Abstract
The crystal structure of the Michaelis complex between the Fab fragment of ferrochelatase antibody 7G12 and its substrate mesoporphyrin has been solved to 2.6-Å resolution. The antibody-bound mesoporphyrin clearly adopts a nonplanar conformation and reveals that the antibody catalyzes the porphyrin metallation reaction by straining/distorting the bound substrate toward the transition-state configuration. The crystal structures of the Fab fragment of the germ-line precursor antibody to 7G12 and its complex with the hapten N-methylmesoporphyrin have also been solved. A comparison of these structures with the corresponding structures of the affinity-matured antibody 7G12 reveals the molecular mechanism by which the immune system evolves binding energy to catalyze this reaction.
Modern theories of biological catalysis date from Haldane's theory of strain: “using Fischer's lock and key simile, the key does not fit the lock perfectly but exercises a certain strain on it” (1). Although the notion that enzymes use binding energy to strain or distort substrates is a fundamental theory of enzyme catalysis (2), it has proven difficult to experimentally validate (3). One approach that has proven effective in testing theories of biological catalysis involves the use of the programmable nature of antibody binding energy to evolve selective catalysts. Antibody catalysis has allowed us to dissect the energetic contributions of transition state stabilization, general base and covalent catalysis, and proximity effects to catalysis (4–9, 29). More recently, we showed that an antibody (7G12) raised against a strained ground-state mimic acted as an efficient porphyrin metallation catalyst (10). We now report the x-ray crystal structure of the Michaelis complex formed between the Fab fragment of the ferrochelatase antibody 7G12 and its substrate mesoporphyrin IX (MP), in which the bound substrate is distorted toward the transition-state conformation for metal insertion. The structure of this complex provides unequivocal structural evidence for the strain theory proposed by Haldane more than 70 years ago. Moreover, the detailed structural and biophysical characterization of the germ-line and affinity-matured antibodies has provided a detailed mechanistic picture of the immunological evolution of this strain mechanism.
The enzyme ferrochelatase catalyzes the insertion of Fe2+ into protoporphyrin IX as the last step in heme biosynthesis pathway (11). It was proposed that the enzyme catalyzes the porphyrin metallation reaction by distorting the porphyrin substrate toward a transition state-like geometry in which the pyrrole nitrogen lone pairs are exposed for metal chelation (12). To test this notion, antibody 7G12 was generated against N-methylmesoporphyrin (NMP) in which the porphyrin macrocycle is distorted due to alkylation at one of the pyrrole nitrogens. The resulting antibody was found to catalyze the metallation of MP by Zn2+ with a catalytic efficiency comparable to that of the natural enzyme (10). The same antibody also catalyzes the insertion of Cu2+ into MP at a lower rate. Determination of the three-dimensional structure of the antibody–substrate MP complex provides the opportunity to characterize the molecular basis for substrate strain in this antibody-catalyzed reaction.
Methods
Crystallization and Structure Determination.
The Fab fragments of the affinity-matured antibody 7G12 and its germ-line precursor had been cloned (13) and were expressed from Escherichia coli 25F2 cells according to the published protocol (14). After isolation from the cell lysates by protein G affinity chromatography, the Fab fragments of 7G12 and some of its mutants showed absorbance at 404 nm, suggesting some of the Fab fragments were associated with heme or heme analogues. The heme-bound and heme-free Fab were separated with cation exchange chromatography on a Mono S 10/10 column (Amersham Pharmacia) by using a gradient from 5% to 15% buffer B (500 mM NaCl/20 mM Mops, pH 7.0). The Fab fragments were further purified by gel-filtration chromatography on a Superdex S-200 column (Amersham Pharmacia) equilibrated with 100 mM NaCl, 10 mM Tris (pH 7.5), and 1 mM methionine. Before crystallization experiments were set up for 7G12 Fab–MP complex, solution of Fab and the crystallization reagents were passed through a column packed with Chelex 100 metal chelating resin (Bio-Rad) to remove traces of bivalent metal ions that might be present in the solution. NMP and MP were purchased from Porphyrin Products (Logan, UT). All crystals were obtained by hanging drop methods. The complex of 7G12 Fab and substrate MP was crystallized from 27% poly(ethylene glycol) (PEG) 2000 monomethylether, 250 mM ammonium sulfate, 0.5 mM MP, and 10 mM Hepes, pH 6.6, at 25°C. The germ-line Fab was crystallized from 22% PEG 2000 monomethylether, 200 mM lithium chloride, and 100 mM sodium citrate, pH 4.0, at 4°C. The complex between the germ-line Fab and hapten NMP was crystallized from 24% PEG 2000 monomethylether, 200 mM ammonium sulfate, 10 mM cadmium sulfate, 1 mM NMP, and 100 mM sodium acetate, pH 4.2, at 4°C. The 7G12 Fab was crystallized from 27% PEG 2000 monomethylether, 200 mM ammonium sulfate, and 100 mM Tris, pH 8.6, at 4°C. Data sets for the 7G12 Fab–MP complex were collected at Stanford Sychrotron Radiation Laboratory, beam-line 9-2, at 100 K. Data sets for the remaining structures were collected in house with an FRD generator and a RAXISIV2+ detector. Data were processed with DENZO and SCALEPACK (15). The structure was determined by molecular replacement techniques using the structure of 7G12 Fab–NMP complex from Protein Data Bank entry 3FCT (13) with the program EPMR (16). Mutation of the molecular replacement model and rebuilding was done by using the program O (17). Refinement was carried out by using positional, simulated annealing and torsional refinement in cns (18), with noncrystallographic symmetry restraints turned on when applicable and bulk solvent corrections applied between 20.0 and 6.0 Å.
Hapten Binding and Porphyrin Metallation Kinetics.
The binding properties (Kd and koff) of 7G12 Fab, germ-line Fab, and all of the mutants with NMP were measured by surface plasmon resonance on a Biacore 3000 biosensor following the published methods (19). BSA was conjugated with NMP and immobilized on a research grade CM5 sensor chip (Biacore). The dissociation constant Kd was measured under equilibrium conditions, and the dissociation rate constant was measured under the kinetic conditions. The association constant (kon) was calculated with the equation kon = koff/Kd. Kinetics of copper ion insertion into MP catalyzed by the Fab fragments of antibody 7G12, its germ-line precursor, and the mutants were measured as reported (13).
Results and Discussion
Crystal Structure of 7G12 Fab–MP Michaelis Complex.
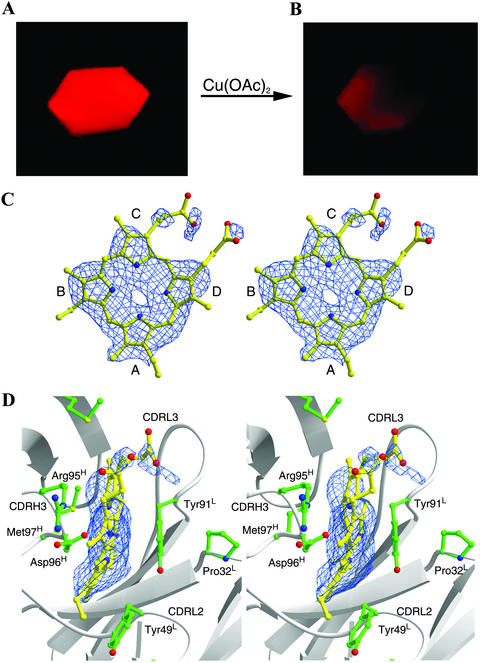
Thin hexagonal plate-like crystals were obtained for the complex of the Fab fragment of antibody 7G12 and its porphyrin substrate, MP, by cocrystallization in the absence of the metal ions. The crystals of the 7G12 Fab–MP complex give intense red fluorescence upon irradiation with green excitation light ≈546 nm on a fluorescence microscope (Fig. 1A). The addition of copper acetate to the crystallization drop to a final concentration of 1 mM leads to a rapid loss of fluorescence due to the formation of the Cu(II)MP complex (Fig. 1B), demonstrating that the antibody–MP cocrystal is catalytically active.
Figure 1.
Fluorescent micrographs of a single crystal of 7G12 Fab–MP complex before (A) and after (B) the addition of copper acetate to a final concentration of 1 mM. Photographs were taken on a Zeiss Axioskop fluorescence microscope equipped with a 546-nm exciter filter. The exposure time was 0.25 s for both pictures. (Magnifications, ×40.) (C and D) Fo − Fc electron density maps contoured at 2.0 σ for the substrate MP molecule bound with 7G12 Fab. (D) Residues making important packing interactions with MP in the antibody-combining site. C and D were drawn with bobscript and RASTER3D (26, 27).
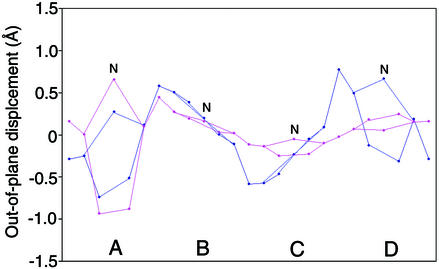
The crystal structure of 7G12 Fab–MP complex was solved to 2.6-Å resolution. The Fo − Fc electron density map clearly shows the substrate MP molecule adopts a nonplanar conformation (Fig. 1 C and D); the out-of-plane displacements for the substrate MP and the hapten NMP are shown in Fig. 2. All pyrroles in MP show significant displacements away from the porphyrin least-squares (PLS) plane. Pyrrole A in MP adopts a similar angle (26°) relative to the PLS plane as was observed in NMP (13); however, the other pyrrole rings form larger angles to the PLS plane than their NMP counterparts (16°, 17°, and 25° for rings B, C, and D, respectively). The porphyrin conformation observed in the crystal structure of Fab–MP complex agrees with prior resonance Raman spectroscopy data, which indicate an up-and-down tilting of the pyrrole rings, based on the observation of specific out-of-plane vibration mode (20). A normal mode decomposition analysis (21), which deconstructs porphyrin deformations into low-frequency normal coordinate displacements, shows a moderate doming (A2u) deformation, as well as strong saddling (B2u) and ruffling (B1u) deformations for the antibody-bound MP.
Figure 2.
Out-of-plane displacement of the porphyrin ring atoms from the porphyrin least-squares plane for MP (blue) and NMP (pink) bound to antibody 7G12. The porphyrin atoms that are involved in the same pyrrole ring are connected to give a pentagon shape. A–D denote the porphyrin pyrroles as in Fig. 1C, and N denotes the pyrrole nitrogen atoms of the porphyrin molecule.
An analysis of the crystal structure of the 7G12 Fab–substrate MP complex reveals those interactions between the residues in the antibody-combining site and MP that lead to substrate distortion. The porphyrin molecule is bound in a cleft, with complementarity-determining region (CDR) L2 and CDR L3 forming one side of the cleft and CDR H3 forming the other side (Fig. 1D). Part of the CDR H3 loop, composed of Arg-95H, Asp-96H, and Met-97H, packs on the macrocyclic ring of the porphyrin. The carboxylic oxygen of Asp-96H points toward the center of the porphyrin ring and is within hydrogen-bonding distance of all pyrrole nitrogen atoms. The guanidino group of Arg-95H forms salt bridges with both carboxylates of the propionic acid side chains of the porphyrin ring. The aromatic side chains of Tyr-49L and Tyr-91Lπ stack on pyrrole rings A and B of the substrate. Whereas the π stacking interaction on one face of pyrrole ring B is balanced by the packing of Asp-96H on the other face, the π stacking interaction with Tyr-49L pushes pyrrole ring A out of plane because of the absence of heavy chain residue contacts on the opposite face. In the crystal structure of Fab–hapten NMP complex, Tyr-49L also packs against the N-methyl pyrrole, which is distorted out of plane of the other pyrrole rings because of the N-methyl substitution (13). Thus, it seems that the distortion of the MP substrate induced by the antibody-combining site is a direct result of the distortion in NMP produced by N-methyl substitution on the free MP.
Comparison of the Antibody-Combining Site in the Germ-Line and Affinity-Matured Antibodies.
To examine the mechanism by which substrate strain was evolved, the germ-line precursor antibody to 7G12 was cloned (13). There are two somatic mutations in the 7G12 VL chain (Ser-14LThr and Ala-32LPro) and three in VH (Arg-50HMet, Ser-76HAsn, and Ser-97HMet). The crystal structures of the unliganded Fab fragment of the germ-line antibody of 7G12, the germ-line Fab–hapten NMP complex, and the unliganded Fab fragment of the affinity-matured antibody 7G12 have been solved to 1.6-, 1.8-, and 2.2-Å resolution, respectively. The final statistics for the structures are shown in Table 1. These structures, in combination with the previously solved crystal structures of the affinity-matured Fab–NMP complex (13) and Fab–MP Michaelis complex, allow a detailed structural analysis of both the germ-line and affinity-matured antibodies with respect to the evolution of binding energy and catalysis.
Table 1.
Data collection and refinement statistics
| Germ-line Fab
|
Affinity-matured Fab
|
|||
|---|---|---|---|---|
| apo | With hapten NMP | apo | With substrate MP | |
| Space group | P1 | P21 | C2221 | P21212 |
| Unit cell dimensions | ||||
| a, Å | 38.9 | 55.5 | 60.8 | 100.7 |
| b, Å | 44.4 | 75.8 | 95.0 | 134.2 |
| c, Å | 57.9 | 60.5 | 155.4 | 73.0 |
| α, ° | 84.2 | 90.0 | 90.0 | 90.0 |
| β, ° | 77.0 | 93.2 | 90.0 | 90.0 |
| γ, ° | 89.4 | 90.0 | 90.0 | 90.0 |
| Number of molecules per asymmetric unit | 1 | 1 | 1 | 2 |
| Refinement resolution, Å | 20.0–1.6 | 20.0–1.8 | 20.0–2.2 | 20.0–2.6 |
| Observations, n | 97,524 | 125,917 | 200,436 | 140,870 |
| Unique reflections, n | 44,964 | 17,466 | 22,861 | 28,954 |
| Completeness to the refined resolution, % | 91.2 (81.9) | 95.5 (67.9) | 98.3 (97.2) | 98.7 (97.7) |
| I/σ | 24.5 (6.7) | 22.5 (4.9) | 29.2 (10.4) | 23.4 (4.8) |
| Rsym (I)*, % | 3.4 (11.2) | 5.6 (22.6) | 6.4 (20.9) | 10.8 (67.2) |
| Rcryst†, % | 21.7 | 20.3 | 23.6 | 26.4 |
| Rfree‡, % | 24.1 | 26.0 | 27.3 | 30.0 |
Values in parentheses correspond to the highest resolution shell.
Rsym = 100 × ∑h∑lIh,i − Ih/∑h∑iIh,i for the intensity I of i observations of reflection h. Ih is the mean intensity of the reflection.
Rcryst = 100 × ∑|Fobs − Fcalc|/∑|Fobs|, where Fobs and Fcalc are the observed and calculated structure factors, respectively.
Rfree is the same as R factor but is calculated from the 10% of the reflection data excluded from refinement.
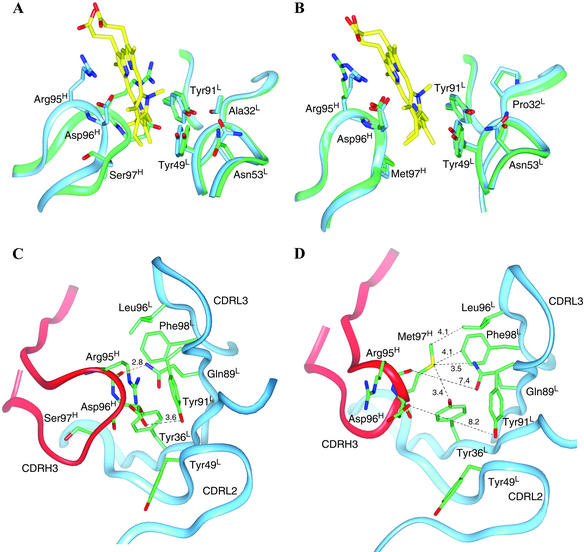
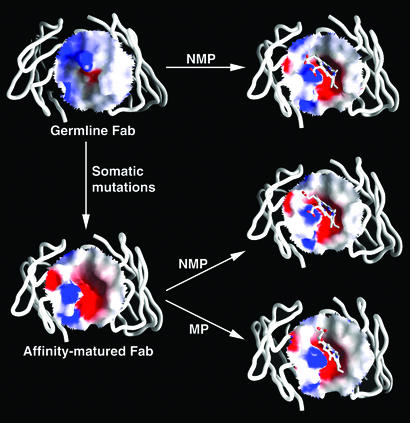
Superposition of the crystal structures of the unliganded germ-line Fab of antibody 7G12 and NMP-bound germ-line Fab shows that there are significant conformational changes in the loop of CDR H3 upon hapten binding (Fig. 3A). In the unliganded germ-line Fab structure, CDR H3 adopts a relaxed and extended conformation. The side chains of Arg-95H and Asp-96H, which are at the tip of the CDR H3 loop, are located in the space occupied by the hapten in the germ-line Fab–NMP complex. There is a hydrogen bond between the backbone carbonyl oxygen of Arg-95H and carboxamide NH of Gln-89L (Fig. 3C). Upon NMP binding, the backbone of CDR H3 loop is pushed backward toward the side of the heavy chain by ≈4 Å (at Cα of Arg-95H) because of the insertion of the NMP molecule into the hapten binding pocket. There are even larger movements for the side chains of Arg-95H (7.0 Å at the guanidino C) and Asp-96H (5.5 Å at the carboxamide C) upon NMP binding (Fig. 3A). In the complex, Arg-95H now packs onto the macrocyclic ring of NMP, and the carboxylate group of Asp-96H is now positioned equidistant from the four pyrrole nitrogen atoms of NMP (2.6–3.0 Å) and may form hydrogen bonds with one or more of the pyrrole NH groups. The structural flexibility of germ-line antibody is also manifested by comparing the electrostatic surface of the antibody-combining site before and after binding of NMP (Fig. 4). In the hapten free form, the germ-line antibody-combining site is flat with minor electrostatic charges distributed on the surface. However, upon hapten binding, the movement of the CDR H3 loop creates a cavity on the surface of the germ-line antibody that complements the shape of the distorted porphyrin molecule. Exposure of residues Arg-95H and Asp-96H also introduces negative charges inside the cavity and positive charges at the rim, which complement the charge distribution of NMP.
Figure 3.
(A) Overlay of the unliganded germ-line Fab (green) and the germ-line Fab–NMP complex (blue). The CDR H3 loop of the germ-line Fab occupies the antibody-combining site in the absence of the hapten NMP and undergoes a conformational change upon the binding of NMP (yellow). (B) Overlay of the unliganded affinity-matured 7G12 Fab (green) and the affinity-matured 7G12 Fab–NMP complex (blue). No significant conformational change occurs upon the binding of NMP (yellow). (C) In the germ-line antibody without NMP bound, CDR H3 extends into the antibody-combining site, and a hydrogen bond (red dotted lines) is formed between the backbone CO of Arg-95H and carboxamide NH of Gln-89L. (D) In the affinity-matured antibody without NMP bound, CDR H3 is kinked because of Ser-97HMet somatic mutation, and the antibody-combining site is preorganized for NMP binding. Dotted lines with numbers show the distances (Å) between the two atoms.
Figure 4.
The difference in electrostatic surface potential of the antibody-combining site in the germ-line and affinity-matured Fab and the changes upon the binding of NMP and MP. The red and blue colors correspond to negative and positive surface potential, respectively. The figure was prepared with grasp (28).
In contrast to the germ-line antibody, comparison of the unliganded and hapten-bound affinity-matured 7G12 structures indicates that minimal changes occur upon hapten binding (Fig. 3B). The shape and the electrostatic characteristics of the antibody-combining site of the affinity-matured Fab do not change significantly before and after the binding of the hapten or the substrate molecule, suggesting the antibody-combining site in the affinity-matured Fab is rigid and preorganized for the binding of distorted porphyrins (Fig. 4). This “lock-and-key” binding mode (22) of the affinity-matured antibody versus “induced-fit” binding mechanism (23) in the germ-line antibody was also observed in the antibodies 48G7, AZ28, and 28B4 (19, 24, 25) and further supports the hypothesis that the structural plasticity of germ-line antibodies is a critical factor in the ability of the immune system to evolve antibodies that bind many distinct chemical structures.
Evolution of Binding and Catalytic Function.
The hapten binding and catalytic properties for the germ-line Fab, affinity-matured Fab, and a number of somatic mutants were measured to determine how the structural changes associated with affinity maturation lead to increased affinity for the distorted NMP and, as a consequence, enhanced catalytic activity (Table 2). The binding affinity of both the affinity-matured and germ-line Fabs to the planar mesoporphyrin substrate are >100 μM.‖ In contrast, the affinity-matured Fab binds NMP with a dissociation constant (Kd) of 20.7 nM, 95-fold lower than the Kd of the germ-line Fab (1.96 μM). The value of kcat/Km for the antibody-catalyzed porphyrin metallation reaction reflects this difference, increasing 92-fold from the germ line to the affinity-matured Fab (the low rate of the germ-line Fab precluded individual measurements of kcat and Km). Thus the catalytic activity for porphyrin metallation seems to have evolved as a consequence of binding affinity for a distorted porphyrin (NMP). This is consistent with the notion that the differential binding energy for the nonplanar versus planar porphyrin is translated into a lower free energy of activation for reaction by binding of the substrate in a higher energy distorted conformation.
Table 2.
NMP binding and catalytic properties of 7G12 Fab, germ-line Fab, and the mutants
| Fab | kon, 103M−1⋅s−1 | koff, 10−3 s−1 | Kd, 10−9 M | kcat h−1 | Km, μM | kcat/Km, μM−1⋅h−1 |
|---|---|---|---|---|---|---|
| 7G12 | 66.7 | 1.38 | 20.7 | 24.8 | 150 | 0.165 |
| Germ-line | 4.4 | 8.62 | 1960 | ND | ND | 0.0018 |
| Germ-line mutants | ||||||
| Ala-32LPro | 6.72 | 5.43 | 808 | ND | ND | 0.0012 |
| Arg-50HMet | 6.34 | 3.77 | 595 | ND | ND | 0.0092 |
| Ser-97HMet | 10.5 | 4.18 | 398 | ND | ND | 0.014 |
| 7G12 mutants | ||||||
| Pro-32LAla | 9.87 | 1.49 | 151 | 37 | 400 | 0.093 |
| Met-50HArg | 9.46 | 2.79 | 295 | 4.5 | 310 | 0.015 |
| Met-97HSer | 4.32 | 1.61 | 373 | 2.2 | 191 | 0.012 |
| Mixed mutants | ||||||
| germ-lineL/7G12H | 8.47 | 1.72 | 203 | 31 | 242 | 0.128 |
| 7G12L/germ-lineH | 4.28 | 4.37 | 1020 | ND | ND | 0.003 |
There are two somatic mutations in the 7G12 VL chain (Ser-14LThr and Ala-32LPro) and three in VH (Arg-50HMet, Ser-76HAsn, and Ser-97HMet). To evaluate the functional role of each somatic mutation, three sets of mutants were created. In the first set, each somatic mutation was introduced individually into the germ-line antibody; this set of mutants is termed the “germ-line mutants.” For the second set of mutations, each residue in the affinity-matured antibody derived from somatic mutation was switched back to its germ-line identity. This set of mutants is termed the “7G12 mutants.” The third set of mutants is termed “mixed mutants,” in which the light and heavy chains of the germ line and the affinity-matured antibodies are cross-combined to evaluate the contribution to binding affinity from the somatic mutations in either the light or heavy chain. ND, not determined.
The somatic mutation Ser-97HMet is located on CDR H3 and leads to a sharp turn in the backbone of CDR H3 in the affinity-matured Fab resulting from the packing interactions of Met-97H with the side chains of Tyr-36L, Gln-89L, Leu-96L, and Phe-98L (Fig. 3D). These interactions maintain the position of CDR H3 loop out of the active site and place residues Arg-95H and Asp-96H in the proper positions to interact with the NMP molecule upon its binding to the affinity-matured antibody. Thus somatic mutation Ser-97HMet fixes the conformation of CDR H3 for NMP binding; in its absence in the germ-line antibody the CDR H3 loop is flexible. The Met-97HSer mutant of the affinity-matured Fab catalyzes the insertion of Cu2+ into MP with a Km of 191 μM and a kcat of 2.2 h−1, corresponding to a virtually unchanged Km and 11-fold lower kcat relative to the affinity-matured Fab (Table 2). Consistent with the kinetic data, the Met-97HSer mutant has a Kd of 373 nM for the binding of NMP, an 18-fold decrease in binding affinity relative to the affinity-matured Fab.
The methionine side chain introduced by the Arg-50HMet somatic mutation packs against the imidazole ring of His-35H and orients the packing of His-35H onto Met-97H, which plays an important role in organizing the conformation of CDR H3 for hapten binding. Thus somatic mutation Arg-50HMet also helps to fix the conformation of CDR H3 in combination with the Ser-97HMet somatic mutation. Finally, the Ala-32LPro somatic mutation increases the steric bulk of the residue at this site in order to reinforce the packing interactions of the antibody on the hapten. In the affinity-matured Fab, Pro-32L packs on Tyr-91L, which packs directly on pyrrole ring B of NMP bound to the antibody and plays a crucial role in distorting the porphyrin ring in the Fab–MP complex (Fig. 3B). Consistent with the suggested roles for the Arg-50HMet and Ala-32LPro somatic mutations in binding the distorted conformation of the porphyrin, mutation of Met-50H or Pro-32L of 7G12 back to the germ-line residue leads to a loss of affinity for the distorted NMP and a decrease in catalytic efficiency (Table 2). The opposite is observed when the Pro-32L and Met-50H somatic mutations are introduced into the germ-line antibody.
Thus the affinity maturation process of the catalytic antibody 7G12 in response to the strained substrate mimic, NMP, resembles the evolution of enzymatic function in which binding energy is translated into a lower free energy of activation. Somatic mutations lead to a rigid antibody-combining site preorganized to bind the substrate MP in a strained, transition state-like conformation. This strain is evident in the crystal structure of the 7G12 Fab–MP Michaelis complex. The ability to bind porphyrin in a distorted conformation is compromised in the germ-line Fab because of the structural flexibility in the antibody-combining site, resulting in correspondingly lower catalytic activity. The structural plasticity found in the germ-line antibody repertoire may have also been a key feature of early proteins. This conformational flexibility may have allowed a limited number of primitive proteins, like germ-line antibodies, to bind a large number of ligands and catalyze a large number of different reactions. Mutations during natural evolution resulted in enhanced binding or catalytic activity by fixing an optimal combining-site conformation, much the same way somatic mutations fix the optimal active-site conformation of this and other catalytic antibodies.
Acknowledgments
This work was supported by National Institutes of Health Grant GM56528. This is manuscript 15240-CH of The Scripps Research Institute.
Abbreviations
- CDR
complementarity-determining region
- MP
mesoporphyrin IX
- NMP
N-methylmesoporphyrin IX
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.rcsb.org (PDB ID codes 1NGZ for the germline Fab, 1N7M for the germline Fab–NMP complex, 1NGY for the affinity-matured Fab, and 1NGW for the affinity-matured Fab–MP complex).
The Kd values of the germ-line and 7G12 Fabs are >100 μM as determined by surface plasmon resonance.
References
- 1.Haldane J B S. Enzymes. London: Longmans Green; 1930. p. 182. [Google Scholar]
- 2.Fersht A. Enzyme Structure and Mechanism. New York: Freeman; 1984. p. 311. [Google Scholar]
- 3.Jencks W P. Catalysis in Chemistry and Enzymology. New York: McGraw–Hill; 1969. p. 263. [Google Scholar]
- 4.Pollack S J, Jacobs J W, Schultz P G. Science. 1986;234:1570–1573. doi: 10.1126/science.3787262. [DOI] [PubMed] [Google Scholar]
- 5.Tramontano A, Janda K D, Lerner R A. Science. 1986;234:1566–1570. doi: 10.1126/science.3787261. [DOI] [PubMed] [Google Scholar]
- 6.Jacobsen J R, Prudent J R, Kochersperger L, Yonkovich S, Schultz P G. Science. 1992;256:365–367. doi: 10.1126/science.256.5055.365. [DOI] [PubMed] [Google Scholar]
- 7.Wagner J, Lerner R A, Barbas C F., III Science. 1995;270:1797–1800. doi: 10.1126/science.270.5243.1797. [DOI] [PubMed] [Google Scholar]
- 8.Shokat K M, Leumann C J, Sugasawara R, Schultz P G. Nature. 1989;338:269–271. doi: 10.1038/338269a0. [DOI] [PubMed] [Google Scholar]
- 9.Wirsching P, Ashley J A, Benkovic S J, Janda K D, Lerner R A. Science. 1991;252:680–685. doi: 10.1126/science.2024120. [DOI] [PubMed] [Google Scholar]
- 10.Cochran A G, Schultz P G. Science. 1990;249:781–783. doi: 10.1126/science.2389144. [DOI] [PubMed] [Google Scholar]
- 11.Dailey H A, Dailey T A, Wu C K, Medlock A E, Wang K F, Rose J P, Wang B C. Cell Mol Life Sci. 2000;57:1909–1926. doi: 10.1007/PL00000672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. McLaughlin, G. M. (1974) J. Chem. Soc. Perkin Trans. 2, 136–140.
- 13.Romesberg F E, Santarsiero B D, Spiller B, Yin J, Barnes D, Schultz P G, Stevens R C. Biochemistry. 1998;37:14404–14409. doi: 10.1021/bi981578c. [DOI] [PubMed] [Google Scholar]
- 14.Ulrich H D, Patten P A, Yang P L, Romesberg F E, Schultz P G. Proc Natl Acad Sci USA. 1995;92:11907–11911. doi: 10.1073/pnas.92.25.11907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Otwinowski Z, Minor W. Methods Enzymol. 1996;276:307–325. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 16.Kissinger C R, Gehlhaar D K, Fogel D B. Acta Crystallogr D. 1999;55:484–491. doi: 10.1107/s0907444998012517. [DOI] [PubMed] [Google Scholar]
- 17.Jones T A, Zou J Y, Cowan S W, Kjeldgaard M. Acta Crystallogr A. 1991;47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 18.Brunger A T, Adams P D, Clore G M, DeLano W L, Gros P, Grosse-Kunstleve R W, Jiang J S, Kuszewski J, Nilges M, Pannu N S, et al. Acta Crystallogr D. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 19.Yin J, Mundorff E C, Yang P L, Wendt K U, Hanway D, Stevens R C, Schultz P G. Biochemistry. 2001;40:10764–10773. doi: 10.1021/bi010536c. [DOI] [PubMed] [Google Scholar]
- 20.Blackwood M E, Jr, Rush T S, III, Romesberg F, Schultz P G, Spiro T G. Biochemistry. 1998;37:779–782. doi: 10.1021/bi972616f. [DOI] [PubMed] [Google Scholar]
- 21.Jentzen W, Song X Z, Shelnutt J A. J Phys Chem. 1997;101:1684–1699. [Google Scholar]
- 22.Fisher E. Ber Dtsch Chem Ges. 1895;28:1429. [Google Scholar]
- 23.Koshland D E., Jr Proc Natl Acad Sci USA. 1958;44:98–104. doi: 10.1073/pnas.44.2.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wedemayer G J, Patten P A, Wang L H, Schultz P G, Stevens R C. Science. 1997;276:1665–1669. doi: 10.1126/science.276.5319.1665. [DOI] [PubMed] [Google Scholar]
- 25.Mundorff E C, Hanson M A, Varvak A, Ulrich H, Schultz P G, Stevens R C. Biochemistry. 2000;39:627–632. doi: 10.1021/bi9924314. [DOI] [PubMed] [Google Scholar]
- 26.Esnouf R M. J Mol Graphics. 1997;15:133–138. doi: 10.1016/S1093-3263(97)00021-1. [DOI] [PubMed] [Google Scholar]
- 27.Merritt E A, Bacon D J. Methods Enzymol. 1997;227:505–524. doi: 10.1016/s0076-6879(97)77028-9. [DOI] [PubMed] [Google Scholar]
- 28.Nicholls A, Sharp K A, Honig B. Proteins Struct Funct Genet. 1991;11:281–296. doi: 10.1002/prot.340110407. [DOI] [PubMed] [Google Scholar]
- 29.Schultz P G, Yin J, Lerner R A. Angew Chem Int Ed Engl. 2002;41:4427–4437. doi: 10.1002/1521-3773(20021202)41:23<4427::AID-ANIE4427>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]