to bile acids. Triol is normally metabolized via CYP27; CYP3A provides an alternate elimination pathway when triol levels are elevated. (b) Elevated hepatic triol levels in the liver of cyp27-null mice. GC-MS was used to measure triol levels in liver extracts from WT or cyp27-null mice (cyp27−/−). (c) An extract from cyp27-null liver activates mPXR. CV-1 cells were transfected with GAL-mPXR as in Fig. 1a. Cells were treated with equal amounts of organic extracts derived from the liver of WT or cyp27-null mice. (d) PXR target genes are induced in the liver of female cyp27-null mice. Northern analysis was performed as in Fig. 1h. (e) Cyp27-null mice are resistant to a xenobiotic challenge. WT and cyp27-null mice received an i.p. injection of tribromoethanol, and their individual sleep times are plotted. WT animals are represented by gray squares; cyp27-null mice (cyp27−/−) are represented by black circles. WT male, n = 6; cyp27−/− male, n = 5; WT female, n = 6; cyp27−/− female, n = 6. **, P < 0.01; ***, P < 0.001. Error bars represent the SEM and are not visible in cases where they are negligible relative to the scale of the figure.
Although triol is a naturally occurring metabolic intermediate, excess levels of this sterol are associated with significant pathologies. Indeed, humans with CYP27 deficiency accumulate and deposit sterols in a variety of tissues. This results in xanthomas, atherosclerosis, gallstones, and neurological dysfunction, the hallmark features of a disease known as cerebrotendinous xanthomatosis (CTX) (13). Excess triol therefore represents an endogenous toxin that must be eliminated to maintain normal physiologic function. Under conditions of CYP27 deficiency, CYP3A becomes critical because it provides an alternate pathway for triol elimination. Indeed, cyp27-null mice induce cyp3a activity and fail to develop the major pathological features of CTX; in contrast, CYP27-deficient patients fail to induce CYP3A and are highly susceptible to the clinical features that define this disease (13). An explanation of the molecular features that underlie this species difference is critical to our understanding of CTX.
We now demonstrate that triol activates mouse PXR (mPXR) and induces cyp3a expression in this species. This provides a salvage pathway for triol degradation that protects mice from the development of CTX-related pathologies. In contrast, triol is not an effective activator of human PXR (hPXR, also known as SXR), thus explaining why humans are susceptible to sterol accumulation during CYP27 deficiency. Our findings provide a rationale for the use of hPXR ligands in the management of CTX and other disorders associated with sterol accumulation.
Materials and Methods
Transient Transfection Assay.
CV-1 cells were transiently transfected with expression vectors, luciferase reporter constructs, and an expression vector for Escherichia coli β-galactosidase as an internal control. Transfection conditions and plasmids were as described (3, 7, 14, 15).
Ligand-Binding Assay.
The SXR ligand-binding domain was expressed in E. coli with an N-terminal His tag and purified. Binding to [3H]SR12813 was performed with a scintillation proximity assay as described (7).
Primary Hepatocyte Cultures and Northern Analysis.
Primary human hepatocyte cultures and human probes were as described (3). Primary mouse hepatocytes were isolated as described (16) from C57BL6/J mice by using collagenase type IV; 5 × 105 cells per well were plated in six-well collagen-coated plates and cultured in DMEM/Ham's F12 media (1:1) containing 10 nM dexamethasone. Seventy hours after plating, the cells were treated with PXR ligands for an additional 24 h. Total RNA was then isolated using Trizol reagent, and Northern blots were prepared with 10 μg of RNA per lane. Probes for mouse genes were as follows: cyp3a11, nucleotides 1,065–1,569 of GenBank accession no. X60452; cyp2c, nucleotides 787–1,193 of accession no. AK008580; oatp2, nucleotides 2,124–2,486 of accession no. NM_021471; and gapdh, nucleotides 590–1,089 of accession no. NM_008094.
Liver Extracts and GC-MS.
WT or cyp27−/− mouse liver tissue (0.5 g) was extracted with 7.5 ml of ethyl acetate/methanol (2:1, vol/vol) along with 5β-cholestane-3β,5α,6β-triol (25 μg) as an internal control for extraction efficiency. This sterol does not occur naturally and does not activate mPXR (Fig. 1a). The extraction was repeated three times, and the organic fraction was pooled and evaporated. To remove the vast excess of cholesterol that interferes with subsequent GC-MS analysis, the evaporated residue was dissolved in 0.5 ml of chloroform/acetone (35:25, vol/vol) and applied to a Bond Elut SI cartridge (Varian, 500 mg). Cholesterol was removed by washing with 6 ml of chloroform/acetone (35:25, vol/vol), and sterols were eluted with 7 ml of chloroform/acetone/methanol (35:25:20, vol/vol/vol). The sterol fraction was evaporated and dissolved in acetonitrile, and a portion was silylated with N,O-bis(trimethylsilyl)trifluoroacetamide containing 1% trimethylchlorosilane (Pierce). The silylated material was analyzed by using a Shimadzu model GC-17A gas chromatograph with a QP5000 mass spectral detector and a Hewlett–Packard Ultra 2 (cross-linked-siloxane) column. The injector port was kept at 250°C, interface temperature was kept at 280°C, and oven temperature was kept at 50°C followed by a gradient of 30°C min−1 up to 300°C. The electron impact ionization source was at 70 eV. Triol levels were determined by comparison to a standard curve. For transfection studies, the sterol fraction was dissolved in DMSO and added to the cell-culture media. The DMSO solution from the WT and cyp27-null liver was normalized to contain equal amounts of the internal control 5β-cholestane-3β,5α,6β-triol.
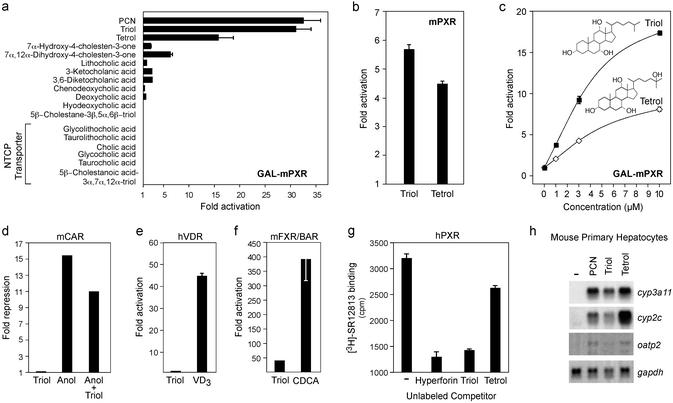
Figure 1.
Triol is an mPXR agonist. (a) Triol and tetrol activate mPXR. CV-1 cells were transiently transfected with a GAL–mPXR expression vector, a GAL4 reporter construct, and a β-galactosidase vector as an internal control. Where noted, the bile acid transporter NTCP was added to the transfection to promote transport of membrane-impermeable bile acids. Transfected cells were exposed to the indicated compounds (10 μM), and fold activation was determined. (b) Same as a except that a full-length mPXR expression vector was used along with a reporter construct containing three copies of the rat cyp3a2 PXR response element. (c) Dose–response analysis of triol and tetrol activity. Cells were treated as in a but with multiple concentrations of each sterol. (d–f) Triol does not effectively activate other nuclear receptors. Experimental conditions as in a except that cells were transfected with mCAR (d), mFXR + hRXRα (e), or hVDR (f) and their corresponding reporter constructs. The following ligands were used as indicated: 10 μM triol, 5 μM 5α-androstan-3α-ol (Anol), 100 nM vitamin D3 (VD3), and 100 μM chenodeoxycholic acid (CDCA). (g) Triol binds directly to PXR. Bacterially expressed hPXR was incubated with [3H]SR12813 in the absence or presence of the following unlabeled competitors: 5 μM hyperforin, 30 μM triol, and 30 μM tetrol. The amount of [3H]SR12813 associated with hPXR is expressed in cpm. (h) Triol and tetrol activate endogenous mPXR target genes. Primary mouse hepatocytes were treated with PXR ligands (10 μM), and Northern analysis was performed using the indicated probes. Transfections were performed in triplicate, and each triplicate experiment was independently repeated three or more times. Error bars represent the standard error of the mean (SEM) from a representative experiment. In some cases, the error bars are not visible because they are negligible relative to the scale of the figure.
Tribromoethanol-Induced Anesthesia.
The sleep test was performed as described (17, 18), except that mice received an i.p. injection of tribromoethanol (0.35 mg/g of body weight). Sleep time was measured until the animals had regained sufficient consciousness to fully right themselves.
Results and Discussion
Endogenous Sterols Activate mPXR.
Previous studies demonstrated that triol, a cholesterol metabolite and bile acid precursor, is an endogenous CYP3A substrate in both human (10) and mouse (11) liver. To determine whether triol activates mPXR, CV-1 cells were transfected with a Gal4 reporter construct and an expression vector containing the ligand-binding domain of mPXR linked to the DNA-binding domain of the yeast transcription factor Gal4 (Gal-mPXR). As expected, the classical PXR inducer pregnenolone-16α-carbonitrile (PCN) (10 μM; ref. 19) was a very effective activator (Fig. 1a), but more importantly, the naturally occurring triol was equally effective (31-fold) at the same concentration. Tetrol (5β-cholestane-3α,7α,12α,25-tetrol), the CYP3A product of triol metabolism (11), also exhibited activity (16-fold) but was less efficacious than triol. Other cholesterol metabolites including lithocholic and 3-ketocholanic acids were inactive at 10 μM concentrations (Fig. 1a). These latter compounds were previously shown to activate PXR at higher concentrations, although they induce hepatic damage before activating PXR (20–22). Thus, the significance of lithocholate as a PXR activator may be limited to extreme pathologic states such as severe cholestasis (22). In contrast, triol was not toxic at the concentrations tested (data not shown).
To confirm the ability of triol and tetrol to activate PXR, the effect of these compounds on full-length mPXR was examined using a reporter construct containing a PXR response element from the rat cyp3a gene (cyp3a2; ref. 15). Similar to the findings with GAL-mPXR, 10 μM triol activated full-length mPXR, whereas tetrol was less effective (Fig. 1b). These findings confirm that triol, a cholesterol metabolite, is an effective activator of mPXR.
Dose–response experiments indicated that triol activated mPXR with an approximate EC50 of ≥3 μM (Fig. 1c). This concentration is close to the Michaelis constant reported for triol as a CYP3A4 substrate (Km = 6 μM; ref. 10), indicating that triol can associate with PXR and CYP3A4 at similar concentrations. Triol is an endogenous substrate for CYP3A4 (10, 11); thus, these findings demonstrate that triol activates PXR at biologically relevant concentrations.
To determine whether triol acts specifically on PXR, we examined its activity on other nuclear receptors. PXR is most closely related to CAR, FXR/BAR, and VDR. Indeed, all three receptors possess a limited overlap in their ligand-binding specificities (8, 20, 21, 23, 24). Nonetheless, 10 μM triol failed to activate or repress the constitutive receptor CAR and had no activity on VDR (Fig. 1 d and e). Triol partially activated FXR but was not nearly as effective as optimal doses of the endogenous FXR/BAR ligand chenodeoxycholic acid (ref. 14; Fig. 1f). Moreover, no activity was seen on other nuclear receptors including the thyroid hormone receptor, retinoic acid receptor, estrogen receptor, PPARα,γ,δ, RXRα, LXRα,β, HNF4α, ERRβ, and RORα (data not shown). Thus, at biologically relevant concentrations, triol is a specific, effective, and potent activator of mPXR.
Triol Is a PXR Ligand.
An in vitro ligand displacement assay was used to determine whether triol interacts directly with the ligand-binding pocket of PXR. Because radiolabeled ligands are available for hPXR but not its rodent ortholog (9), we asked whether triol could compete for binding of hPXR to a tritiated ligand. As expected, hPXR bound to [3H]SR12813, and binding was effectively displaced by unlabeled hyperforin (refs. 9 and 25; Fig. 1g), a high-affinity ligand for hPXR. The endogenous PXR agonist triol was an equally effective competitor, whereas tetrol was less active. A variety of related compounds that failed to activate PXR (Fig. 1a) also failed to displace the radiolabeled ligand. These inactive compounds include 7α-hydroxy-4-cholesten-3-one, 7α,12α-dihydroxy-4-cholesten-3-one, 5β-cholestanoic acid-3α,7α,12α-triol, chenodeoxycholic acid, and cholic acid (data not shown). Taken together, these findings demonstrate that triol activates PXR via direct interactions with this receptor.
Triol Activates mPXR in Vivo.
Because triol is a potent and effective agonist of transfected PXR, we asked whether it modulates expression of endogenous target genes. Primary mouse hepatocytes were treated with PCN, triol, or tetrol (10 μM), and RNA levels were monitored by Northern analysis. As expected, the synthetic ligand PCN activated expression of the PXR target genes cyp3a11 (18, 20), cyp2c (3), and oatp2 (20) but had no effect on the gapdh control (Fig. 1h). More importantly, both triol and tetrol specifically induced expression of all three PXR target genes. Because hepatocytes rapidly convert triol to bile acids, the intracellular levels of triol would be expected to decrease during the course of this experiment. This precludes an accurate analysis of relative efficacies, because triol-mediated responses would be underestimated in hepatocyte cultures. Indeed, whereas triol was more efficacious in activating PXR in CV-1 cells (Fig. 1 a and c), tetrol was the more effective activator of hepatocyte-specific genes (Fig. 1h). Although relative efficacies cannot be determined, these studies clearly demonstrate that triol and tetrol are both effective inducers of endogenous PXR target genes.
We next sought to determine the effects of triol on PXR in vivo. Triol is produced as an intermediate in the hepatic conversion of cholesterol to bile acids (12). Under physiological conditions, triol metabolism is predominantly controlled by the activity of CYP27, a sterol-27-hydroxylase. In addition to hydroxylating cholesterol (Fig. 2a), CYP27 also introduces a hydroxyl group at position 27 of triol (Fig. 2a). This 27-hydroxyl is further oxidized to a metabolite (5β-cholestanoic acid-3α,7α,12α-triol) that is destined for conversion to cholic acid. Animals lacking CYP27 activity would not be expected to convert cholesterol to 27-hydroxycholesterol, thereby shunting cholesterol toward the triol pathway (Fig. 2a). This shunt would be expected to enhance triol synthesis. Moreover, the loss of CYP27 activity would also eliminate the predominant route of triol catabolism. As a result, triol levels should be highly elevated in the livers of CYP27-deficient animals. Indeed, previous studies have reported that triol and tetrol levels are elevated in hepatic microsomes of cyp27-null mice and in CTX patients with CYP27 deficiency (13). Thus, we asked whether triol levels were elevated in whole liver extracts from cyp27−/− mice. Using a quantitative gas chromatography–mass spectroscopy approach, we found that triol levels were 16-fold higher in the livers of cyp27-null mice compared with their WT counterparts (Fig. 2b). These liver extracts were then examined for their ability to activate PXR in a cell-based transactivation assay. As seen in Fig. 2c, the extract from cyp27-null mice induced a 13-fold activation of PXR, whereas extracts from WT mice were inactive. Taken together, these findings confirm that cyp27-null mice provide a useful model to study the in vivo effects of triol on PXR activity.
PXR can respond to a variety of synthetic compounds (1, 2); thus, an important question is whether triol acts as an endogenous PXR ligand. Because triol levels are elevated in cyp27-null mice, one would predict that PXR target gene expression would also be elevated. To test this notion, RNA was isolated from the livers of WT and cyp27-null female mice, and Northern analysis was used to measure the expression of a battery of PXR target genes. When compared with WT mice, the expression of cyp3a11, cyp2c, and oatp2 were all dramatically enhanced in the liver of cyp27-null mice (Fig. 2d). This effect was specific as the gapdh control transcript was unaffected. Similar results were seen with male mice (data not shown). These findings demonstrate that endogenous triol activates mPXR in vivo.
Because PXR is a master regulator of small-molecule clearance, continuous activation of PXR in cyp27-null mice should produce a physiologic state of enhanced resistance to endogenous and exogenous toxins. To test this in a physiological setting, mice were challenged with the anesthetic agent tribromoethanol, and their rate of recovery from drug-induced anesthesia was recorded. Previous studies have demonstrated that sensitivity to tribromoethanol is decreased in mice treated with PXR ligands (17) or in transgenic mice with liver-specific expression of a constitutively active PXR chimera (18). Thus, the tribromoethanol-induced sleep test provides a direct and quantitative measure of hepatic PXR activity in live animals. As shown in Fig. 2e, male cyp27-null mice were highly resistant to tribromoethanol; they awoke 30.6 ± 2.9 min (mean ± SEM) after exposure compared with 42.2 ± 1.9 min for WT controls (P < 0.01). In female mice, the difference was even more significant: 35.8 ± 1.6 min for cyp27-null mice compared with 49.6 ± 2.0 min for WT (P < 0.001). These results confirm that PXR clearance pathways are highly active in mice with elevated levels of hepatic triol.
Resistance to tribromoethanol demonstrates that triol-induced PXR activation effectively protects mice from certain small-molecule toxins. When triol itself is accumulating to pathological levels, the ability of this sterol to activate cyp3a11 (Fig. 2d) becomes highly significant as CYP3A establishes an alternate or salvage pathway for the elimination of excess triol (refs. 10 and 11; Fig. 2a). Based on the reported Km of triol for CYP3A (6 μM; ref. 10), this salvage pathway would be initiated as triol levels approach the low micromolar range. We show that triol activates mPXR at these same concentrations (Fig. 1c, EC50 ≥3 μM), and this in turns leads to enhanced expression of cyp3a11 (Fig. 2d). Thus, our findings define a regulatory loop that minimizes triol accumulation, thereby protecting cyp27-null mice from the pathological consequences of excess sterol accumulation.
hPXR Fails to Respond to Excess Sterols.
In sharp contrast to cyp27-null mice, triol accumulates to much higher levels in humans with CYP27 deficiency (13). These patients develop CTX, a disease characterized by sterol deposits that produce xanthomas, atherosclerosis, gallstones, and neurological dysfunction. The appearance of CTX in CYP27-deficient humans, but not mice, implies that humans are unable to induce PXR-mediated pathways that are required for sterol elimination. This is unexpected in that triol is a substrate for both mouse and human CYP3A, and both species of PXR effectively activate the promoters of their corresponding CYP3A genes (15, 18, 20, 26). Instead, we wondered whether the defective response in humans could be explained by an inability of the human receptor to respond to triol. In control experiments, 10 μM triol activated mPXR with the same efficiency as optimal amounts of the synthetic agonist PCN (Fig. 3a). In sharp contrast, triol displayed very weak activity on the human receptor, which can be fully activated by synthetic ligands such as hyperforin (Fig. 3a). Similarly, the triol precursor 7α,12α,-dihydroxy-4-cholesten-3-one and the tetrol metabolite retained activity on mPXR but failed to activate hPXR. These findings demonstrate that specific sterol metabolites that accumulate in CYP27 deficiency fail to activate hPXR. Furthermore, an extract obtained from cyp27-null livers also failed to activate the human receptor (Fig. 3a). Taken together, these findings demonstrate that hPXR fails to respond to the pool of sterols that accumulate in CYP27 deficiency.
Figure 3.
Triol does not activate hPXR-mediated clearance pathways. (a) Triol is a weak agonist of hPXR. CV-1 cells were transfected as in Fig. 1a with either GAL-mPXR (Left) or GAL-hPXR (Right). Ligands were as follows: 2.5 μM hyperforin and 10 μM PCN, triol, tetrol, and 7α,12α-dihydroxy-4-cholesten-3-one. For mPXR, the data are plotted as percent of maximal fold-activation achieved with PCN; for hPXR, it is relative to hyperforin. (b) Triol is a partial agonist/antagonist of hPXR. Experimental conditions were as in a using hPXR. (c) Triol fails to activate human CYP3A4 expression. Northern analysis was performed as in Fig. 1h but using primary human hepatocytes and the following ligands: 2.5 μM hyperforin and 10 μM rifampicin, triol, and tetrol. Error bars represent the SEM and are not visible in cases where they are negligible relative to the scale of the figure.
The inability of triol to activate hPXR was unexpected because triol can bind to the human receptor (Fig. 1g). This apparent discrepancy implies that triol may function as a partial agonist or weak antagonist of hPXR. Indeed, whereas hyperforin maximally activates hPXR, the combination of hyperforin and triol results in suboptimal levels of activation (Fig. 3b). These findings confirm that triol is a poor activator of the human receptor and suggest that triol would not effectively activate CYP3A4-mediated clearance pathways in humans. We explored this directly by measuring CYP3A4 expression in primary human hepatocytes treated with either triol or the synthetic agonists rifampicin and hyperforin. Indeed, triol or its tetrol metabolite had no effect (Fig. 3c), whereas the synthetic agonists rifampicin and hyperforin (9) strongly induced CYP3A4 expression.
Previous studies have identified PXR as a master regulator of xenobiotic clearance. This designation reflects the receptor's ability to detect a wide variety of foreign compounds and to promote their elimination via a tightly regulated network of xenobiotic metabolizing genes (1, 2). This regulatory paradigm provides an efficient means to protect the body from potentially toxic foreign compounds. However, it has been unclear whether endogenous PXR ligands exist and what their biological functions might be. We now demonstrate that excess levels of a naturally occurring cholesterol metabolite (triol) functions as a PXR ligand in mice. These findings extend the role of PXR as an endogenous sterol sensor. Interestingly, triol is a key intermediate in the classical pathway of bile acid synthesis, which provides the major route for cholesterol degradation in vivo (12). This pathway converts cholesterol to triol, which is subsequently catabolized via the enzymatic activity of CYP27 (Fig. 2a). Thus, individuals that are deficient in CYP27 accumulate high levels of triol and ultimately develop the clinical features of CTX (13). Fortunately, triol can be metabolized, albeit less efficiently (low Vmax; ref. 10), via an alternative CYP3A-mediated pathway. Indeed, our data demonstrate that mice respond to excess triol by activating mPXR and its cyp3a11 target gene. This minimizes the amount of triol that accumulates in mice and protects them from CTX-related pathologies (11).
In contrast, we show that excess triol fails to activate hPXR and fails to induce CYP3A4 expression in human hepatocytes. This defines a molecular deficiency that prevents CTX patients from disposing of excess triol. Our findings raise the possibility that the diminished PXR responsiveness in CTX could be overcome by drugs (e.g., rifampicin) or herbal agents (e.g., St. John's Wort/hyperforin; ref. 25) that activate the human receptor. We therefore propose that hPXR ligands should be considered for the treatment and/or prevention of CTX.
Finally, our data raise the possibility that PXR may contribute more broadly to cholesterol homeostasis. By activating CYP3A4 expression, PXR has the potential to enhance flux through the alternate, CYP3A4-mediated pathway of sterol degradation. Thus, PXR agonists may provide an additional strategy for cholesterol lowering in the context of hypercholesterolemia and atherosclerosis.
Acknowledgments
We thank Steven R. Lear for his contributions to these studies and the Gonda family for support of City of Hope facilities. The Cell Culture Core of the University of Southern California Liver Disease Research Center (DK48522) helped in the preparation of mouse hepatocytes for this study. This work was supported by a National Institutes of Health grant (to B.M.F.) and by National Institutes of Health and Veterans Affairs Hospital Research Service awards (to S.K.E. and G.S.).
Abbreviations
- CTX
cerebrotendinous xanthomatosis
- PCN
pregnenolone-16α-carbonitrile
- PXR
pregnane X receptor
- mPXR
mouse PXR
- hPXR
human PXR
- triol
5β-cholestane-3α,7α,12α-triol
- tetrol
5β-cholestane-3α,7α,12α,25-tetrol
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Dussault I, Forman B M. Crit Rev Eukaryotic Gene Expression. 2002;12:53–64. doi: 10.1615/critreveukaryotgeneexpr.v12.i1.30. [DOI] [PubMed] [Google Scholar]
- 2.Xie W, Evans R M. J Biol Chem. 2001;276:37739–37742. doi: 10.1074/jbc.R100033200. [DOI] [PubMed] [Google Scholar]
- 3.Synold T W, Dussault I, Forman B M. Nat Med. 2001;7:584–590. doi: 10.1038/87912. [DOI] [PubMed] [Google Scholar]
- 4.Maglich J M, Stoltz C M, Goodwin B, Hawkins-Brown D, Moore J T, Kliewer S A. Mol Pharmacol. 2002;62:638–646. doi: 10.1124/mol.62.3.638. [DOI] [PubMed] [Google Scholar]
- 5.Rae J M, Johnson M D, Lippman M E, Flockhart D A. J Pharmacol Exp Ther. 2001;299:849–857. [PubMed] [Google Scholar]
- 6.Sonoda J, Xie W, Rosenfeld J M, Barwick J L, Guzelian P S, Evans R M. Proc Natl Acad Sci USA. 2002;99:13801–13806. doi: 10.1073/pnas.212494599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dussault I, Lin M, Hollister K, Wang E H, Synold T W, Forman B M. J Biol Chem. 2001;276:33309–33312. doi: 10.1074/jbc.C100375200. [DOI] [PubMed] [Google Scholar]
- 8.Moore L B, Parks D J, Jones S A, Bledsoe R K, Consler T G, Stimmel J B, Goodwin B, Liddle C, Blanchard S G, Willson T M, et al. J Biol Chem. 2000;275:15122–15127. doi: 10.1074/jbc.M001215200. [DOI] [PubMed] [Google Scholar]
- 9.Jones S A, Moore L B, Shenk J L, Wisely G B, Hamilton G A, McKee D D, Tomkinson N C, LeCluyse E L, Lambert M H, Willson T M, et al. Mol Endocrinol. 2000;14:27–39. doi: 10.1210/mend.14.1.0409. [DOI] [PubMed] [Google Scholar]
- 10.Furster C, Wikvall K. Biochim Biophys Acta. 1999;1437:46–52. doi: 10.1016/s0005-2760(98)00175-1. [DOI] [PubMed] [Google Scholar]
- 11.Honda A, Salen G, Matsuzaki Y, Batta A K, Xu G, Leitersdorf E, Tint G S, Erickson S K, Tanaka N, Shefer S. J Biol Chem. 2001;276:34579–34585. doi: 10.1074/jbc.M103025200. [DOI] [PubMed] [Google Scholar]
- 12.Chiang J Y L. Front Biosci. 1998;3:D176–D193. doi: 10.2741/a273. [DOI] [PubMed] [Google Scholar]
- 13.Honda A, Salen G, Matsuzaki Y, Batta A K, Xu G, Leitersdorf E, Tint G S, Erickson S K, Tanaka N, Shefer S. J Lipid Res. 2001;42:291–300. [PubMed] [Google Scholar]
- 14.Wang H, Chen J, Hollister K, Sowers L C, Forman B M. Mol Cell. 1999;3:543–553. doi: 10.1016/s1097-2765(00)80348-2. [DOI] [PubMed] [Google Scholar]
- 15.Blumberg B, Sabbagh W, Jr, Juguilon H, Bolado J, Jr, van Meter C M, Ong E S, Evans R M. Genes Dev. 1998;12:3195–3205. doi: 10.1101/gad.12.20.3195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bissell D M, Guzelian P S. Ann NY Acad Sci. 1980;349:85–98. doi: 10.1111/j.1749-6632.1980.tb29518.x. [DOI] [PubMed] [Google Scholar]
- 17.Selye H. J Pharm Sci. 1971;60:1–28. doi: 10.1002/jps.2600600102. [DOI] [PubMed] [Google Scholar]
- 18.Xie W, Barwick J L, Downes M, Blumberg B, Simon C M, Nelson M C, Neuschwander-Tetri B A, Brunt E M, Guzelian P S, Evans R M. Nature. 2000;406:435–439. doi: 10.1038/35019116. [DOI] [PubMed] [Google Scholar]
- 19.Kliewer S A, Moore J T, Wade L, Staudinger J L, Watson M A, Jones S A, McKee D D, Oliver B B, Willson T M, Zetterstrom R H, et al. Cell. 1998;92:73–82. doi: 10.1016/s0092-8674(00)80900-9. [DOI] [PubMed] [Google Scholar]
- 20.Staudinger J L, Goodwin B, Jones S A, Hawkins-Brown D, MacKenzie K I, LaTour A, Liu Y, Klaassen C D, Brown K K, Reinhard J, et al. Proc Natl Acad Sci USA. 2001;98:3369–3374. doi: 10.1073/pnas.051551698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xie W, Radominska-Pandya A, Shi Y, Simon C M, Nelson M C, Ong E S, Waxman D J, Evans R M. Proc Natl Acad Sci USA. 2001;98:3375–3380. doi: 10.1073/pnas.051014398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schuetz E G, Strom S, Yasuda K, Lecureur V, Assem M, Brimer C, Lamba J, Kim R B, Ramachandran V, Komoroski B J, et al. J Biol Chem. 2001;276:39411–39418. doi: 10.1074/jbc.M106340200. [DOI] [PubMed] [Google Scholar]
- 23.Makishima M, Lu T T, Xie W, Whitfield G K, Domoto H, Evans R M, Haussler M R, Mangelsdorf D J. Science. 2002;296:1313–1316. doi: 10.1126/science.1070477. [DOI] [PubMed] [Google Scholar]
- 24.Xie W, Barwick J L, Simon C M, Pierce A M, Safe S, Blumberg B, Guzelian P S, Evans R M. Genes Dev. 2000;14:3014–3023. doi: 10.1101/gad.846800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Moore L B, Goodwin B, Jones S A, Wisely G B, Serabjit-Singh C J, Willson T M, Collins J L, Kliewer S A. Proc Natl Acad Sci USA. 2000;97:7500–7502. doi: 10.1073/pnas.130155097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bertilsson G, Heidrich J, Svensson K, Asman M, Jendeberg L, Sydow-Backman M, Ohlsson R, Postlind H, Blomquist P, Berkenstam A. Proc Natl Acad Sci USA. 1998;95:12208–12213. doi: 10.1073/pnas.95.21.12208. [DOI] [PMC free article] [PubMed] [Google Scholar]