Abstract
MurG is an essential glycosyltransferase that forms the glycosidic linkage between N-acetyl muramyl pentapeptide and N-acetyl glucosamine in the biosynthesis of the bacterial cell wall. This enzyme is a member of a major superfamily of NDP-glycosyltransferases for which no x-ray structures containing intact substrates have been reported. Here we present the 2.5-Å crystal structure of Escherichia coli MurG in complex with its donor substrate, UDP-GlcNAc. Combined with genomic analysis of other superfamily members and site-specific mutagenesis of E. coli MurG, this structure sheds light on the molecular basis for both donor and acceptor selectivity for the superfamily. This structural analysis suggests that it will be possible to evolve new glycosyltransferases from prototypical superfamily members by varying two key loops while maintaining the overall architecture of the family and preserving key residues.
Glycosyltransferases (GTases) are one of the most diverse groups of enzymes in nature. They are involved in the biosynthesis of glycolipids, glycoproteins, polysaccharides, and a huge range of secondary metabolites, including many biologically active natural products. Although the acceptor substrates of GTases vary widely and include many classes of oxygen and nitrogen nucleophiles, the donor substrates are almost always activated sugars, with the most common activated species being NDP sugars.
There are currently >7,000 known or putative GTase sequences in the databanks, the majority of which are UDP/TDP-GTases. These enzymes have low sequence homology, and because there was no structural information on them until recently, they have been classified into dozens of different families (1) (http://afmb.cnrs-mrs.fr/∼cazy/CAZY/index.html). In the past few years, 11 different UDP/TDP-GTase structures from 10 different families have been reported (2–15), but all belong to only two different structural superfamilies, GT-A and GT-B (16). Eight of the eleven crystal structures obtained to date belong to the GT-A superfamily, which includes most of the GTases found in the Golgi apparatus and the endoplasmic reticulum. This superfamily is characterized by the presence of a DXD motif and depends on metal ions for activity (16, 17). The GT-B superfamily, which does not require metal ions for enzymatic activity (3, 11, 18), is functionally diverse. It includes most of the GTases involved in the biosynthesis of biologically active secondary metabolites, such as glycopeptides, polyketides, and anthracyclines (11); some of the GTases involved in the biosynthesis of eukaryotic cell-surface glycolipid receptors (19); and many of the GTases that glycosylate lipophilic compounds for clearance from the body (20). This superfamily is also proposed to include O-linked GlcNAc transferase, a GTase that posttranslationally modifies transcription factors and other proteins in eukaryotes (21, 22). Despite its evident importance, this superfamily is not as well understood as the GT-A superfamily, in part because there have been no crystal structures of any GT-B enzymes with intact glycosyl donor or acceptor molecules (23). Escherichia coli MurG belongs to the GT-B superfamily, and the x-ray structure of the MurG:UDP-GlcNAc complex presented here, combined with genomic analysis of the superfamily, provides information on the origins of substrate selectivity and how it can be manipulated.
Materials and Methods
Purification.
Protein expressed from pET21b and containing the C-terminal tag LEHHHHHH (designated C1 MurG) was purified as described previously (8) and concentrated to 10 mg/ml in a storage buffer consisting of 20 mM Tris⋅HCl (pH 7.9), 150 mM NaCl, and 50 mM EDTA. The protein concentrate was stored at −70°C in 50- or 100-μl aliquots and thawed on use.
Site-Directed Mutagenesis via PCR.
Mutations were introduced into the cloned murG gene by using the QuikChange Site-Directed Mutagenesis Kit (Stratagene). Mutations were confirmed by sequencing. The mutated MurG genes were expressed in BL21(DE3)pLysS following standard methods. The mutants were purified as described previously for the protein and stored at −70°C.
Determination of Kinetic Parameters.
The kinetic parameters of the enzymes were measured by using the continuous fluorescence assay described previously (23). The apparent Km for the donor was determined at a fixed concentration of Lipid I analogue (100 μM) because of the limited supply of the synthetic substrate. The Km of the acceptor analogue was determined at donor concentrations estimated to be at least 10 times its Km.
Crystallization and Data Collection.
Crystals were grown at room temperature by using the hanging-drop vapor-diffusion method. In a typical experiment, 2 μl of the protein stock (10 mg⋅ml−1 protein and 10 mM UDP-GlcNAc) were mixed with 2 μl of a reservoir buffer consisting of 0.1 M NaMES (pH 6.5), 10–14% polyethylene glycol (PEG) 4000, 0.4% Triton X-100, and 10 mM DTT. Crystals formed within 2 days with a typical size of 200 × 200 × 50 μm. The crystals were soaked in the buffer containing 16% PEG 4000, 0.1% Triton X-100, 10 mM DTT, 25% glycerol, and 10 mM UDP-GlcNAc, and flash-frozen in a liquid nitrogen stream. The cocrystals belong to the orthorhombic space group P212121, and the unit cell has dimensions of 66.703 × 83.651 × 146.245 Å. Data were collected at the Advanced Photon Source (BioCARS-14BMC) at a wavelength of 0.9 Å. Data were reduced by using denzo and scalepack (24). The structure was solved by using molecular replacement, as implemented in the program cns (25). Rotation and translation were carried out by using the structure of free enzyme as a search model. The initial Rfree after molecular replacement was 37.6%. A model was built with the program o (26) and refined further by simulated annealing and B factor refinement by using the program cns (25). Water molecules were gradually added to the structure during the refinement with cns. The final Rfree was 28.3% at 2.5 Å for the MurG:UDP-GlcNAc model including 121 water molecules. The statistics are given in Table 1. The N-terminal six residues and the C-terminal His-tag plus one additional residue had no electron density and were not included in the model.
Table 1.
Statistics from crystallographic analysis
| Data collection | |
| Resolution, Å | 20–2.5 |
| Measured intensities | 195,355 |
| Unique reflections | 26,835 |
| Rsym* (last shell†) | 0.062 (0.254) |
| Completeness, % (last shell) | 92.6 (82.6) |
| I/I(δ) | 18.6 |
| Structure refinement | |
| Reflections | 25,853 |
| Rcryst, %‡ | 22.6 |
| Rfree, %§ | 28.3 |
| Protein atoms | 5,264 |
| Ligand atoms | 78 |
| Solvent atoms | 145 |
| Average B factor, Å2 | 51 |
| rms deviations | |
| Bonds, Å | 0.0066 |
| Angles, ° | 1.33 |
| Ramachandran plot, %¶ | |
| Most favored | 91.5 |
| Additionally allowed | 7.8 |
| Generally allowed | 0.5 |
| Disallowed | 0.2 |
Rsym = Σ |Ii − 〈I〉|/ΣIi, where Ii is the intensity of a reflection and 〈I〉 is the average intensity of that reflection.
Last shell is 2.59–2.50 Å.
R-factor = Σ ∥Fobs| − |Fcalc ∥/Σ |Fobs|.
Rfree is the R-factor calculated by using 10% of the reflection data chosen randomly and omitted from the start of refinement.
Calculated with program PROCHECK.
Results and Discussion
Overall Structure.
The x-ray structure of the MurG:UDP-GlcNAc complex was solved by molecular replacement by using the free enzyme as a search model (Table 1) (8). As in the free enzyme, there are two protein molecules in the asymmetric unit. The two molecules are related by a 2-fold axis in the asymmetric unit and have very similar structures, with a rms deviation of 0.63 Å over 350 aligned residues. The final 2.5-Å structure includes residues 7–356 and UDP-GlcNAc in both proteins, as well as four glycerols and 121 water molecules. The proteins consist of two α/β domains separated by a cleft 20 Å deep and 16 Å across at its widest point, which is ≈2 Å narrower than in the free enzyme. Comparing the UDP-GlcNAc:MurG complex with the free protein structure reveals that the individual N and C domains are similar in the presence and absence of substrate. The N domains have a rms deviation of 0.59 Å for all Cα atoms, whereas C domains have a Cα atom rms deviation of 0.91 Å. However, there is a change in the relative orientation of the two domains so that in the presence of UDP-GlcNAc, MurG adopts a more closed conformation. The conformational change results mostly from a rigid body domain movement, in which the entire C-terminal domain rotates ≈10° relative to its position in the free protein. The hinge around which the rotation takes place corresponds approximately to residue N161 in the linker between domains. Torsion angles in T343 at the end of helix Cα7 also change significantly, presumably so that helix Nα6 can maintain contact with the rest of the N domain.
The UDP-GlcNAc moieties in the two proteins of the asymmetric unit exist in two different conformers around the glycosidic linkage. The A conformer is presumed to be the Michaelis complex, because it is consistent with the enzymatic reaction, which requires the incoming nucleophile to displace the UDP group with inversion of configuration. Furthermore, the hexose in the A conformer contacts invariant residue Q289, and mutation of this residue increases the Km of UDP-GlcNAc significantly (Table 2). The nucleotide in the B conformer makes the same contacts to the protein as that in the A conformer, but the glycosidic bond is in a conformation that places the hexose sugar in the cleft between the domains where the anomeric carbon would not be accessible to the incoming nucleophile. Here we focus on the A conformer only.
Table 2.
Kinetic parameters for MurG and mutants
| UDP-GlcNAc Km, mM* | Lipid I Km, mM†‡ | kcat (min−1) | |
|---|---|---|---|
| WT | 0.053 ± 0.003 | 0.053 ± 0.006 | 837 |
| E269D | 1.21 ± 0.31 | 0.081 ± 0.011 | 604 |
| E269A | 0.89 ± 0.14 | 0.047 ± 0.012 | 518 |
| S192A | 1.33 ± 0.21 | 0.179 ± 0.062 | 20.5 |
| T16A | 0.13 ± 0.026 | 0.055 ± 0.0069 | 2.16 |
| H19A | 0.084 ± 0.006 | 0.026 ± 0.0063 | 0.72 |
| Q289A | 0.127 ± 0.016 | 0.041 ± 0.006 | 7.29 |
| N128A | 0.061 ± 0.006 | 0.08 ± 0.004 | 13 |
Determined at [Lipid I] = 100 μM.
Determined at [UDP-GlcNAc] > 10 times apparent Km measured for each construct.
An analogue of Lipid I containing a tetraprenyl lipid chain (23) was used for all experiments.
The UDP-GlcNAc-Binding Site.
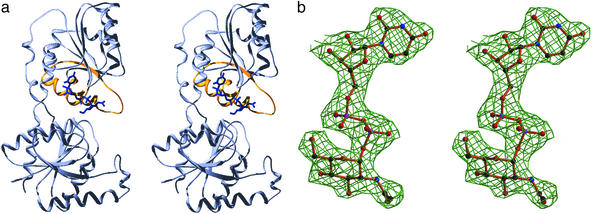
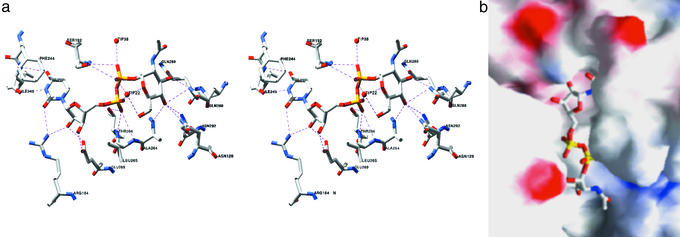
The structure of the UDP-GlcNAc:MurG complex (Fig. 1) shows that the UDP-GlcNAc moiety mostly contacts the C-terminal domain, which was previously proposed to be the donor-binding site based on the presence of a sequence motif that is found in most members of the GT-B superfamily (8, 18). This sequence motif consists of a pattern of prolines and glycines on which are grafted other characteristic residues. The structure of MurG in the absence of substrate has shown that this proline-glycine motif encodes an α/β/α subunit in which the two α helices are located near the cleft between the two domains (Fig. 1a) (8). UDP-GlcNAc makes several contacts to these helices as well as to the loops connecting them to the adjacent β strands (Fig. 2). Notable contacts include hydrogen bonds between the invariant residue E269 and the ribose 2′ and 3′ hydroxyls; between the backbone amides of L265 and T266 to one of the α phosphate oxygens; from the backbone amide of A264 to the C4 hydroxyl of GlcNAc; from the side chain amide of Q288 to both the C3 and C4 hydroxyls of GlcNAc; and from Q289 to the C3 hydroxyl of GlcNAc. The importance of some of these contacts in binding UDP-GlcNAc has been confirmed by mutational analysis (Table 2).
Figure 1.
Overall structure of the MurG:UDP-GlcNAc complex. (a) Stereo view showing the conserved α/β/α motif displayed in gold and UDP-GlcNAc in blue. The figure was produced with SWISS-PDBVIEWER (32) and rendered by POV-RAY (downloaded from www.povray.org). (b) Final 2Fo−Fc electron density map for UDP-GlcNAc at 2.5 Å, contoured at 1.0 σ. The figure was generated with BOBSCRIPT (33, 34).
Figure 2.
Substrate-binding site. (a) Stereo view of contacts between UDP-GlcNAc and MurG. Amino acids 264, 265, 266, 269, 288, 289, and 292 are located in the α/β/α motif; amino acid 192 is located in the invariant GGS loop; amino acid 128 is located in the invariant HEQN loop. (b) Electrostatic surface representation of the binding cleft (GRASP) (35). F244 stacks over the uracil ring, obscuring it. The HEQN loop partially obscures the hexose.
In addition to the numerous contacts to the conserved α/β/α subdomain, there are contacts from the β phosphate to the GGS loop between Cβ1 and Cα1. Although the overall structure of the C domain is very similar in the free and bound enzyme structures, there are notable changes in the position of the GGS loop. It moves down toward the α/β/α motif, and the movement appears to be mediated by contacts between the β phosphate and S192. Mutation of S192 confirms the importance of the GGS loop, which is conserved in all MurG homologs as well as other GT-B superfamily members. Replacing S192 with alanine affects all kinetic parameters, including the Km for Lipid I (Table 2). MurG utilizes a sequential ordered mechanism in which UDP-GlcNAc binds first (23), and it is possible that conformational changes in the GGS loop play a role in the adjustments required for Lipid I binding, as well as contributing directly to UDP-GlcNAc binding.
The uracil base is bound in a pocket flanked on one edge by Cβ3 and the strand connecting the N- and C-terminal domains and on the other edge by the GGS loop. Hydrogen bonds from the backbone amide of I245 to N3H and O4 anchor the base; contacts from R164 to uracil O2 may also contribute to binding. In addition, the aromatic ring of F244 has rotated from its position in the free enzyme to stack over the uracil ring, capping the binding pocket (Fig. 2). Many other GTases in the GT-B superfamily contain an aromatic residue in approximately the same position as F244, suggesting that this capping interaction is conserved.
Donor Sugar Specificity.
Members of the GT-B superfamily use UDP or TDP sugars as donors, and it is generally believed that the enzymes do not discriminate between the two nucleotides (27). Although this may be true for some GT-B family members, we have found that TDP-GlcNAc is not a donor for MurG (Table 3). Thus, MurG strongly prefers UDP to CDP, GDP, ADP (28), and even TDP. The structure of the complex reveals the basis for the UDP leaving group selectivity. The uracil-binding pocket is too small to accommodate a purine, whereas the hydrogen bonds to I245 that stabilize uracil would destabilize cytosine. UDP appears to be selected over TDP for a combination of electronic and steric reasons. For example, the crystal structure shows that the side chain of E269 contacts both the 2′ and 3′ ribose hydroxyls of UDP. The distance between the carboxylate oxygen of E269 (E269 OE2) and the 2′-hydroxyl is quite close at 2.7 Å, suggesting a functionally important interaction. Consistent with this, when the 2′-deoxy ribosyl analogue of UDP-GlcNAc was used as a donor, the Km was 30 times higher (Table 3) than for UDP-GlcNAc itself, although the kcat changed only modestly. Furthermore, mutation of E269 to either alanine or aspartate causes the Km of UDP-GlcNAc to increase by more than an order of magnitude, again with minimal change in kcat (Table 2). These results suggest that the contact between the C2′ ribose hydroxyl and E269 plays a key role in the ability of MurG to discriminate between UDP and TDP. The complex also reveals that a C5 methyl substituent would clash with the GGS loop, explaining why MurG can process 2′-deoxy UDP-GlcNAc, albeit poorly, but not TDP-GlcNAc.
Table 3.
Kinetic parameters of MurG with UDP-GlcNAc and donor analogues
| Donor analogues | Donor analogues Km, mM* | Lipid I Km, mM† | kcat (min−1) |
|---|---|---|---|
| UDP-GlcNAc | 0.053 ± 0.003 | 0.053 ± 0.006 | 837 |
| 2′-Deoxy | 1.5 ± 0.17 | ND‡ | 674 |
| UDP-GlcNAc | |||
| TDP-GlcNAc | No activity§ |
Determined at [Lipid I] = 100 μM.
An analogue of Lipid I containing a tetraprenyl lipid chain (23) was used for all experiments.
Not determined.
No detectable activity at [TDP-GlcNAc] = 1 mM.
In addition to its high selectivity for UDP, MurG also shows good selectivity for GlcNAc. We have previously reported that MurG does not use UDP-GalNAc as a donor, indicating that the equatorial hydroxyl at C4 of GlcNAc is critical (28). The crystal structure of the complex shows that at least three side chain amides and one backbone amide are close to this C4 hydroxyl: Q288, N292, N128, and A264 (Fig. 2). The three side chain amides are on residues conserved in all MurG homologues, whereas the backbone amide is located at the N terminus of the first helix in the conserved α/β/α motif. The C4 hydroxyl of UDP-GalNAc cannot form hydrogen bonds to A264 or N292. Although it would be able to contact N128, the direction of the H bond would be different. Because N128 is a critical residue for catalysis (Table 2), it is likely very sensitive to hydrogen bond geometry in the active site.
Comparison to Other GT-B Members.
Peptidoglycan biosynthesis is remarkably conserved in bacteria, and there are MurG homologues in all bacteria that make peptidoglycan. Although the sequence homology between E. coli MurG and other homologues varies from <30 to >90%, it is presumed that the structures of the enzymes are similar because they catalyze the same reaction using similar or identical substrates. Therefore, by comparing the sequences of MurG homologues from a wide range of microorganisms that have diverged over millions of years, it is possible to identify the residues that are most critical for binding and catalysis. These residues turn out to be confined to two regions in the C domain, the α/β/α motif and the GGS loop, as well as three regions in the N domain, one of which, the HEQN loop, is directly across from the hexose-binding site (Figs. 1–3). This loop is proposed to form a key part of the acceptor-binding site (8).
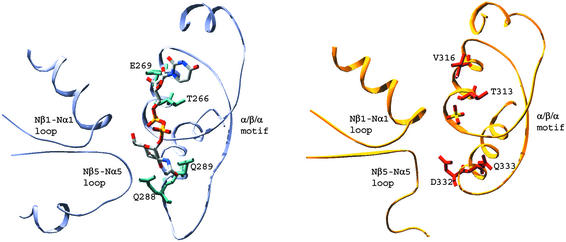
Figure 3.
Comparison of MurG (light gray) with bound UDP-GlcNAc and GtfB (gold) with bound sulfate (PDB ID code 1IIR). Selected invariant regions in MurG are shown (Gloop 1 = Nβ1-Nα1 loop, HEQN loop = Nβ5-Nα5 loop, and α/β/α motif) along with the corresponding regions of GtfB. The loop between Cβ5 and Cα5 in MurG (part of the α/β/α motif) is characteristically longer than in other GT-B family members including GtfB. The Nβ5-Nα5 loop of GtfB, the proposed acceptor-binding site, contains a long proline-rich polypeptide and the electron density is not continuous to Nα5, so only a portion of this region is shown. The figures were generated by using SWISS-PDBVIEWER (32) and rendered by POV-RAY (www.povray.org).
Structures of two other GT-B NDP-GTases have been reported (2–4, 11), although only one of the two is typical of the superfamily in that it contains the characteristic pro-gly sequence motif. This structure is of chloroeremomycin GTase (GtfB), a GTase that attaches glucose to the aglycone of chloroeremomycin (11, 29). There are several homologues of GtfB that attach sugars to other glycopeptide aglycones, or to different positions on chloroeremomycin. These enzymes have remarkably similar sequences, and the differences among them are presumed to be related to the differences in substrate selectivity. Because the basic architecture of GT-B family GTases is highly conserved, comparisons between MurG homologues, which reveal the residues that play key roles in binding and catalysis, and between the GtfB homologues, which reveal the regions that can vary depending on the structure of the substrates, shed considerable light on the superfamily.
As shown in Fig. 3, the α−β−α subdomain of GtfB superimposes extremely well on that of MurG. The polar residues that anchor the C3 and C4 hydroxyls of the hexose in MurG (Q288 and Q289) have conserved counterparts at virtually identical positions in GtfB (D332 and Q333), suggesting that the hexoses are bound in a similar manner. Furthermore, where the α phosphate is bound in MurG, GtfB contains a sulfate ion, suggesting a similar mode of α phosphate binding. We have, therefore, concluded that the α/β/α motif, which can be identified for most GT-B family members based on sequence analysis (Fig. 4) (16, 30), plays a key role stabilizing the α phosphate in the absence of metal ions and in anchoring both the ribose and the hexose sugars.
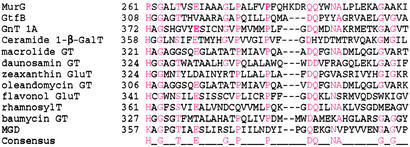
Figure 4.
Sequence alignments of selected GT-B superfamily members showing the conserved proline- and glycine-rich motif that corresponds to the α/β/α fold. Conserved residues are in red. The GTases shown represent a range of prokaryotic and eukaryotic GTases that play roles in both primary and secondary metabolic processes. The sequences referred to are: MurG (UDP-N-acetyl-glucosaminyltransferase, E. coli, P17443); GtfB (chloroeremomycin gtfB, CAA11775.1); GnT 1A (glucuronosyltransferase 1A, Homo sapiens, P22309); ceramide 1-β-GalT (ceramide 1-β-galactosyltransferase, H. sapiens, O00196); macrolide GT (macrolide GTase, Streptomyces lividans, Q54387); daunosamin GT (daunosamine transferase, Stretomyces peucetius, Q54824); zeaxanthin GluT (zeaxanthin glucosyltransferase, Erwinia ananus, P21686); oleandomycin GT (oleandomycin GTase, Streptomyces antibioticus, Q53685); flavonol GluT (flavonol O3-glucosyltransferase, Perilla frutescens, O04114); rhamnosylT (UDP rhamnose/anthocyanidin-3-glucoside rhamnosyltransferase, Petunia hybrida, Q43716); baumycin GT (baumycin GTase, Streptomyces sp. C5, Q53881); and MGD (monogalactosyldiacylglycerol synthase, Arabidopsis thaliana, O82730). In E. coli MurG, the consensus E residue plays a role in binding to the hydroxyls on the ribose sugar of the UDP group. The conserved threonine located at the N terminus of the first α helix helps anchor the α phosphate (along with backbone amides from adjacent residues), and the polar residues (consensus DQ) located in the loop immediately preceding the second α helix anchor the hexose sugar.
Within the α/β/α region, there are some differences between MurG and GtfB that may be related to substrate discrimination. For example, in GtfB there is a valine (316) in place of E269, which plays a key role in MurG's ability to discriminate UDP over TDP. Unlike MurG, GtfB does not discriminate between UDP and TDP, and it has a much higher donor Km than MurG. The catalytic efficiency of GtfB with respect to its natural glycosyl donor is thus much lower than that of MurG (≈25 min−1⋅mM−1 versus 15,800 min−1⋅mM−1). Unlike MurG, which is a key enzyme involved in a primary metabolic pathway, GtfB is not subject to selective pressures that maintain high efficiency. It is tempting to speculate that replacing V316 with a suitable acidic residue will improve the catalytic efficiency of UDP donors relative to TDP donors, which might in turn increase the utility of GtfB as a tool for enzymatic synthesis (29, 31). If so, this modification could be extended to other Gtf family members, which also have low catalytic efficiency.
In addition to the similarities in the donor-binding C domains, MurG and GtfB bear unexpected and remarkable similarities in the N domains. For example, despite the differences in the glycosyl acceptors and minimal sequence homology (≈20%), there is a 1.4-Å rms deviation over 82 aligned Cα atoms. Furthermore, the invariant HEQN loop of MurG, the proposed acceptor-binding site, is located at the same position as the variable loop of GtfB. A comparison of GtfB and closely related GTases shows that this loop varies in length and composition depending on the structure of the acceptor (11). The comparison of MurG and GtfB combined with sequence information thus implies that nature has been able to use the same two-domain α/β scaffold to accomplish glycosyltransfer from TDP and/or UDP sugar donors to a wide range of different nucleophiles. By varying the length and composition of the loop between Nβ5 and Nα5 while maintaining the overall architecture of the scaffold, it is evidently possible to alter the acceptor selectivity significantly. We suggest that it will be possible to evolve new GTases by varying this loop as well as the loop between Cβ5 and Cα5 while maintaining the overall architecture of the superfamily and preserving certain key residues.
Because MurG is an enzyme that is conserved in all bacteria that make peptidoglycan, it is a potential antibiotic target. The structure reported here may be a useful starting point for the design of inhibitors. Beyond that, the structure is of significant interest because it sheds light on a major superfamily of GTases and provides insight into the origins of substrate selectivity as well as how catalytic efficiency might be improved for some family members. The ability to block GTases and/or to engineer them to use other substrates will be useful both for probing biological roles of GTases and their products, and for exploiting GTases in enzymatic synthesis of glycoconjugates with new biological activities. Additional structural information on other members of this superfamily would be invaluable for accomplishing these goals.
Acknowledgments
We thank K. Brister and the staff at BioCARS 14-C (Advanced Photon Source, Argonne, IL) for help with data collection. We are indebted to Y. Shi (Princeton) and his laboratory for technical assistance with all aspects of x-ray crystallography and for helpful comments. Support has been provided by a grant from the National Institutes of Health (R01A144854 to S.W.).
Abbreviations
- GTase
glycosyltransferase
- GtfB
chloroeremomycin GTase
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, www.rcsb.org (PDB ID code 1NLM).
References
- 1.Campbell J A, Davies G J, Bulone V, Henrissat B. Biochem J. 1997;326:929–939. doi: 10.1042/bj3260929u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vrielink A, Ruger W, Driessen H P, Freemont P S. EMBO J. 1994;13:3413–3422. doi: 10.1002/j.1460-2075.1994.tb06646.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Morera S, Imberty A, Aschke-Sonnenborn U, Ruger W, Freemont P S. J Mol Biol. 1999;292:717–730. doi: 10.1006/jmbi.1999.3094. [DOI] [PubMed] [Google Scholar]
- 4.Morera S, Lariviere L, Kurzeck J, Aschke-Sonnenborn U, Freemont P S, Janin J, Ruger W. J Mol Biol. 2001;311:569–577. doi: 10.1006/jmbi.2001.4905. [DOI] [PubMed] [Google Scholar]
- 5.Charnock S J, Davies G J. Biochemistry. 1999;38:6380–6385. doi: 10.1021/bi990270y. [DOI] [PubMed] [Google Scholar]
- 6.Gastinel L N, Cambillau C, Bourne Y. EMBO J. 1999;18:3546–3557. doi: 10.1093/emboj/18.13.3546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ünligil U M, Zhou S, Yuwaraj S, Sarkar M, Schachter H, Rini J M. EMBO J. 2000;19:5269–5280. doi: 10.1093/emboj/19.20.5269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ha S, Walker D, Shi Y, Walker S. Protein Sci. 2000;9:1045–1052. doi: 10.1110/ps.9.6.1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gastinel L N, Bignon C, Misra A K, Hindsgaul O, Shaper J H, Joziasse D H. EMBO J. 2001;20:638–649. doi: 10.1093/emboj/20.4.638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rao M, Tvaroska I. Proteins. 2001;44:428–434. doi: 10.1002/prot.1108. [DOI] [PubMed] [Google Scholar]
- 11.Mulichak A M, Losey H C, Walsh C T, Garavito R M. Structure (Cambridge, UK) 2001;9:547–557. doi: 10.1016/s0969-2126(01)00616-5. [DOI] [PubMed] [Google Scholar]
- 12.Persson K, Ly H D, Dieckelmann M, Wakarchuk W W, Withers S G, Strynadka N C. Nat Struct Biol. 2001;8:166–175. doi: 10.1038/84168. [DOI] [PubMed] [Google Scholar]
- 13.Ramakrishnan B, Balaji P V, Qasba P K. J Mol Biol. 2002;318:491–502. doi: 10.1016/S0022-2836(02)00020-7. [DOI] [PubMed] [Google Scholar]
- 14.Pedersen L C, Darden T A, Negishi M. J Biol Chem. 2002;277:21869–21873. doi: 10.1074/jbc.M112343200. [DOI] [PubMed] [Google Scholar]
- 15.Gibbons B J, Roach P J, Hurley T D. J Mol Biol. 2002;319:463–477. doi: 10.1016/S0022-2836(02)00305-4. [DOI] [PubMed] [Google Scholar]
- 16.Bourne Y, Henrissat B. Curr Opin Struct Biol. 2001;11:593–600. doi: 10.1016/s0959-440x(00)00253-0. [DOI] [PubMed] [Google Scholar]
- 17.Ünligil U M, Rini J M. Curr Opin Struct Biol. 2000;10:510–517. doi: 10.1016/s0959-440x(00)00124-x. [DOI] [PubMed] [Google Scholar]
- 18.Ha S, Gross B, Walker S. Curr Drug Targets Infect Disord. 2001;1:201–213. doi: 10.2174/1568005014606116. [DOI] [PubMed] [Google Scholar]
- 19.Kapitonov D, Yu R K. Glycobiology. 1999;9:961–978. doi: 10.1093/glycob/9.10.961. [DOI] [PubMed] [Google Scholar]
- 20.King C D, Rios G R, Green M D, Tephly T R. Curr Drug Metab. 2000;1:143–161. doi: 10.2174/1389200003339171. [DOI] [PubMed] [Google Scholar]
- 21.Wrabl J O, Groshin N V. J Mol Biol. 2001;314:365–374. doi: 10.1006/jmbi.2001.5151. [DOI] [PubMed] [Google Scholar]
- 22.Vosseller K, Wells L, Hart G W. Biochimie. 2001;83:575–581. doi: 10.1016/s0300-9084(01)01295-0. [DOI] [PubMed] [Google Scholar]
- 23.Chen L, Men H, Ye X-Y, Brunner L, Hu Y, Walker S. Biochemistry. 2002;41:6824–6833. doi: 10.1021/bi0256678. [DOI] [PubMed] [Google Scholar]
- 24.Ötwinowski Z, Minor W. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 25.Brunger A T, Adams P D, Glore G M, DeLano W L, Gros P, Grosse-Kunstleve R W, Jiang J S, Kuszewski J, Nilges M, Pannu N S, et al. Acta Crystallogr D. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 26.Jones T A, Zou J Y, Cowan S W, Kjeldgaard M. Acta Crystallogr A. 1991;47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 27.Öhrlein R. Top Curr Chem. 1999;200:227–254. [Google Scholar]
- 28.Ha S, Chang E, Lo M-C, Men H, Park P, Ge M, Walker S. J Am Chem Soc. 1999;121:8415–8426. [Google Scholar]
- 29.Losey H C, Peczuh M W, Chen Z, Eggert U S, Dong S D, Pelczer I, Kahne D, Walsh C T. Biochemistry. 2001;40:4745–4755. doi: 10.1021/bi010050w. [DOI] [PubMed] [Google Scholar]
- 30.Abdian P L, Lellouch A C, Gautier C, Ielpi L, Garemia R A. J Biol Chem. 2000;275:40568–40575. doi: 10.1074/jbc.M007496200. [DOI] [PubMed] [Google Scholar]
- 31.Dong S, Öbethur M, Losey H C, Anderson J W, Eggert U S, Peczuh M W, Walsh C T, Kahne D. J Am Chem Soc. 2002;124:9064–9065. doi: 10.1021/ja026342h. [DOI] [PubMed] [Google Scholar]
- 32.Guex N, Peitsch M C. Electrophoresis. 1997;18:2714–2723. doi: 10.1002/elps.1150181505. [DOI] [PubMed] [Google Scholar]
- 33.Kraulis P J. J Appl Crystallogr. 1991;24:946–950. [Google Scholar]
- 34.Esnouf R M. J Mol Graphics. 1997;15:132–134. doi: 10.1016/S1093-3263(97)00021-1. [DOI] [PubMed] [Google Scholar]
- 35.Nicholls A, Sharp K A, Honig B. Proteins. 1991;11:281–296. doi: 10.1002/prot.340110407. [DOI] [PubMed] [Google Scholar]