Abstract
The ability to bind immunosuppressive drugs such as cyclosporin and FK506 defines the immunophilin family of proteins, and the FK506-binding proteins form the FKBP subfamily of immunophilins. Some FKBPs, notably FKBP12 (the 12-kDa FK506-binding protein), have defined roles in regulating ion channels or cell signaling, and well established structures. Other FKBPs, especially the larger ones, participate in important biological processes, but their exact roles and the structural bases for these roles are poorly defined. FKBP51 (the 51-kDa FKBP) associates with heat shock protein 90 (Hsp90) and appears in functionally mature steroid receptor complexes. In New World monkeys, FKBP51 has been implicated in cortisol resistance. We report here the x-ray structures of human FKBP51, to 2.7 Å, and squirrel monkey FKBP51, to 2.8 Å, by using multiwavelength anomalous dispersion phasing. FKBP51 is composed of three domains: two consecutive FKBP domains and a three-unit repeat of the TPR (tetratricopeptide repeat) domain. This structure of a multi-FKBP domain protein clarifies the arrangement of these domains and their possible interactions with other proteins. The two FKBP domains differ by an insertion in the second that affects the formation of the progesterone receptor complex.
The immunophilins were named for their ability to bind immunosuppressive drugs, and there are two subfamilies: the FK506-binding proteins (FKBPs) and the cyclosporin A-binding cyclophilins. In addition to binding potent immunosuppressive drugs, both the FKBPs and cyclophilins are peptidylprolyl isomerases (PPIase) that catalyze the cis-trans conversion of peptidylprolyl bonds. In FKBP12, which mediates the therapeutic effect of FK506 (1, 2), FK506 binds in a deep pocket in a structurally well characterized fashion (3). The FKBP12–FK506 complex forms a complementary binding surface for calcineurin (Cn) (4), and the FKBP12-FK506–Cn complex cannot dephosphorylate nuclear factor of activated T cells, which results in immunosuppression (5). FKBP12 also binds the immunosuppressant rapamycin, but this complex inhibits the serine/threonine kinase activity of FRAP (FKBP12-rapamycin associated protein)/mTOR (mammalian target of rapamycin) (6–8). The ability of FK506 and rapamycin to enforce the association of two different proteins has led to schemes by which these small molecules or analogs can control cellular functions such as gene expression (9) and affinity modulation (10).
FKBP12's intrinsic roles involve regulation of signaling pathways. For example, FKBP12 inhibits transforming growth factor β (TGF β) receptor I's signaling until it is released by TGF β receptor II phosphorylation of receptor I (11). FKBP12 also regulates signaling by the ryanodine receptor (12, 13).
The roles of the larger FKBPs are less well known, but FKBP51 and FKBP52 participate in the formation of high-affinity steroid receptor complexes. These receptors for glucocorticoids (GR) or progesterone (PR) must be assembled through an ordered pathway involving Hsp40, Hsp70, Hsp90, and an assortment of Hsp-binding cochaperones (14). A physiological consequence of FKBP51 incorporation into GR complexes was recently observed in New World primates such as squirrel monkeys (15–18). These animals are hyposensitive to cortisol because of lowered GR hormone-binding affinity, similar to some clinical patients. Whereas a GR mutation has been shown to underlie cortisol resistance in some patients, the increased incorporation of squirrel monkey FKBP51 in GR complexes leads to reduced hormone binding. In human cells, typically low FKBP51 gene expression is highly induced by steroids (19–22), suggesting a possible autoregulatory mechanism for altering secondary hormonal responses (23, 24).
To provide a structural basis for understanding the biological roles of FKBP51, we determined the three-dimensional structures for human and squirrel monkey (Saimiri sciureus) FKBP51 to 2.7 and 2.8 Å, respectively. The structures are similar and contain three domains, two FKBP-like domains and a three-unit TPR domain.
Materials and Methods
Protein Expression and Purification.
For high-level expression of intein fusion proteins in bacteria, the plasmid vector pDS50 was generated in a pET28a (Stratagene) backbone by inserting the intein/chitin-binding domain cassette from pCYB4 (New England Biolabs). Either human or squirrel monkey cDNA encoding the entire ORF of FKBP51 was inserted in pDS50 in-frame with the downstream intein-coding sequences. The resulting plasmids were transformed into bacterial strain BL-21 for protein expression. Cells, typically from 18-liter culture volumes, were induced for 3 h with 1 mM isopropyl β-d-thiogalactoside, harvested, and washed twice with cold PBS. The resulting cell pellet was suspended in cell lysis buffer (20 mM Tris, pH 8/500 mM NaCl/0.1 mM EDTA/0.1% Triton X-100) containing a protease inhibitor mixture (Sigma, P-8465) and disrupted by sonication. Cleared lysate was applied to a chitin affinity resin (New England Biolabs), and FKBP51 was cleaved from the intein/chitin-binding domain according to manufacturer's instructions. Peak fractions were pooled and concentrated by using a Centriprep 30 concentrator (Amicon).
For selenomethionine incorporation into FKBP51, minor modifications were made to a published protocol (25). Briefly, BL21 cells transformed with 51-intein/pDS50 were cultured overnight in 36 ml of LB medium. Pelleted cells were resuspended in 1 liter of M9++ [M9 minimal media supplemented with glucose (4% wt/vol), MgSO4 (75 mg/liter), and kanamycin (30 mg/liter)]. Cells were grown at 37°C with shaking at 250 rpm until reaching ≈0.6 optical density at 600 nm. l-Amino acids were added (50 mg of selenomethionine; 10 mg each of lysine, threonine, and phenylalanine; 5 mg each of leucine, isoleucine, and valine) and, after shaking at 37°C for 15 min, the culture was adjusted to 1 mM isopropyl β-d-thiogalactoside and grown an additional 12–14 h at 37°C. Cells were lysed and protein was purified as described above.
PR Complex Assembly.
Human FKBP51 or FKBP52 cDNA in pSPUTK (Stratagene) was used for in vitro expression of wild-type and mutant proteins. FKBP51 variants were created by means of site-directed mutagenesis (QuikChange kit by Stratagene). The Δ3 mutant, lacking codons 195–197, was generated with a mutagenic primer in which the corresponding 9 bases were deleted (5′-GGCGAAGGAGAA_ATTCCAATTGG; the underline indicates deleted base). Radiolabeled immunophilins were prepared by expressing each of the cDNAs in an in vitro transcription–translation system (Promega, TnT kit) containing [35S]methionine. Equivalent amounts of radiolabeled proteins were added to normal rabbit reticulocyte lysate (Green Hectares, Oregon, WI) for cell-free assembly of PR complexes as described (26). Washed complexes were extracted into SDS sample buffer, separated by SDS/PAGE, and visualized by Coomassie blue staining and autoradiography.
Crystallization and Data Collection.
FKBP51 crystals were obtained in sitting-drop vapor diffusion experiments. Drops contained equal parts of 8 mg/ml FKBP51 (20 mM Tris⋅HCl, pH 8.0/50 mM NaCl/0.1 mM EDTA/10 mM DTT) and a reservoir solution of 15–18% polyethylene glycol 5000 monomethyl ether (Fluka)/100 mM Mes (pH 5.8)/0.2 M ammonium sulfate. Selenomethionine-containing crystals were obtained under similar conditions with decreased precipitant concentration (12%). The crystals grew to 0.15 × 0.15 × 0.35 mm within 10–14 days. To improve the diffraction quality and resolution, a mild dehydration treatment was used; the reservoir was exchanged with a similar reservoir of 18–25% polyethylene glycol 5000 monomethyl ether and allowed to equilibrate for 14 days. The crystals were extracted from the drops with a nylon loop and frozen in liquid nitrogen.
Data collection and statistics are given in Tables 1 and 2. The multiwavelength anomalous dispersion data were collected on beam line F2 at the Cornell High Energy Synchrotron Source (CHESS) by using the inverse beam technique and a single selenomethionine-containing crystal. Indexing and integration were done by using mosflm [from the Data Processing Suite (27)], scaled by using scala and truncated by using truncate from the Collaborative Computational Project No. 4 package (28).
Table 1.
Crystallographic data for human FKBP51
| Edge | Peak | Remote | Native | |
|---|---|---|---|---|
| Data collection | ||||
| Wavelength, Å | 0.9794 | 0.9792 | 0.9679 | 0.9134 |
| Unit cell, Å | ||||
| a and b | 88.22 | 87.58 | ||
| c | 132.05 | 132.03 | ||
| Resolution, Å | 20–3.2 | 25–2.7 | ||
| No. of reflections | ||||
| Observed | 72,028 | 72,015 | 72,141 | 80,664 |
| Unique | 9.963 | 9,984 | 9,982 | 16,517 |
| Completeness, % | 94.6 | 94.6 | 94.6 | 100 |
| Rsym,* % | 8.2 | 8.5 | 8.3 | 6.6 |
| Refinement statistics | ||||
| Resolution, Å | 25–2.7 | |||
| R factor, % | ||||
| Working | 28.4 | |||
| Free | 37.5 | |||
| rms† | ||||
| Lengths, Å | 0.01 | |||
| Angles, ° | 1.58 | |||
| Backbone | 3.3 | |||
| Side chain | 5.8 | |||
| No. of nonhydrogen atoms | ||||
| Proteins | 2,828 | |||
| Ions | 15 | |||
| Water | 49 |
Rsym = Σ | I0 − 〈I〉 |/_ I0, where 〈I〉 = average intensity obtained from multiple observations of symmetry-related reflections.
rms deviation from ideal geometry and rms variation in the B factors of bonded atoms.
Table 2.
Crystallographic data for squirrel monkey FKBP51
| Edge | Peak | Remote | Native | |
|---|---|---|---|---|
| Data collection | ||||
| Wavelength, Å | 0.9793 | 0.9791 | 0.9679 | 0.9251 |
| Unit cell, Å | ||||
| a and b | 91.24 | 91.04 | ||
| c | 131.98 | 132.65 | ||
| Resolution, Å | 20–3.2 | 45–2.8 | ||
| No. of reflections | ||||
| Observed | 101,060 | 101,030 | 101,029 | 95,940 |
| Unique | 10,908 | 10,908 | 10,908 | 16,161 |
| Completeness, % | 99.9 | 99.9 | 99.9 | 100 |
| Rsym,* % | 8.0 | 8.0 | 8.0 | 8.7 |
| Refinement statistics | ||||
| Resolution, Å | 45–2.8 | |||
| R factor, % | ||||
| Working | 25.5 | |||
| Free | 31.4 | |||
| rms† | ||||
| Lengths, Å | 0.01 | |||
| Angles, ° | 1.56 | |||
| Backbone | 2.7 | |||
| Side chain | 4.7 | |||
| No. of nonhydrogen atoms | ||||
| Proteins | 2,956 | |||
| Ions | 10 | |||
| Water | 31 |
Rsym = Σ | I0 − 〈I〉 |/_ I0, where 〈I〉 = average intensity obtained from multiple observations of symmetry-related reflections.
rms deviation from ideal geometry and rms variation in the B factors of bonded atoms.
Structure Determination and Refinement of FKBP51.
SHELXL-97 (29) revealed 9 of the 14 possible selenium positions for human FKBP51 and 7 of the 11 selenium positions for squirrel monkey FKBP51. The refinement and phasing were done with sharp (30), solomon (31) was used for density modification within sharp, and this map was readily interpretable. The initial model was built by using o (32) and refined against the native data for each species by using cns (33), with rebuilding done in o.
Results and Discussion
Overall Architecture of FKBP51.
The final refined models (Fig. 1A) contain residues 33–412 of 457 with a few small gaps (38–45, 62–66, 70–75, and 382–385) for the human structure and residues 28–421 with gaps (38–41, 64–75, 108–111) for the squirrel monkey structure. The first two domains, residues 33–138 and residues 147–251, are typical FKBP domains consisting of an antiparallel five-stranded β sheet wrapped around a central α-helix (3). The third domain, residues 261 to ≈400, is similar to the Hsp90-binding TPR domains of protein phosphatase 5 (PP5; ref. 36), Hop (Hsp70/90 organizing protein; ref. 37), and Cyp40 (cyclophilin 40; ref. 38). In general, TPR domains are all-helical structures consisting of 2–16 units of a consensus 34-aa motif (36). A single unit consists of two consecutive α-helices of 12– 15 residues each crossing at an angle of ≈20° to each other. The FKBP51 TPR domain contains three such units, plus an additional helix.
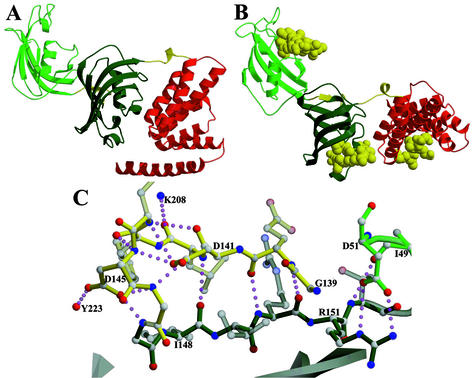
Figure 1.
Crystal structure of FKBP51. (A) The overall structure is as follows: light green is the FK1 domain, dark green is the FK2 domain, red is the TPR domain, and yellow regions are the linkers between domains. (B) In each case, coordinates of a similar domain (FKBP12, FKBP12, and the Hop TPR 2a domain) bound to a ligand [rapamycin (39), rapamycin, and Hsp90 fragment MEEVD (37), respectively] were superposed on FKBP51s domains (FK1, FK2, and TPR). The resulting ligand coordinates are shown in yellow space-filling atoms. (C) Extensive hydrogen bonding of the linker region between FK1 and FK2. The yellow bonds are the linking regions, dark-green bonds are FK2, and light-green bonds are FK1. Figs. 1–5 were rendered with molscript (34) and RASTER3D (35).
The orientation of the binding pockets of the FKBP domains and the binding groove of the TPR domain suggests a plausible domain orientation for FKBP51 in multiprotein complexes. The binding pocket of the first FKBP domain, FK1, as seen in Fig. 1B, is oriented ≈180° from the putative binding pocket of the second FKBP domain, FK2. The connection between FK1 and FK2 is only eight residues, and this short link has extensive interactions that appear to reduce its flexibility (Fig. 1C). The link begins with a small three-amino acid (residues 140–42) antiparallel β-strand interaction with β1 of FK2. The next four amino acids (residues 143–146) form a tight loop stabilized by hydrogen bonding of the side-chain carboxylates of Asp-141 and Asp-145 with backbone nitrogens in the loop.
The concave face of the TPR domain is facing the same direction and 26 Å away from the binding pocket of the FK2 domain. The connecting region, residues 252–260, extends 16.7 Å from Cα to Cα with a single helical turn (256–258), and α1 of the TPR domain is 21 Å from the last residue of FK2's β3. The first two αhelices then come back toward the FK2 domain, forming two direct hydrogen bonds between FK2 and TPR α1, and a salt bridge involving Asp-195, Lys-274, and Tyr-278, and two water-mediated hydrogen bonds.
FK1 Domain.
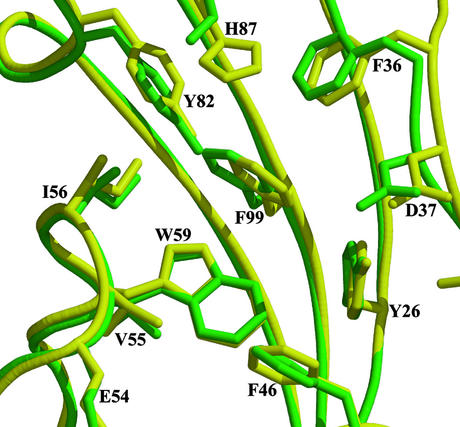
This first FKBP domain consists of five antiparallel β-strands curved around a central α-helix, the usual FK fold (Fig. 1 A and B). Several of the loops are disordered, notably the loop between β1 and β4 and the “40s” loop. FK1 and FKBP12 share 48% sequence identity (60% similarity), and the overall structures have a 0.7-Å rms deviation over all Cα atoms available in the FKBP51 structure and 0.9-Å rms deviation over all atoms in identical residues of the binding pockets (Fig. 2). The major difference in the structures occurs in the “80s” loop (FKBP51 residues 110–125), a region known to show the most variations among the characterized FKBP domains (40). The PPIase activity of FKBP51 is 0.48 × 106 M−1⋅s−1 (41), comparable to that of FKBP12, 0.64 × 106 M−1⋅s−1 (42). FKBP51 Ki values for FK506 and rapamycin are ≈10 and <5 nM, respectively (41), compared with ≈1 nM for FKBP12 (42). A double point mutation in FK1 decreased the enzymatic activity of human FKBP51 by >90% (26), arguing that this domain is the measurably active PPIase domain in FKBP51.
Figure 2.
Binding pocket of FK1. FKBP12 (yellow with black labels) coordinates were superposed with FK1 of FKBP51 (green with green labels). Only one amino acid differs, at H87 FKBP51, which instead has a serine. The remaining side chains superpose well.
Two well ordered waters sit deep in the FK1 binding pocket. Water-519 sits ≈4.2 Å above Trp-90 (Trp-59 in FKBP12) surrounded by hydrophobic residues and forms a single hydrogen bond to water-531. Water-531 forms a hydrogen bond with Tyr-113 (Tyr-82 of FKBP12), mimicking the hydrogen bonding seen in FK506 and rapamycin complexes. A third water, 522, sits on the outside of the binding pocket forming a hydrogen bond with the carbonyl oxygen of Gly-84, reproducing the hydrogen bonding of rapamycin and FK506.
FK2 Domain.
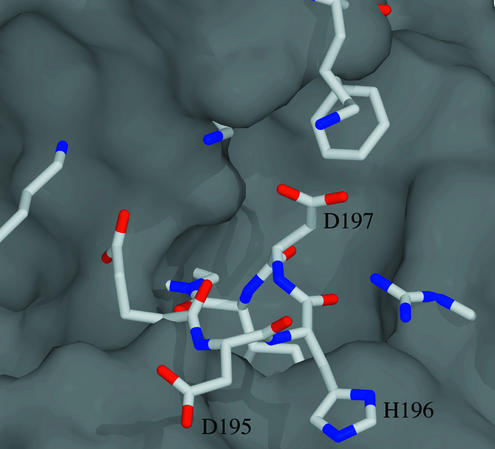
FK2 is also structurally similar to FKBP12 despite having only 26% sequence identity and 44% similarity (Fig. 1A). The deletion of the 40s loop results in a continuous β5 strand. Of the 12 residues forming the binding pocket, only 3 are identical (F181, D182, and E194) and 3 are conserved (V55/I198, Y82/F225, and F99/Y243; Table 3). An insertion of D195 H196 D197 between β5 and the α helix pushes into the binding pocket as shown in Fig. 3. Both the lack of sequence conservation and the binding pocket insertion agree with previous work that suggested FK2 lacks measurable PPIase activity (26). We have confirmed with FKBP51 truncation mutants lacking FK1 that neither FK2 nor the mutant Δ3, deletion of D195, H196, and D197, the residues that fill the FK2 pocket, have detectable PPIase activity (data not shown).
Table 3.
Sequence conservation in the binding pocket of the FKBPs
| Domains | Conserved residues | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| human_ FKBP12 | Y26 | F36 | D37 | R42 | F46 | E54 | V55 | I56 | W59 | Y82 | H87 | I91 | F99 |
| human_ FKBP13 | Y | F | D | Q | F | Q | V | I | W | Y | A | I | F |
| human_ FKBP25 | Y | F | D | T | L | K | V | I | W | Y | Q | I | F |
| FKBP51_ FK1 | Y57 | F67 | D68 | R73 | F77 | Q85 | V86 | I87 | W90 | Y113 | S118 | I122 | F130 |
| FKBP51_ FK2 | L172 | F181 | D182 | — | V186 | E194 | I198 | P199 | I202 | F225 | K230 | G234 | Y243 |
| FKBP52_ FK1 | F | F | D | R | F | E | V | I | W | Y | S | I | F |
| FKBP52_ FK2 | L | F | D | — | L | E | L | P | L | F | K | Q | Y |
Figure 3.
Binding pocket of FK2. The molecular surface of FKBP12, calculated with spock (43), was superposed on FK2 of FKBP51. The binding pocket of FKBP12 is clearly seen in the surface. The FK2 residues, shown as stick models, show how the 3-aa insertion (D195, H196, and D197) and other mutations obstruct the binding pocket.
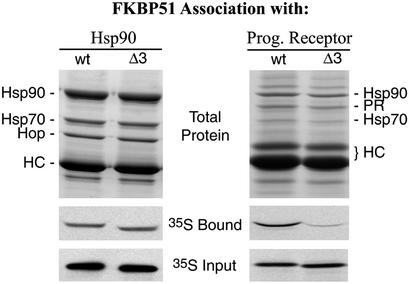
Because the FK2-binding pocket is so near the binding groove of the TPR domain, we used Δ3 to test for possible involvement of the FK2 insertion loop with Hsp90 binding or receptor association. Wild-type and Δ3 proteins were compared for interactions with either Hsp90 or PR complexes by using established coimmunoprecipitation approaches (26). As seen in Fig. 4, the deletion had no apparent effect on the binding of FKBP51 to Hsp90; in contrast, the deletion reduced FKBP51 recovery in PR complexes to a level approximating that of FKBP52. Deletion of the corresponding FK2 insertion loop from FKBP52 affected neither Hsp90 binding nor PR association (results not shown), suggesting that the related FKBPs have different interactions with the receptor. An attractive possibility is that FK2 in FKBP51 is distinctly oriented and brought into proximity by Hsp90 for direct interaction with PR. A unique interaction between receptor and the FK2 domain of FKBP51 would help explain why FKBP51 is competitively preferred over FKBP52 in PR complexes (44).
Figure 4.
FK2 involvement in PR association but not Hsp90 binding. Both wild-type human FKBP51 (wt) and a deletion mutant lacking D195, H196, and D197 from the FK2 domain (Δ3) were tested for association with Hsp90 (Left) or PR (Right) complexes. Hsp90 complexes were immunoprecipitated from rabbit reticulocyte lysate that had been supplemented with radiolabeled wt or Δ3, and washed beads were extracted and the proteins were separated by SDS/PAGE. (Top) An image of the Coomassie blue-stained gel with labeled bands from the predominate Hsp90-Hop-Hsp70 and the heavy chain (HC) of antibody used for immunoprecipitating complexes. (Middle and Bottom) Autoradiograms that show the levels of radiolabeled FKBP51 recovered in the gel samples (35S Bound), or representative amounts of the two FKBP forms included in the incubation mixture (35S Input), respectively. Similarly, PR complexes were assembled in reticulocyte lysate mixtures and analyzed for the association of radiolabeled FKBP51. For reference, FKBP51 comigrates with antibody heavy chains.
TPR Domain.
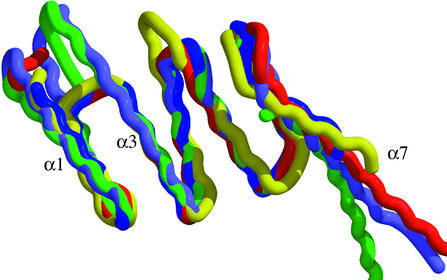
The TPR domain of FKBP51 has three repeats of the TPR motif and is structurally very similar to the Hsp90-binding domains of PP5 (36), Hop (37), and Cyp40 (38). At 21 and 25 residues, respectively, α1 and α3 are longer than the usual 12– 15 residues, but α2 and α4-α6 are within the average size for TPR helices. Similar to the PP5 and Cyp40 structures, where the domains were not shortened by digestion, FKBP51 also contains a seventh helix that extends beyond the final TPR motif. The TPR backbones overlay well (Fig. 5) and there are few differences in the core of the TPR domains. Although the seventh extended helix does vary in the angle with which it protrudes from the TPR domain, this variation could have more to do with crystal packing than with any interactions between α5 or α6 and α7.
Figure 5.
TPR domains. The known TPR domains were superposed onto the TPR domain of FKBP51 and color-coded according to their proteins: green, FKBP51; red, PP5; lavender, Cyp40; blue, Hop TPR1; and yellow, Hop TPR2a. The backbones of the six helices forming the TPR domain superpose well, whereas α7 continues out from the TPR domains at slightly differing angles.
The structure of the Hop TPR domains (37) shows binding in an antiparallel orientation to α1, α3, α5, and α7, which form the groove for both Hsp70 and Hsp90 fragments. The report of the Cyp40 structure (38) does not have an Hsp90 fragment bound, but instead uses the crystal packing to model Hsp90 binding, which results in a binding parallel, not antiparallel, to the direction of the helices lining the binding groove. The structure of the FKBP51 TPR domain gives no indication of the binding orientation of Hsp90. However, when the structures of the Hop, Cyp40, and PP5 TPR domains are superimposed on the FKBP51 domain, an alanine conserved throughout the Hop (Ala-46 and Ala-267) and Cyp40 (Ala-281) domains is a leucine in PP5 (Leu-70) and a methionine (M325) in FKBP51. The current binding models for Hsp are incompatible with these leucine and methionine residues, which would clash with an Hsp bound as suggested. Studies on a full or larger segment of Hsp with a full TPR containing protein will be needed to unravel the complexities of Hsp binding.
Significance of the FKBP51 Structure.
The structure of Mip, a small FKBP with a dimerization and linkage domains, was recently reported (45), but the FKBP51 structures reported here are for a large FKBP with multiple FK506 binding domains. Although the resolution is limited and some residues are missing, the structures provide an important initial insight into the relative orientations of the FK1, FK2, and TPR domains, which are important for biological function. Based on our PR assembly study, there appear to be cooperative protein–protein interactions involving TPR and, surprisingly, FK2 rather than the active PPIase FK1. Presumably, FK2 resulted from an FK domain duplication event; yet, while FK2 has lost its PPIase activity, it appears to have gained a protein interaction ability.
Glucocorticoid Hyposensitivity.
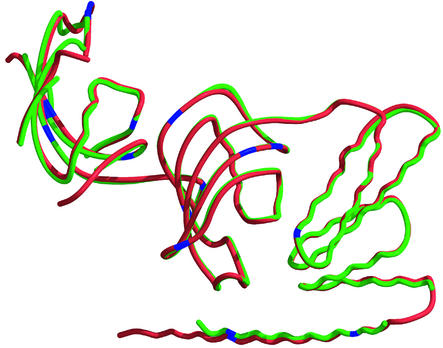
Reynolds et al. (16) showed that lymphocytes from squirrel monkeys, compared with human, have 13 times higher FKBP51 protein levels and incorporate more FKBP51 into GR complexes. Studies with transfected cells and cell-free approaches have clearly shown that squirrel monkey FKBP51 reduces GR hormone-binding affinity and expression of a reporter gene (16, 17). Expressing human FKBP51 to similar levels resulted in some cortisol resistance, but to only one-fifth the extent caused by monkey FKBP51. The two proteins are 94% identical (97% similar), and a superposition of the crystal structures (Fig. 6) shows that the overall architecture is the same. There are 15 substitutions in residues contained within the crystal structure, most of which are conservative. Outside the resolved structures, there are six additional substitutions in the N-terminal 32 aa and five in the C-terminal 30 aa, most of which are not conservative. Although not apparent from the crystal structures, these regions may indirectly influence FK2 or other interactions that, in the context of Hsp90–steroid receptor complexes, modulate receptor-hormone-binding affinity.
Figure 6.
Structural comparison of human and squirrel monkey FKBP51s. The smFKBP51 (red) was superposed onto huFKBP51 (green). The two proteins have very little difference in their overall structures, with the “80s” loop of the FK1 domains showing the most variability. Blue marks the residues that are different; most of these 15 mutations are conservative.
Acknowledgments
We thank M. Cristina Nonato, Shenping Liu, Damon Carr, and Donna Valentine for their valuable assistance in this work. This work was supported in part by a National Institutes of Health Chemistry/Biology Interface Training Grant (to C.R.S.), National Institutes of Health Grants R01 CA59021 (to J.C.) and R01 DK48218 (to D.F.S.), and National Center for Research Resources Grant RR-13200 (to J.G.S.). This work is based in part on research conducted at the Cornell High Energy Synchrotron Source (CHESS), which is supported by the National Science Foundation under Award DMR-9311772, using the Macromolecular Diffraction at CHESS (MacCHESS) facility, which is supported by Award RR-01646 from the National Institutes of Health.
Abbreviations
- Cyp40
Cyclophilin 40
- FKBP
FK506-binding protein
- FK1
first FKBP domain of FKBP51
- FK2
second FKBP domain of FKBP51
- GR
glucocorticoid receptor
- Hsp
heat shock protein
- Hop
Hsp70/90 organizing protein
- PP5
protein phosphatase 5
- PPIase
peptidylprolyl isomerase
- PR
progesterone receptor
- TPR
tetratricopeptide repeat
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, www.rcsb.org [PDB ID codes 1KT0 (human FKBP51) and 1KT1 (squirrel monkey FKBP51)].
References
- 1.Siekierka J J, Hung S H Y, Poe M, Lin C S, Sigal N H. Nature. 1989;341:755–757. doi: 10.1038/341755a0. [DOI] [PubMed] [Google Scholar]
- 2.Harding M W, Galat A, Uehling D E, Schreiber S L. Nature. 1989;341:758–760. doi: 10.1038/341758a0. [DOI] [PubMed] [Google Scholar]
- 3.Van Duyne G D, Standaert R F, Karplus P A, Schreiber S L, Clardy J. Science. 1991;252:839–842. doi: 10.1126/science.1709302. [DOI] [PubMed] [Google Scholar]
- 4.Liu J. Cell. 1991;66:807–815. doi: 10.1016/0092-8674(91)90124-h. [DOI] [PubMed] [Google Scholar]
- 5.O'Keefe S J, Tamura J, Kincaid R F, Tocci M J, O'Neill M A. Nature. 1992;357:692–694. doi: 10.1038/357692a0. [DOI] [PubMed] [Google Scholar]
- 6.Brown E J, Albers M W, Shin T B, Ichikawa K, Keith C T, Lane W S, Schreiber S L. Nature. 1994;369:756–758. doi: 10.1038/369756a0. [DOI] [PubMed] [Google Scholar]
- 7.Brown E J, Beal P A, Keith C T, Chen J, Shin T B, Schreiber S L. Nature. 1995;377:441–446. doi: 10.1038/377441a0. [DOI] [PubMed] [Google Scholar]
- 8.Sabers C J, Martin M M, Brunn G J, Williams J M, Dumont F J, Wiederrecht G, Abraham R T. J Biol Chem. 1995;270:815–822. doi: 10.1074/jbc.270.2.815. [DOI] [PubMed] [Google Scholar]
- 9.Spencer D M, Wandless T J, Schreiber S L, Crabtree G L. Science. 1993;262:1019–1024. doi: 10.1126/science.7694365. [DOI] [PubMed] [Google Scholar]
- 10.Briesewitz R, Ray G T, Wandless T J, Crabtree G R. Proc Natl Acad Sci USA. 1999;96:1953–1958. doi: 10.1073/pnas.96.5.1953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Aghdasi B, Ye K, Resnick A, Huang A, Ha H C, Guo X, Dawson T M, Dawson V L, Snyder S H. Proc Natl Acad Sci USA. 2001;98:2425–2430. doi: 10.1073/pnas.041614198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Timerman A B, Ogunbumni E, Freund E, Wiederrecht G, Marks A R, Fleischer S. J Biol Chem. 1993;268:22992–22999. [PubMed] [Google Scholar]
- 13.Barg S, Copello J A, Fleischer S. Am J Physiol. 1997;272:C1726–C1733. doi: 10.1152/ajpcell.1997.272.5.C1726. [DOI] [PubMed] [Google Scholar]
- 14.Pratt W B, Toft D O. Endocr Rev. 1997;18:306–360. doi: 10.1210/edrv.18.3.0303. [DOI] [PubMed] [Google Scholar]
- 15.Reynolds P D, Pittler S J, Scammell J G. J Clin Endcrinol Metab. 1997;82:465–472. doi: 10.1210/jcem.82.2.3755. [DOI] [PubMed] [Google Scholar]
- 16.Reynolds P D, Ruan Y, Smith D F, Scammell J G. J Clin Endcrinol Metab. 1999;84:663–669. doi: 10.1210/jcem.84.2.5429. [DOI] [PubMed] [Google Scholar]
- 17.Denny W B, Valentine D L, Reynolds P D, Smith D F, Scammell J G. Endocrinology. 2000;141:4107–4113. doi: 10.1210/endo.141.11.7785. [DOI] [PubMed] [Google Scholar]
- 18.Scammell J G, Denny W B, Valentine D L, Smith D F. Gen Comp Endocrinol. 2001;124:152–165. doi: 10.1006/gcen.2001.7696. [DOI] [PubMed] [Google Scholar]
- 19.Baughman G, Wiederrecht G J, Campbell N F, Martin M M, Bourgeois S. Mol Cell Biol. 1995;15:4395–4402. doi: 10.1128/mcb.15.8.4395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kester H A, van der Leede B M, van der Saag P T, van der Burg B. J Biol Chem. 1997;272:16637–16643. doi: 10.1074/jbc.272.26.16637. [DOI] [PubMed] [Google Scholar]
- 21.Amler L C, Agus D B, LeDuc C, Sapinoso M L, Fox W D, Kern S, Lee D, Wang V, Leysens M, Higgins B, et al. Cancer Res. 2000;60:6134–6141. [PubMed] [Google Scholar]
- 22.Zhu W, Zhang J S, Young C Y. Carcinogenesis. 2001;22:1399–1403. doi: 10.1093/carcin/22.9.1399. [DOI] [PubMed] [Google Scholar]
- 23.Cheung J, Smith D F. Mol Endocrinol. 2000;14:939–946. doi: 10.1210/mend.14.7.0489. [DOI] [PubMed] [Google Scholar]
- 24.Scammell J G. ILAR J. 2000;41:19–25. doi: 10.1093/ilar.41.1.19. [DOI] [PubMed] [Google Scholar]
- 25.Van Duyne G D, Standaert R F, Karplus P A, Schreiber S L, Clardy J. J Mol Biol. 1993;229:105–124. doi: 10.1006/jmbi.1993.1012. [DOI] [PubMed] [Google Scholar]
- 26.Barent R L, Nair S C, Carr D C, Ruan Y, Rimerman R A, Fulton J, Zhang Y, Smith D F. Mol Endocrinol. 1998;12:342–354. doi: 10.1210/mend.12.3.0075. [DOI] [PubMed] [Google Scholar]
- 27.Leslie A G W. mosflm User Guide. Cambridge, U.K.: Medical Research Council (MRC) Laboratory of Molecular Biology; 1994. . Version 5.30. [Google Scholar]
- 28.Collaborative Computational Project No. 4. Acta Crystallogr D. 1994;50:760–763. [Google Scholar]
- 29.Sheldrick G M. Methods Enzymol. 1997;276:628–641. doi: 10.1016/S0076-6879(97)76083-X. [DOI] [PubMed] [Google Scholar]
- 30.de La Fortelle E, Bricogne G. Methods Enzymol. 1997;276:472–494. doi: 10.1016/S0076-6879(97)76073-7. [DOI] [PubMed] [Google Scholar]
- 31.Abrahams J, Leslie A. Acta Crystallogr D. 1996;52:30–42. doi: 10.1107/S0907444995008754. [DOI] [PubMed] [Google Scholar]
- 32.Jones T A, Kjeldgaard M. Methods Enzymol. 1997;277:173–208. doi: 10.1016/s0076-6879(97)77012-5. [DOI] [PubMed] [Google Scholar]
- 33.Brunger A T, Adams P D, Clore G M, DeLano W L, Gros P, Grosse-Kunstleve R W, Jiang J S, Kuszewski J, Nilges M, Pannu N S, et al. Acta Crystallogr D. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 34.Kraulis P J. J Appl Crystallogr. 1991;24:946–950. [Google Scholar]
- 35.Merritt E A, Bacon D J. Methods Enzymol. 1997;277:505–524. doi: 10.1016/s0076-6879(97)77028-9. [DOI] [PubMed] [Google Scholar]
- 36.Das A K, Cohen P T, Barford D. EMBO J. 1998;17:1192–1199. doi: 10.1093/emboj/17.5.1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Scheufler C, Brinker A, Bourenkov G, Pegoraro S, Moroder L, Bartunik H, Hartl F U, Moarefi I. Cell. 2000;101:199–210. doi: 10.1016/S0092-8674(00)80830-2. [DOI] [PubMed] [Google Scholar]
- 38.Taylor P, Dornan J, Carrello A, Minchin R F, Ratajczak R, Walkinshaw M D. Structure (London) 2001;9:431–438. doi: 10.1016/s0969-2126(01)00603-7. [DOI] [PubMed] [Google Scholar]
- 39.Van Duyne G D, Standaert R F, Schreiber S L, Clardy J. J Am Chem Soc. 1991;113:7433–7434. [Google Scholar]
- 40.Wilson K P, Yamashita M M, Sintchak M D, Rotstein S H, Murcko M A, Boger J, Thomson J A, Fitzgibbon M J, Black J R, Navia M A. Acta Crystallogr D. 1995;51:511–521. doi: 10.1107/S0907444994014514. [DOI] [PubMed] [Google Scholar]
- 41.Yeh W-C, Li T-K, Bierer B E, McKnight S L. Proc Natl Acad Sci USA. 1995;92:11081–11085. doi: 10.1073/pnas.92.24.11081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Harrison R K, Stein R L. Biochemistry. 1990;29:3813–3816. doi: 10.1021/bi00468a001. [DOI] [PubMed] [Google Scholar]
- 43.Christopher J A. spock: The Structural Properties Observation and Calculation Kit (Program Manual) Texas A&M University, College Station: Center for Macromolecular Design; 1998. [Google Scholar]
- 44.Nair S C, Rimerman R A, Toran E J, Chen S, Prapapanich V, Butts R N, Smith D F. Mol Cell Biol. 1997;17:594–603. doi: 10.1128/mcb.17.2.594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Riboldi-Tunnicliffe A, König B, Jessen S, Weiss M S, Rahfeld J, Hacker J, Fischer G, Hilgenfeld R. Nat Struct Biol. 2001;8:779–783. doi: 10.1038/nsb0901-779. [DOI] [PubMed] [Google Scholar]