Abstract
Animal mtDNAs are typically small (≈16 kbp), circular-mapping molecules that encode 37 or fewer tightly packed genes. Here we investigate whether similarly compact mitochondrial genomes are also present in the closest unicellular relatives of animals, i.e., choanoflagellate and ichthyosporean protists. We find that the gene content and architecture of the mitochondrial genomes of the choanoflagellate Monosiga brevicollis, the ichthyosporean Amoebidium parasiticum, and Metazoa are radically different from one another. The circular-mapping choanoflagellate mtDNA with its long intergenic regions is four times as large and contains two times as many protein genes as do animal mtDNAs, whereas the ichthyosporean mitochondrial genome totals >200 kbp and consists of several hundred linear chromosomes that share elaborate terminal-specific sequence patterns. The highly peculiar organization of the ichthyosporean mtDNA raises questions about the mechanism of mitochondrial genome replication and chromosome segregation during cell division in this organism. Considering that the closest unicellular relatives of animals possess large, spacious, gene-rich mtDNAs, we posit that the distinct compaction characteristic of metazoan mitochondrial genomes occurred simultaneously with the emergence of a multicellular body plan in the animal lineage.
Keywords: multichromosome mitochondrial genome‖linear mtDNA‖ancestors of animals‖protists
Human mtDNA was the first mitochondrial genome to be completely sequenced (1). Since then, >270 complete animal (metazoan) mtDNA sequences have become available, primarily from chordates, but also from most of the invertebrate phyla (see the complete organelle genome section of the National Center for Biotechnology Information, www.ncbi.nlm.nih.gov/PMGifs/Genomes/euk_o.html). The majority of animal mtDNAs are relatively small (≈13–19 kbp in size), map as circular molecules, and comprise a mix of covalently closed circular monomers and varying proportions of concatenated dimers and higher oligomers (for reviews, see refs. 2 and 3). Animal mitochondrial genes are predominantly encoded on both strands and are compactly arrayed, with coding segments separated by no or very short (only a few base pairs) noncoding spacers; in rare cases, genes even overlap. The set of mitochondrial genes is nearly identical throughout the Metazoa, specifying 12–13 proteins involved in electron transport and oxidative phosphorylation and 24–25 structural RNAs (small and large subunit rRNAs and tRNAs). Moreover, metazoan mitochondrial genes are generally not interrupted by introns. Exceptions to this general pattern are found within the phylum Cnidaria. For example, only one or two tRNA genes are present in mtDNAs of the cnidarian class Anthozoa, which includes sea anemones and corals (4–6). In addition, one or two mitochondrial group I introns have been discovered in mtDNAs of sea anemones and a scleratinian coral (4, 5); in addition, an extra mitochondrial gene resembling the bacterial mismatch repair gene, mutS, has been identified in an alcyonarian coral (6). Finally, relatively large (20–42 kbp) mtDNAs have been found in some mollusks, nematodes, and insects (7–10). These expanded animal mitochondrial genomes owe their large size either to the presence of several copies of a repeated element, an extended duplication, or expansion of a single A+T-rich region.
The observed uniformity of animal mitochondrial genomes stands in stark contrast to the situation in plants, fungi, and numerous protists (amoebae, flagellates, and algae). Protist and plant mtDNAs are generally much larger in size than their animal counterparts and encode a substantially greater number of proteins. The most gene-rich mitochondrial genome known is that of Reclinomonas americana (11), a member of a group of flagellate protozoans termed jakobids (12). The ≈70-kbp circular-mapping mtDNA of R. americana encodes a total of nearly 100 genes (11), i.e., three to six times more than animal mtDNAs. This raises the question as to whether the marked gene loss and tight packing of mtDNAs found in extant animals only occurred with the emergence of Metazoa, or whether these features existed before that time, in the unicellular protist ancestors of animals.
Until recently, there was considerable controversy and uncertainty about the nature of the single-celled animal ancestors. As early as a century ago, it was proposed that the predecessor of Metazoa was a choanoflagellate-like unicellular protist (13). This supposition has been both corroborated and challenged by numerous molecular phylogenetic studies, but was confirmed with compelling evidence only recently (ref. 14 and references therein). At the same time, a second unicellular eukaryotic group, the Ichthyosporea, also termed Mesomycetozoa (15) or DRIPs (16), was recognized as comprising specific relatives of multicellular animals (14). These mostly parasitic protists, whose evolutionary affiliation has been controversial for a number of years (15–18), are now known to represent the first offshoot within the specific lineage leading to Metazoa (14).
To address the question as to when in evolutionary history the distinctive compaction of the animal mitochondrial genome occurred, we investigated mtDNA structure in representatives of the two known phyla of unicellular relatives of animals, i.e., the choanoflagellate Monosiga brevicollis and the ichthyosporean Amoebidium parasiticum. Unlike most members of Choanoflagellata and Ichthyosporea, the two chosen species are experimentally amenable for the kind of study reported here.
Materials and Methods
M. brevicollis was obtained from the American Type Culture Collection (ATCC 50154). The organism was grown in batch cultures at ≈25°C in sterile natural seawater and fed with live bacteria (Enterobacter aerogenes, ATCC 13048). A. parasiticum JAP-7-2 was obtained from R. W. Lichtwardt (Department of Botany, University of Kansas, Lawrence) and cultured in liquid medium (1% yeast extract/3% glycerol) under shaking conditions.
Cells of M. brevicollis and A. parasiticum were suspended in sorbitol buffer (0.6 M sorbitol/5 mM EDTA/50 mM Tris, pH 7.4) and broken mechanically by shaking with glass beads. Mitochondria were collected by differential centrifugation and subsequently lysed in the presence of 1% SDS and 100 μg/ml proteinase K. SDS was eliminated by NaCl precipitation. To exclude potential DNA fragmentation during cell disruption, A. parasiticum mtDNA was also prepared without mechanical treatment, which did not change the mtDNA pattern. Nucleic acids were fractionated by isopycnic centrifugation in CsCl/Hoechst 33258 dye. The upper (A+T-rich) band was recentrifuged. Random clone libraries were constructed by nebulization of the purified mtDNA (fragment sizes 1–3 kbp) and cloning into pBluescript (Stratagene). The corresponding protocol is available at the web site of the organelle genome megasequencing program, ogmp (http://megasun.bch.umontreal.ca/ogmp/).
Clones were sequenced by a combination of automated Li-Cor (Lincoln, NE) and manual methods. Sequence readings were assembled and proofread using the gap software suite (19). The fasta program (20) was used for searches in local databases, the blast network service (21) for similarity searches in GenBank at the NCBI, and clustalw (22) for multiple protein alignments. Custom-made batch utilities used for submitting queries and browsing the results are available at the OGMP web site.
Results and Discussion
The 76,568-bp mtDNA of M. brevicollis is circular-mapping and has an exceptionally high overall A+T content (86%). Fig. 1 depicts the gene map inferred from the mtDNA sequence. This mitochondrial genome contains a total of 55 different genes (protein-coding, rRNA and tRNA combined, and two unidentified ORFs), i.e., 1.5–3 times as many as are found in animals. The extra genes of M. brevicollis mtDNA, otherwise absent from animal mtDNAs, specify 11 ribosomal proteins, ATP synthase subunit 9, and the MttB protein, which is involved in Sec-independent membrane targeting and protein translocation (23). Intergenic regions of M. brevicollis mtDNA are extremely A+T-rich (93%), several hundred to several thousand base pairs long, and constitute more than half (53%) of the Monosiga mitochondrial genome. The proportion of spacer regions is unusually high compared with mtDNA of other protists, fungi, and, especially, animals, being more characteristic of that observed in the spacious land plant mitochondrial genome (24, 25). Intergenic regions in the M. brevicollis mtDNA encompass various types of repeat motifs (up to 137 bp in length) that consist nearly exclusively of A and T. The several hundred direct repeats are mostly ≈25 nt long and often arranged in tandem arrays of up to 12 motifs, whereas the hundred or so inverted repeats are generally ≈50 bp long and dispersed. In addition to these repeats, four group I introns contribute to the large size of M. brevicollis mtDNA. All these features of the choanoflagellate mtDNA are in sharp contrast to the compactness of animal mitochondrial genomes. Table 1 summarizes the characteristics of M. brevicollis mtDNA, showing that it most closely resembles a typical protist mitochondrial genome in overall size and gene content.
Figure 1.
Genetic map of M. brevicollis mtDNA. Exons are depicted by black boxes, group I introns by open boxes, and intronic ORFs (coding for potential endonucleases) by shaded boxes. All genes are transcribed in a clockwise direction. Black labels denote genes common to animal and fungal mtDNAs, blue labels represent genes typically not found in either animal or fungal mtDNAs, and green labels denote unique ORFs of unknown function.
Table 1.
Features of the mitochondrial genomes of A. parasiticum, M. brevicollis, and other eukaryotes
| Taxon | Size, kbp | Coding portion | No. of tRNAs | Genes coding for
|
No. of
|
UGA (Trp) | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| rRNAs | Respiratory chain subunits | Ribosomal proteins | Other | ORFs | Introns | |||||
| A. parasiticum | >200 | ≈20% | ≥25 (47) | rnl, rns | atp6,8,9 cob, cox1-3nad1Δ,2Δ3,4Δ, 4L,5,6Δ | rps3,4,13 | — | ≥24 | ≥21 (I) ≥2 (II) | + |
| M. brevicollis | 76.6 | 46.9% | 25 | rnl, rns | atp6,8,9cob, cox1-3nad1–4,4L,5,6 | rps3,4,8,12–14,19rpl2,5,14,16 | mttB | 2 | 4 (I) | + |
| Animals | 13–22 | 62–95% | 2–23 | rnl, rns | atp6,(8) cob, cox1-3nad1–4,4L,5,6 | — | (mutS) | — | 0–2 | + |
| Fungi | 19–94 | 41–89% | 7–26 | rnl, rns | atp6,8,(9) cob, cox1-3 (nad1–4,4L,5,6) | (rps3) | (rnpB) | 0–36 | 0–37 | (+) |
| Plants | 187–367 | 46–65% | 22–27 | rnl, rns, rrn5 | atp1,6,8,9 cob, cox1-3 nad1–4,4L,5,6, (7),9 (sdh3,4) | rps(1,2),3,4,7, (8,10,11), 12,13, (14,19) rpl(2),5,(6,16) | mttByeiR,U,(V) ymf39 | ≈50–100 | 20–32 | − |
| R. americana | 69.0 | 91.3% | 26 | rnl, rns, rrn5 | atp1,3,6,8,9cob, cox1-3nad1-4,4L,5-11 sdh2-4 | rps1-4,7,8,10-14,19rpl1,2,5,6,10,11,14,16, 18–20,27,31,32,34 | cox11, mttBrnpB, rpoA-DsecY, tufAyejR,U,V,Wymf39 | 3 | 1 (II) | − |
Data for fungi, animals, and plants have been compiled from completely sequenced mtDNAs and are taken from NCBI's complete genome section, www.ncbi.nlm.nih.gov/PMGifs/Genomes/euk_o.html, and GOBASE, http://megasun.bch.umontreal.ca/gobase. Sizes for mtDNA larger than those indicated have been reported, e.g., in animals, but complete sequences are not available for these particular cases.
Data for A. parasiticum and M. brevicollis are reported here. Data for R. americana are from ref. 11 and GenBank accession no. NC_001823. +, Feature present; −, feature absent; (+), feature present only in some taxa of the particular group; Δ, gene fragment. In the case of A. parasiticum tRNA genes, the larger number in parentheses includes tRNA gene duplicates. Genes enclosed in parentheses are present only in some taxa of the particular group. Roman numerals (I and II) indicate the intron group. Genes and corresponding gene products are: atp1–9, ATP synthase subunits; cob, cytochrome b apoprotein; cox1–3, cytochrome c oxidase subunits; cox11, cytochrome oxidase assembly protein; mttB, Sec-independent protein translocase; nad1–11, NADH dehydrogenase subunits; rnl, LSU rRNA; rns, SSU rRNA; rrn5, 5S rRNA; rnpB, RNAse P RNA; rpl1–34, LSU ribosomal proteins; rps1–19, SSU ribosomal proteins; rpoA-D, RNA polymerase subunits and sigma factor; sdh2–4, succinate dehydrogenase subunits; secY, SecY-type preprotein translocase subunit; tufA, elongation factor A; yejR-W, ABC transporter involved in cytochrome c biogenesis; ymf39, conserved membrane protein of unknown function. Coding portion, the percentage of sequence having a coding function, including identified genes, gene fragments, unidentified ORFs, introns, and intron ORFs. ORFs, only free-standing ORFs, not those included in introns. Only ORFs ≥ 100 amino acids are enumerated, except for fungi and R. americana, where ORFs ≥60 residues are listed.
Analysis of A. parasiticum mtDNA revealed that it has a most unusual structure. Electrophoretic separation on an agarose gel indicates that the isolated mtDNA consists of a large set of quite small (0.3–8.3 kbp) chromosomes comprising several hundred distinct molecular species (Fig. 2). The uneven band intensities observed after gel electrophoresis suggest that the different chromosomes are present in nonstoichiometric ratio and/or comigrate.
Figure 2.
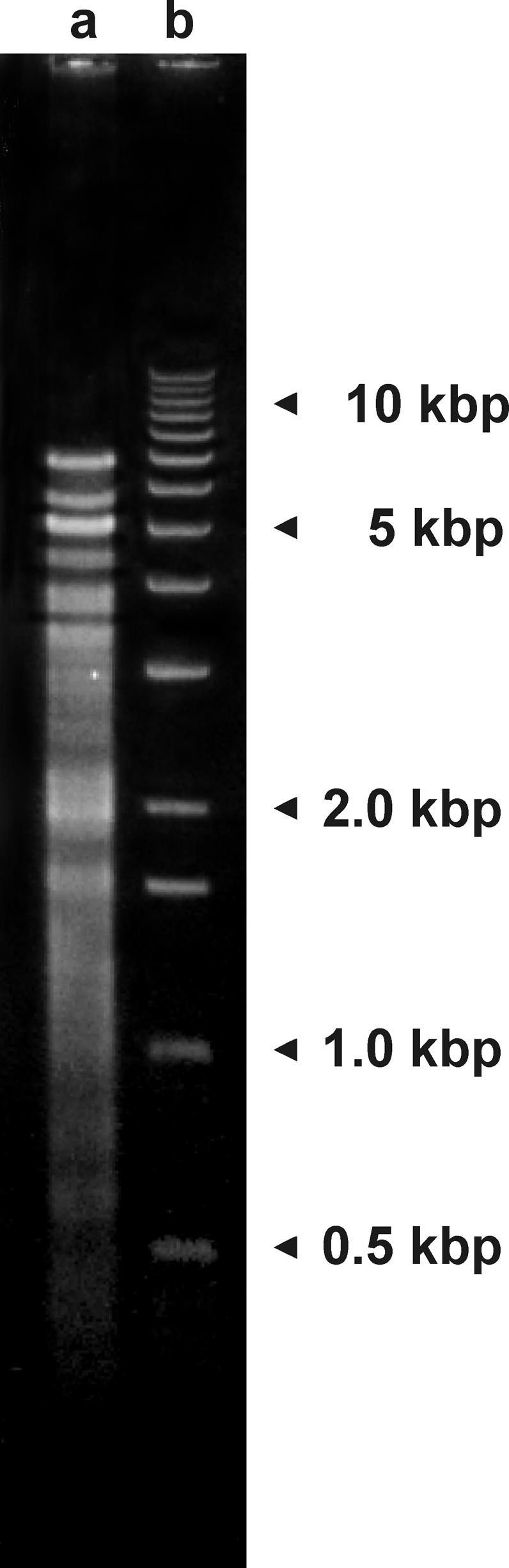
Mitochondrial chromosomes of A. parasiticum, separated by agarose gel electrophoresis. Lane a, mtDNA fraction, as isolated. Lane b, A 1-kbp ladder (Stratagene) included as a size marker.
The Amoebidium mitochondrial chromosomes are linear, as determined by electrophoresis in agarose gels of differing concentration (data not shown), and as confirmed by sequence analysis. Their extremities appear to lack sophisticated decorations, such as covalently closed single-stranded termini or covalently attached proteins. Such structures, which are found in several other linear mitochondrial genomes (26), are notorious for significantly reducing the cloning efficiency of terminal regions (27), a difficulty that was not encountered in the case of A. parasiticum mtDNA.
The organization of the A. parasiticum mitochondrial genome is unprecedented. Although a dozen or so mitochondrial genomes are known to exist as linear chromosomes, these consist of only a single type of DNA molecule (25–27). Exceptions are found in four cnidarian classes, including Hydra, whose mtDNA is in two linear pieces (28, 29). A few additional cases of multiple (>2) but circular-mapping mitochondrial chromosomes have been reported. For example, the mitochondrial genome consists of three circular-mapping molecules in Spizellomyces punctatus, a chytridiomycete fungus (GenBank accession nos. AF404303–AF404305), and at least that many in Globodera, a parasitic nematode (30). Moreover, single-gene minicircles were detected in mitochondria of Dicyema, an enigmatic multicellular organism (probably a highly derived animal; ref. 31). Interestingly, single-gene minicircles carrying chloroplast genes have been isolated from dinoflagellates, whereas chloroplast DNAs from other eukaryotic groups are large, circular, multigene molecules (32).
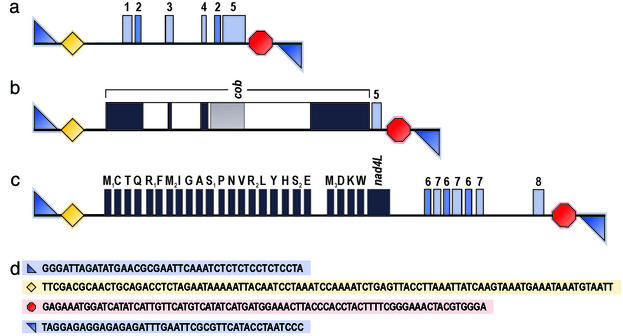
To date, we have sequenced 39 chromosomes of A. parasiticum mtDNA to completion, focusing on the larger ones that contain genes. Together with partial sequences from an additional ≈40 chromosomes, a total of 161,071 bp of A. parasiticum mtDNA have been determined, an amount sufficient to provide us with a global picture of the unique nature of this organelle genome. Fig. 3 depicts the three types of chromosomes that can be discerned in A. parasiticum mtDNA: (i) small molecules without identified coding function (Fig. 3a), (ii) medium-sized molecules carrying a single gene (Fig. 3b), and (iii) large molecules harboring multiple genes (Fig. 3c). The size range of sequenced chromosomes is listed in Tables 2 and 3.
Figure 3.
Structure and genetic maps of three representative mitochondrial chromosomes of A. parasiticum. Genes, introns, and ORFs are represented by boxes, as in Fig. 1. Terminal repeat motifs are represented by a blue triangle (repx), a yellow lozenge (repa), and a red octagon (repb), and internal repeat motifs by blue rectangles. The numerals denote different internal repeat motifs. Depicted chromosomes have the following identifiers and GenBank accession nos.: (a) 0316riay49 (AF538050); (b) 2902riaz91 (AF538047); and (c) 2153iay75 (AF538045). (d) Sequence of the terminal repeats of chromosome 0316riay49 (GenBank accession no. AF538050). The background colors blue, yellow, and red correspond to repx, repa, and repb, respectively.
Table 2.
Mitochondrial chromosomes of A. parasiticum containing genes and gene fragments
| Chromosome | Size, kbp | Genes and gene fragments* encoded | Completely sequenced | GenBank accession no. |
|---|---|---|---|---|
| 0681iay42 | 8.36 | rnl (6 introns), 1 trn | + | AF538042 |
| 0555iay58 | 7.26 | cox1 (9 introns) | + | AF538043 |
| 0283riay48 | >6.91 | atp8, cox3 (1 intron), rns (2 introns) | − | AF538044 |
| 2153iay75 | 5.07 | nad4L, 24 trn | + | AF538045 |
| 0277iay24 | >4.29 | atp9, 9 trn | − | AF538046 |
| 0229riay36 | 4.12 | cox1Δ, rnlΔ | + | — |
| 0242riay48 | 3.94 | 9 trn | + | — |
| 0150iay48 | 3.65 | atp9Δ | + | — |
| 2902riaz91 | 3.34 | cob (3 introns) | + | AF538047 |
| 0356iay27 | 3.24 | nad5 (2 introns) | + | AF538048 |
| 0403iay27 | 2.95 | cox2 | + | AF538049 |
| 0512iay29 | 2.00 | rps3 | + | — |
| 0101riay43 | 1.75 | nad2Δ | + | — |
| 1403riay71 | 1.68 | nad3 | + | AF538051 |
| 0417iay60 | 1.68 | atp6, rps13 | + | AF538052 |
| 0090riay34 | 1.66 | nad6Δ | + | — |
| 0389iay27 | 1.50 | rps13Δ | + | — |
| 2380riay71 | >1.38 | rps4 | − | — |
| 1206iay44 | >1.21 | atp6Δ, nad4Δ | − | — |
| 0829riaz14 | >1.02 | nad1Δ | − | — |
Gene fragments are marked by Δ. Fragments of atp6, cox1, nad1, nad2, nad4, rnl, rns, rps13, and a group II intron were found in incompletely sequenced chromosomes (see Table 3); only two of the corresponding unassembled contigs (i.e., 1206iay44 and 0829riaz14) are listed in this table. +, yes; −, no. —, unpublished.
Table 3.
Coding content of sequenced mitochondrial chromosomes of A. parasiticum
| Complete chromosomes
|
Unassembled contigs*
|
|||||
|---|---|---|---|---|---|---|
| No. | Size range | GenBank accession no. | No. | Size range | GenBank accession no. | |
| Genes + gene fragments | 10 | 1.68–8.36 | AF538042, AF538043, AF538045, AF538047, AF538048, AF538049, AF538051, AF538052 | 5 | >1.18–>6.90 | AF538044, AF538046 |
| Gene fragments only | 5 | 1.21–4.12 | — | 9 | >1.20–>3.31 | — |
| No identified genes/gene fragments | 24 | 1.37–2.00 | AF538050 | 33 | >0.17–>2.14 | — |
—, unpublished.
Some of these contigs may derive from the same chromosome.
The linear mitochondrial chromosomes known from other eukaryotes carry 1- to 3-kbp-long inverted terminal repeats, which either (i) do not contain any recognizable coding sequence, as in the fungus Hyaloraphidium curvatum (27), (ii) include ORFs, as in the golden alga Ochromonas danica (GenBank accession no. NC_002571), or (iii) harbor identified genes, as in the ciliate Tetrahymena pyriformis (33). In addition to the gene-containing terminal repeats, the linear mitochondrial chromosome of this ciliate carries at its extremities telomeric sequences that consist of tandem arrays of 31-bp-long repeat motifs. In contrast to all other instances described to date, the linear chromosomes of A. parasiticum are characterized by a common pattern of short terminal repeat structures. The extremities consist invariably of a 43- to 45-bp terminal sequence motif in inverted orientation (termed repx), followed by one subterminal copy of repa (≈100 bp) at one end of the chromosomes and one subterminal copy of repb (≈65 bp) at the other (Fig. 3d). Whereas sequence variation within the repx repeat family is ≈10%, the two copies of repx in any given chromosome are identical. [So far, only one exception to this rule has been detected, in chromosome 2153iay75 (GenBank accession no. AF538045), whose two repx motifs differ by three positions in their central portion.] Intriguingly, the first five residues of repx are identical to the terminal sequence of the partially analyzed 8.1-kbp linear molecule from Hydra mtDNA (29). It is conceivable that the repx sequence, which is G-rich between positions 1 and 20 and C-rich between positions 29 and 42 (Fig. 3d), has the propensity to form a guanine quadruplex structure like that known to stabilize nuclear chromosome ends (34, 35).
The two subterminal repeats in A. parasiticum mtDNA (repa and repb) are less well conserved, with a length variation of ≈30% in both cases and sequence identity of >65% for repa and >55% for repb. Interestingly, in all gene-carrying chromosomes, transcription is in the direction repa → repb, strongly suggesting that these motifs play a role in transcription initiation and/or termination. Finally, numerous other repeats distinct from the end-localized repx, repa, and repb motifs are found in the central portions of the chromosomes. These internal repeats belong to at least 20 different classes, are 50–250 bp long, and are arranged in dispersed single units and tandem arrays.
In the currently determined portion of the A. parasiticum mitochondrial genome, we have identified a total of 44 distinct genes and an exceptionally large number (>100) of coding regions, including multiple gene copies and gene fragments (Tables 2 and 3). All genes of the basic set common to fungal and animal mtDNAs are present, with four exceptions. The genes nad1, nad2, nad4, and nad6 (coding for NADH dehydrogenase subunits) are so far only represented by incomplete fragments. We expect that the missing nad genes are located on other, as-yet-unsequenced mitochondrial chromosomes. Among the multicopy genes, those encoding tRNAs are the most abundant. Half of the 25 distinct tRNA gene species occur in up to four copies of essentially identical sequence. Moreover, A. parasiticum mtDNA contains at least four protein-coding genes in addition to the basic animal set, namely atp9 and the ribosomal protein genes rps3, rps4, and rps13, as well as numerous ORFs without recognizable counterparts in other genomes (Table 1). We presume that some of these ORFs might code for additional ribosomal proteins whose sequences have diverged too much to be identified as such. A well documented case of a barely recognizable mitochondrion-encoded ribosomal protein gene is rps11 of the golden alga Chrysodidymus synuroideus. This gene was only discovered through gene order and sequence comparison including closely related species (36). Finally, with its 23 or more introns (predominantly of group I but at least two of group II), A. parasiticum mtDNA is very intron-rich. To date, the largest numbers of mitochondrial introns have been found in mtDNAs of land plants and some fungi; e.g., 32 and 37 introns, respectively, in the mtDNA of the liverwort, Marchantia polymorpha (37), and in the chytridiomycete fungus, Rhizophydium (GenBank accession no. NC_003053).
The results described above graphically illustrate that mitochondrial genome architecture is markedly different among A. parasiticum, M. brevicollis, and multicellular animals. Few features of their mtDNAs corroborate the close phylogenetic relationship of these organisms evident from sequence-based phylogenetic tree construction (14). The only common characteristic of all three taxa is that the mitochondrial UGA (“stop”) codon specifies tryptophan, and is even preferred over UGG(Trp) codons (Table 1); in contrast, lower fungi and many protists do not use the UGA codon in their mitochondrial protein-coding regions (24, 38). Moreover, A. parasiticum, M. brevicollis, and perhaps all hexacoral animals (4, 5) share an intron in their nad5 genes that is inserted at the same point. [Note, however, that a positionally equivalent intron has also been found in certain fungal groups and is seen sporadically in plants and protists (37, 39, 40).] In A. parasiticum and M. brevicollis, the mtDNA-encoded trnS(gcu) sequences share highly unusual structural features, although homologous tRNA genes are generally among the least well conserved mitochondrial genes. In both protists, nucleotide 8, connecting the aminoacyl and D stems, is missing; the second nucleotide in the D loop is a U instead of a canonical A; and position 26 is pyrimidine instead of purine. Remarkably, and similar to the situation in cnidarian animals (4–6, 29), none of the mitochondrial tRNAs from A. parasiticum or M. brevicollis has a truncated D or T loop structure, features that are otherwise widespread in animal mitochondria.
In both A. parasiticum and M. brevicollis, the secondary structures of the mtDNA-encoded large subunit (LSU) and small subunit (SSU) rRNAs retain numerous features that are shared with eubacteria and minimally derived eukaryotes, and that set them apart from the highly reduced secondary structures of their animal relatives (including cnidarians). Notably, these protist mitochondrial SSU and LSU rRNA sequences closely approximate the sizes of their eubacterial homologs (1,596 and 2,878 nt in M. brevicollis; 1,385 and 3,053 nt in A. parasiticum, compared with 1,542 and 2,904 nt in E. coli). Moreover, the LSU rRNA structures contain the typical 5′-terminal 5.8S-like and 3′-terminal 4.5S-like domains that are completely missing in their metazoan mitochondrial counterparts. (Secondary structure diagrams of these protist mitochondrial rRNAs can be found in the GOBASE database, http://megasun.bch.umontreal.ca/gobase.)
Conclusions
Systematic and comprehensive investigation of protist mitochondrial genomes has proven a powerful tool for new discoveries (24, 25, 41), including the finding reported here of an extraordinary mitochondrial genome architecture in A. parasiticum mtDNA, which consists of several hundred linear chromosomes. The large number of chromosomes and apparent absence of the centromeric structures typical of nuclear chromosomes raise questions about the mechanism of concerted replication and equal segregation of mtDNA during mitochondrial division and subsequent cell division in A. parasiticum. A similar dilemma exists in kinetoplastid protists (Trypanosoma, Leishmania, Crithidia, and Bodo), whose mitochondria contain up to a few dozen maxicircles and up to several thousand minicircles of equal length [0.5–10 kbp, depending on the species (42)]. The minicircle DNAs do not code for genes but instead specify guide RNAs that are involved in posttranscriptional editing of mitochondrial mRNAs (for a review, see ref. 43). In one subgroup of the Kinetoplastida, i.e., the Trypanosomatina, the DNA circles are concatenated and interlocked, forming a giant network (kDNA). This network structure is believed to facilitate, by an as-yet-unknown mechanism, the segregation of minicircles. However, other kinetoplastid protists, such as Bodo caudatus, contain a large number of minicircles that do not form a network. As documented here, the linear mitochondrial chromosomes of A. parasiticum also exist as separate monomers. In organisms such as A. parasiticum and B. caudatus, whose mitochondrial genome consists of many physically separate molecules, the mechanism of mitochondrial chromosome segregation remains obscure.
Our findings suggest that the last common ancestor of the Holozoa [multicellular animals plus their closest unicellular relatives, Ichthyosporea and Choanoflagellata (14)] possessed a gene-rich mtDNA. This was unexpected from a phylogenetic point of view, because Fungi, the sister clade of Holozoa (14), have virtually the same highly reduced gene content as do animals. Before the description of mitochondrial genomes from Choanozoa and Ichthyosporea reported here, the most parsimonious hypothesis would have predicted that the shared missing mitochondrial genes in Fungi and animals had already been lost in their common predecessor. However, the data presented here imply that the loss of mitochondrial genes, especially those coding for ribosomal proteins, has occurred independently in the Holozoa and Fungi.
Our data also suggest that in ancestral Holozoa, mitochondrial genome architecture underwent substantial remodeling by three radically different routes: one involving genome fragmentation and rampant expansion of genome size through the accumulation of repeat sequences (ichthyosporean lineage); a second pathway characterized by a marked increase in the size of intergenic regions but otherwise retaining features of an ancestral protist mtDNA (choanoflagellate lineage); and a third route featuring extensive gene loss coupled with size contraction, resulting in an extremely tight gene arrangement (metazoan lineage). As mitochondrial genome evolution in the two clades of unicellular relatives of animals follows an expansionary trend, mtDNA streamlining likely occurred relatively recently in the evolutionary history of the Metazoa, concomitant with the emergence of a multicellular body plan. To further test this hypothesis, investigation of mitochondrial genome structure in the most early diverging animal group, the sponges, is underway.
Acknowledgments
We thank R. W. Lichtwardt for supplying an axenic culture of A. parasiticum, S. Teijeiro for critically reading the manuscript, I. Plante for clone library construction, Z. Wang and S. Cagna for DNA sequencing, and M. Coulombe for data compilations and graphical art work. This project was supported by Canadian Institutes for Health Research Grants MSP-14226 and MOP-42475 and equipment grants from Sun Microsystems (Palo Alto, CA) and Li-Cor (Lincoln, NE). Salary and interaction support from the Canadian Institute for Advanced Research (G.B., M.W.G., and B.F.L.) and the Canada Research Chairs Program (to M.W.G.) is gratefully acknowledged.
Footnotes
References
- 1.Anderson S, Bankier A T, Barrell B G, de Bruijn M H, Coulson A R, Drouin J, Eperon I C, Nierlich D P, Roe B A, Sanger F, et al. Nature. 1981;290:457–465. doi: 10.1038/290457a0. [DOI] [PubMed] [Google Scholar]
- 2.Wolstenholme D R. Int Rev Cytol. 1992;141:173–216. doi: 10.1016/s0074-7696(08)62066-5. [DOI] [PubMed] [Google Scholar]
- 3.Boore J L. Nucleic Acids Res. 1999;27:1767–1780. doi: 10.1093/nar/27.8.1767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beagley C T, Okimoto R, Wolstenholme D R. Genetics. 1998;148:1091–1108. doi: 10.1093/genetics/148.3.1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Van Oppen M J H, Catmull J, McDonald B J, Hislop N R, Hagerman P J, Miller D J. J Mol Evol. 2002;55:1–13. doi: 10.1007/s00239-001-0075-0. [DOI] [PubMed] [Google Scholar]
- 6.Pont-Kingdon G, Okada N A, Macfarlane J L, Beagley C T, Watkins-Sims C D, Cavalier-Smith T, Clark-Walker G D, Wolstenholme D R. J Mol Evol. 1998;46:419–431. doi: 10.1007/pl00006321. [DOI] [PubMed] [Google Scholar]
- 7.Rigaa A, Monnerot M, Sellos D. J Mol Evol. 1995;41:189–195. doi: 10.1007/BF00170672. [DOI] [PubMed] [Google Scholar]
- 8.Fuller K M, Zouros E. Curr Genet. 1993;23:365–369. doi: 10.1007/BF00310901. [DOI] [PubMed] [Google Scholar]
- 9.Beck J L, Hyman B C. Curr Genet. 1988;14:627–636. doi: 10.1007/BF00434089. [DOI] [PubMed] [Google Scholar]
- 10.Boyce T M, Zwick M E, Aquadro C F. Genetics. 1989;123:825–836. doi: 10.1093/genetics/123.4.825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lang B F, Burger G, O'Kelly C J, Cedergren R, Golding G B, Lemieux C, Sankoff D, Turmel M, Gray M W. Nature. 1997;387:493–497. doi: 10.1038/387493a0. [DOI] [PubMed] [Google Scholar]
- 12.O'Kelly C J. J Eukaryotic Microbiol. 1993;40:627–636. [Google Scholar]
- 13.James-Clark H. Am J Sci. 1866;1:113–114. [Google Scholar]
- 14.Lang B F, O'Kelly C, Nerad T, Gray M W, Burger G. Curr Biol. 2002;12:1773–1778. doi: 10.1016/s0960-9822(02)01187-9. [DOI] [PubMed] [Google Scholar]
- 15.Herr R A, Ajello L, Taylor J W, Arseculeratne S N, Mendoza L. J Clin Microbiol. 1999;37:2750–2754. doi: 10.1128/jcm.37.9.2750-2754.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ragan M A, Goggin C L, Cawthorn R J, Cerenius L, Jamieson A V C, Plourde S M, Rand T G, Söderhäll K, Gutell R R. Proc Natl Acad Sci USA. 1996;93:11907–11912. doi: 10.1073/pnas.93.21.11907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ustinova I, Krienitz L, Huss V A R. Protist. 2000;151:253–262. doi: 10.1078/1434-4610-00023. [DOI] [PubMed] [Google Scholar]
- 18.Benny G L, O'Donnell K. Mycologia. 2000;92:1133–1137. [Google Scholar]
- 19.Staden R. Mol Biotechnol. 1996;5:233–241. doi: 10.1007/BF02900361. [DOI] [PubMed] [Google Scholar]
- 20.Pearson W R. Methods Enzymol. 1990;183:63–98. doi: 10.1016/0076-6879(90)83007-v. [DOI] [PubMed] [Google Scholar]
- 21.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 22.Thompson J D, Higgins D G, Gibson T J. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Weiner J H, Bilous P T, Shaw G M, Lubitz S P, Frost L, Thomas G H, Cole J A, Turner R J. Cell. 1998;93:93–101. doi: 10.1016/s0092-8674(00)81149-6. [DOI] [PubMed] [Google Scholar]
- 24.Gray M W, Lang B F, Cedergren R, Golding G B, Lemieux C, Sankoff D, Turmel M, Brossard N, Delage E, Littlejohn T G, et al. Nucleic Acids Res. 1998;26:865–878. doi: 10.1093/nar/26.4.865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lang B F, Gray M W, Burger G. Annu Rev Genet. 1999;33:351–397. doi: 10.1146/annurev.genet.33.1.351. [DOI] [PubMed] [Google Scholar]
- 26.Nosek J, Tomaska L, Fukuhara H, Suyama Y, Kovac L. Trends Genet. 1998;14:184–188. doi: 10.1016/s0168-9525(98)01443-7. [DOI] [PubMed] [Google Scholar]
- 27.Forget L, Ustinova J, Wang Z, Huss V A R, Lang B F. Mol Biol Evol. 2002;19:310–319. doi: 10.1093/oxfordjournals.molbev.a004084. [DOI] [PubMed] [Google Scholar]
- 28.Warrior R, Gall J. Arch Sci (Geneva) 1985;38:439–445. [Google Scholar]
- 29.Pont-Kingdon G, Vassort C G, Warrior R, Okimoto R, Beagley C T, Wolstenholme D R. J Mol Evol. 2000;51:404–415. doi: 10.1007/s002390010103. [DOI] [PubMed] [Google Scholar]
- 30.Armstrong M R, Blok V C, Phillips M S. Genetics. 2000;154:181–192. doi: 10.1093/genetics/154.1.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Watanabe K I, Bessho Y, Kawasaki M, Hori H. J Mol Biol. 1999;286:645–650. doi: 10.1006/jmbi.1998.2523. [DOI] [PubMed] [Google Scholar]
- 32.Zhang Z, Green B R, Cavalier-Smith T. Nature. 1999;400:155–159. doi: 10.1038/22099. [DOI] [PubMed] [Google Scholar]
- 33.Burger G, Zhu Y, Littlejohn T G, Greenwood S J, Schnare M N, Lang B F, Gray M W. J Mol Biol. 2000;297:365–380. doi: 10.1006/jmbi.2000.3529. [DOI] [PubMed] [Google Scholar]
- 34.Sen D, Gilbert W. Nature. 1988;28:364–366. doi: 10.1038/334364a0. [DOI] [PubMed] [Google Scholar]
- 35.Spackova N, Berger I, Sponer J. J Am Chem Soc. 2001;123:3295–3307. doi: 10.1021/ja002656y. [DOI] [PubMed] [Google Scholar]
- 36.Chesnick J M, Goff M, Graham J, Ocampo C, Lang B F, Seif E, Burger G. Nucleic Acids Res. 2000;28:2512–1518. doi: 10.1093/nar/28.13.2512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Oda K, Yamato K, Ohta E, Takemura Y N M, Nozato N, Akashi K, Ogura T K Y, Kohchi T, Ohyama K. J Mol Biol. 1992;223:1–7. doi: 10.1016/0022-2836(92)90708-r. [DOI] [PubMed] [Google Scholar]
- 38.Paquin B, Lang B F. J Mol Biol. 1996;255:688–701. doi: 10.1006/jmbi.1996.0056. [DOI] [PubMed] [Google Scholar]
- 39.Paquin B, Roewer I, Wang Z, Lang B F. Can J Bot. 1995;21:S180–S185. [Google Scholar]
- 40.Kroymann J, Zetsche K. J Mol Evol. 1998;47:431–440. doi: 10.1007/pl00006400. [DOI] [PubMed] [Google Scholar]
- 41.Gray M W, Burger G, Lang B F. Science. 1999;283:1476–1481. doi: 10.1126/science.283.5407.1476. [DOI] [PubMed] [Google Scholar]
- 42.Lukes J, Guilbride D L, Votypka J, Zikova A, Benne R, Englund P T. Eukaryotic Cell. 2002;1:495–502. doi: 10.1128/EC.1.4.495-502.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Simpson L. In: RNA World. Gesteland R, Atkins J, Cech T, editors. Plainview, NY: Cold Spring Harbor Lab. Press; 1999. pp. 585–608. [Google Scholar]