Abstract
The Gram-negative bacterial pathogen Yersinia delivers six effector proteins into the host cells to thwart the host innate immune response. One of the effectors, YopT, causes the disruption of the actin cytoskeleton and contributes to the inhibition of phagocytosis of the pathogen. YopT functions as a cysteine protease to cleave Rho family GTPases. We have analyzed the YopT cleavage products of Rho GTPases by TLC and determined their chemical structure by MS. Amino acid labeling experiments were performed to locate the exact site in RhoA where the YopT cleavage occurs. Our data unambiguously demonstrate that YopT cleaves N-terminal to the prenylated cysteine in RhoA, Rac, and Cdc42 and that the cleavage product of the GTPases is geranylgeranyl cysteine methyl ester. YopT cleaves GTP- and GDP-bound forms of RhoA equally, suggesting that the cleavage does not depend upon the conformation status of the GTPases. YopT also cleaves both farnesylated and geranylgeranylated forms of RhoA. The polybasic sequence in the C terminus of RhoA is essential for YopT substrate recognition and cleavage.
The causative agent of plague is Yersinia pestis, whereas Yersinia pseudotuberculosis and Yersinia enterocolitica cause gastrointestinal disorders in humans. All three pathogenic species of Yersinia harbor an extrachromosomal plasmid of 70 kb that is essential for Yersinia pathogenicity. This plasmid encodes the genes of a type III secretion system, a sophisticated translocation apparatus highly conserved among a number of Gram-negative pathogenic bacteria (1, 2). On contact with the host cell, the Yersinia type III secretion system delivers a set of effector proteins termed Yops (Yersinia outer proteins) into the host cell. Six Yop effectors (YopH, YopE, YopJ/P, YpkA/YopO, YopT, and YopM) have been identified to date, and they function to attenuate the host immune response during infection (3–5). Five of the six Yops have catalytic activities that seem to be essential for their pathogenic functions (6). YopH, a protein tyrosine phosphatase (7), inhibits the tyrosine phosphorylation signaling that is essential for the assembly of focal adhesion complexes (8–10), resulting in the inhibition of macrophage phagocytosis. YpkA/YopO (a serine/threonine kinase) and YopE (a Rho GTPase-activating protein) contribute to the disruption of the host cytoskeleton, also preventing phagocytosis of the pathogen (11–14). YopJ blocks proinflammatory cytokine production through disrupting host NFκB and mitogen-activated protein kinase kinase signaling (15, 16). YopJ has been shown to share amino acid sequence identity with the adenovirus cysteine protease. The catalytic triad of YopJ is required for its antiinflammatory functions (16, 17).
YopT was recently identified as a cytotoxin that causes the disruption of the actin cytoskeleton and results in rounding up of host cells (18). Disruption of the host cell cytoskeleton by YopT contributes to the antiphagocytic effect of Yersinia (19). Infection of host cells with a mutant Y. enterocolitica strain secreting only YopT also induces an isoelectric point shift of RhoA (20), a small GTPase known to regulate the actin cytoskeleton (21). YopT causes the release of RhoA from cell membranes or artificial vesicles (22). Additional insights into the function of YopT were obtained from the observation that Rho family GTPases, including RhoA, Rac, and Cdc42, are all known to undergo sequential posttranslational modifications at their C-terminal CaaX box (C, cysteine; a, aliphatic residue; X, any residue) (23). The CaaX box provides the recognition elements for prenylation of the cysteine, followed by endoprotease removal of the aaX tripeptide and carboxylmethylation of the now terminal prenylated cysteine. These modifications allow for membrane anchorage of the GTPases (23). We have recently demonstrated that YopT carries out a proteolytic cleavage near the C termini of RhoA/Rac1/Cdc42 in vitro and in vivo and that this cleavage results in the removal of the lipid modification from the GTPases and their subsequent membrane detachment (24). Cleavage of the Rho GTPases by YopT is responsible for the disruption of actin cytoskeleton. The proteolytic activity of YopT is dependent upon the invariant Cys/His/Asp residues that are conserved in a novel family of cysteine proteases involved in both animal and plant bacterial pathogenesis (24). In the present study, we have identified the YopT cleavage site in Rho GTPases using specific radiolabeling experiments, as well as MS. Our analysis suggests that YopT recognizes a sequence of basic amino acids along with the lipid-modified cysteine methyl ester in the C termini of Rho GTPases. Cleavage of the GTPases by YopT removes the geranylgeranyl cysteine methyl ester (GGCM) from the GTPases, irreversibly releasing the GTPases from their membrane attachment.
Materials and Methods
Plasmids and Protein Expression.
The YopT gene was amplified by PCR from the Y. pseudotuberculosis virulence plasmid provided by James Bliska (State University of New York, Stony Brook). Mammalian GST-Rho GTPases and GST-H-Ras fusion proteins were expressed in the pEBG-3X vector, and the template constructs have been described (24). GST-CLVL, GST-ARRGKKKSGCLVL (C-terminal 13 residues in RhoA), GST-RhoAL63 [K(R)5Q], GST-RhoAL63(V192Y), GST-RhoAL63(L193M), and GST-H-Ras (1–166)-ARRGKKKSGCLVL expression constructs were generated by a standard PCR cloning strategy. The bacterial GST-RhoA expression plasmid was obtained by inserting the RhoA coding sequence into the pGEX-KG vector via the BamHI/EcoRI sites (25). All other constructs used have been previously described (24). Recombinant GST-YopT protein was expressed and purified as previously described (24). The protocol for recombinant GST-RhoA expression and purification was essentially the same as that for recombinant GST-YopT except that the Escherichia coli DH5α strain was used. Protein concentrations were estimated by Coomassie blue staining of SDS/PAGE gels using BSA standards.
Cell Culture, Transfection, and Metabolic Labeling.
HEK293T cells were maintained in DMEM containing 10% (vol/vol) FBS, 2 mM glutamine, and 100 μg/ml penicillin/streptomycin (Life Technologies, Grand Island, NY) at 37°C in a 5% CO2 incubator. Transfection was performed by using the FuGENE 6 transfection kit (Roche Molecular Biochemicals) as recommended by the manufacturer. [3H]Mevalonate labeling was carried out as previously described (24). For in vivo labeling, 6 × 106 HEK293T cells were transfected with 5 μg of GST-RhoAL63 constructs. Sixteen hours after transfection, cells were labeled with either 0.06 mCi/ml [35S]Cys/[35S]Met labeling mix (NEN) or 0.0375 mCi/ml [14C]Gly (American Radiolabeled Chemicals, St. Louis) in cysteine-free (Cys labeling) or normal DMEM (Gly labeling) medium for 12 h. Cells were then lysed in 1 ml of buffer containing 10 mM Hepes (pH 7.5), 50 mM NaCl, 1% Triton, 2 mM EDTA, and a protease inhibitor mixture (Roche), and the lysates were subjected to GST-pulldown as described below.
GST-Pulldown Assay, in Vitro Cleavage, and Lipid Extraction.
The procedures for these experiments have been described in detail previously (24). To examine the binding between YopT and RhoA, Flag-tagged catalytically inactive YopT (C139S) constructs were cotransfected with the indicated GST-RhoA constructs, and the proteins were isolated by GST affinity chromatography and separated by SDS/PAGE. GST pulldowns were then subjected to anti-Flag (Sigma) and anti-GST (Santa Cruz Biotechnology) Western blot analysis. The amount of 3H-GST-RhoA before or after YopT cleavage was visualized by autoradiography, and the total amount of GST-RhoA was measured by Western blot using anti-RhoA antibody (26C4, Santa Cruz Biotechnology). The lipid cleavage products were isolated by chloroform extraction. The resulting chloroform extracts were subjected to scintillation counting to detect the presence of [35S]Cys and [14C]Gly, TLC analysis to identify the lipid cleavage products and measure their TLC mobility, or MS analysis to determine chemical structures.
Chemical Synthesis of GGCM.
Synthesis of GGCM was accomplished by following a previously described protocol (26). Briefly, 28.3 μmol of geranylgeranyl bromide (American Radiolabeled Chemicals) was reacted with 28.3 μmol of cysteine methyl ester (Sigma) for 1.5 h on ice with stirring in a solution containing 84.9 μmol of zinc acetate in 1 ml of dimethylformamide/acetonitrile/0.025% trifluoroacetic acid [2:1:1 (vol/vol)]. The solutions were loaded onto preparative silica gel TLC plates (Whatman PF5LK), and the plates were developed in 90% isopropanol. The reaction products were detected with phosphomolybdic acid (Sigma) staining. The silica containing the putative GGCM product was removed and extracted with 6 ml of 2:1 (vol/vol) chloroform/MeOH. The solution was dried in a speed vacuum, and the sample was resuspended in isopropanol. The identity of the synthetic GGCM was confirmed by electrospray MS. The same procedure was followed to synthesize and purify 3H-labeled GGCM except that the reaction scale was lowered to 10 nmol.
TLC Analysis.
The chloroform extracts of the YopT cleavage reaction were dried down to ≈15–20 μl in a speed vacuum. The synthetic GGCM (or the corresponding radiolabel product) was dissolved in isopropanol. The samples were then spotted onto a glass-backed TLC plate (Silica gel 60, 20 × 20 cm, EM Separations Technology, Gibbstown, NJ). The TLC plate was developed in a solvent system consisting of n-propyl alcohol/ammonium hydroxide/H2O (60:30:10) as described previously (27) and dried. The YopT cleavage product with 3H-labeled lipid was visualized by autoradiography using a transcreen LE intensifying screen (Kodak).
MS.
Nanoelectrospray tandem MS (MS/MS) was performed on the chloroform extracts of the YopT cleavage reaction using a Picoview nanospray source (New Objectives, Cambridge, MA) coupled to an LCQ DECA quadrupole ion trap mass spectrometer (ThermoFinnigan, San Jose, CA). The chloroform-extracted material was loaded on a 75-μm picofrit C18 column that terminated in a 15-μm id needle (New Objectives) by direct infusion with a mobile phase consisting of 1% formic acid and 10% acetonitrile in water. The samples were eluted into the mass spectrometer at a flow rate of 200 nl/min with a mobile phase consisting of 1% formic acid and 50% acetonitrile in water. Conditions for nanoelectrospray were 1.7 kV spray voltage, 38 V capillary voltage, 170°C capillary temperature, and 35% relative collision energy. Mass spectra were acquired using the triple play mode of operation where the most abundant parent ion in full MS mode was selected for high resolution zoom scan analysis and MS/MS fragmentation to obtain a product ion spectrum.
In Vitro Geranylgeranylation of Recombinant RhoA.
To modify RhoA in vitro with a geranylgeranyl group, 10 μg of highly purified GST-RhoA was incubated with 2.4 μg of rat geranylgeranyl transferase-I (Calbiochem) and 6 μCi of 3H-geranylgeranyl pyrophosphate (Amersham Pharmacia) in a buffer containing 50 mM Hepes (pH 7.5), 2 mM DTT, 10 mM MgCl2, 0.05 mM ZnCl2, and 0.02% Triton X-100. The reaction was carried out at 30°C for 3 h. Aliquots of the reaction were loaded onto an SDS/PAGE gel, and geranylgeranylation of RhoA was confirmed by autoradiography of the gel. The geranylgeranylated GST-RhoA was then immobilized to glutathione beads and subjected to YopT cleavage following the procedures described above.
Results and Discussion
Identification of the Lipid Cleavage Products and the YopT-Cleavage Site in RhoA/Rac1/Cdc42.
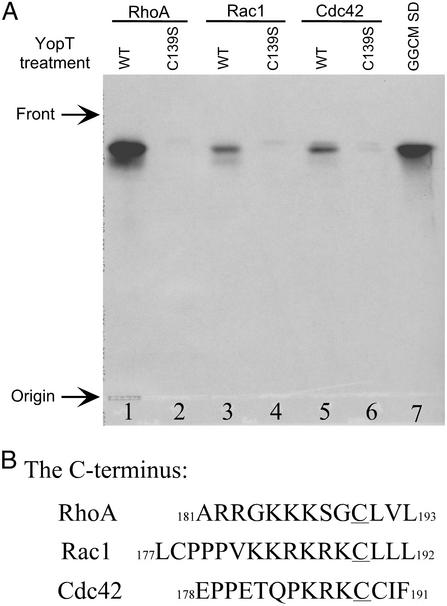
We have previously demonstrated that YopT cleaves near the carboxyl termini of Rho family GTPases by following the loss of the radiolabeled prenylated cysteine from the GTPases (24). To directly assess the proteolytic cleavage site, we used TLC to examine the 3H-geranylgeranylated cleavage product. In vitro cleavage reactions were performed initially with GST-tagged RhoAL63 (a constitutive GTP-bound form of RhoA) containing the tritium-labeled geranylgeranyl modification. The lipid cleavage product was extracted into chloroform and separated on a TLC plate. As shown in Fig. 1A, a radiolabeled product with the same Rf value as the synthetic standard, GGCM, was obtained when wild-type YopT was incubated with GST-RhoAL63 (lanes 1 and 7). In contrast, this product was absent when a protease-deficient YopT mutant protein (C139S) was used (lane 2). Equal amounts of tritium-labeled GTPase substrates were present in both reactions (data not shown). We were also interested in determining whether YopT cleaves Rac1 and Cdc42 at the same site as that of RhoA. An examination of RhoA, Rac1, and Cdc42 C termini indicates that their sequences diverge upstream of the CaaX box (Fig. 1B). Thus, if YopT cleaves N-terminal to the lipid-modified Cys in Rac1 and Cdc42, a common product (GGCM) should be produced from each GTPase. In contrast, if cleavage occurs elsewhere in the C terminus, the labeled cleavage product will be generated as di, tri (etc.) peptides, containing the lipid modification, with properties different from the synthetic standard, GGCM. We compared the mobility of the lipid cleavage products from Rac1 and Cdc42 with that of RhoA and the synthetic standard GGCM (Fig. 1A). All of the lipid cleavage products have the same Rf value as observed for the GGCM synthetic standard. Similar results were obtained using different solvent systems (data not shown). Collectively, these results suggest that YopT cleaves Rho family GTPases (RhoA/Rac1/Cdc42) at a common site that is likely located N-terminal to the prenylated cysteine.
Figure 1.
Identification of YopT cleavage products by TLC. (A) TLC separation of lipid cleavage products. [3H]Mevalonate-labeled RhoAL63 (lanes 1 and 2), Rac1L61 (lanes 3 and 4), and Cdc42L61 (lanes 5 and 6) were incubated with either wild-type (lanes 1, 3, and 5) or YopT C139S mutant protein (lanes 2, 4, and 6) in vitro. The chloroform extracts of each cleavage reaction were spotted on a TLC plate. The lipid cleavage products were visualized by autoradiography of the TLC plate. GGCM refers to the 3H-labeled synthetic GGCM standard (lane 7). (B) The C-terminal sequence of Rho family GTPases RhoA, Rac1, and Cdc42.
To confirm that YopT removes only the prenylated cysteine methyl ester from Rho GTPases, we carried out additional amino acid labeling experiments followed by YopT cleavage. A glycine (Gly-189) residue is located immediately N-terminal to the prenylated cysteine (Cys-190) in RhoA, so we labeled GST-RhoAL63 either with [14C]glycine or [35S]cysteine. Radiolabeled GST-RhoA was isolated from HEK293T cells and incubated with either wild-type YopT or the C139S mutant. Fig. 2A Upper shows the amount of radioactivity present in the chloroform-extractable material. When [35S]Cys-labeled GST-RhoA was incubated with wild-type YopT, a 9-fold increase in radioactivity in the chloroform phase was observed over that seen with treatment of the same labeled substrate with the YopT C139S mutant. When similar experiments were carried out using RhoA labeled with [14C]glycine, negligible amounts of radioactivity were extracted into chloroform phase following treatment with either wild-type or C139S YopT. Equal amounts of radiolabeled RhoA (Fig. 2A Lower Left) as well as total RhoA protein (Fig. 2A Lower Right) were used in the cleavage assays. The labeling efficiency for [14C]glycine and [35S]cysteine is also comparable (Fig. 2A Lower). This result indicates that the glycine residue adjacent to the prenylated cysteine in RhoA is not present in the lipid cleavage product, suggesting that the cleavage occurs between the Gly-189 and Cys-190 residue of RhoA.
Figure 2.
Determination of the YopT cleavage site in RhoA. (A) The prenylated [35S]Cys, but not the [14C]Gly (N-terminal to the prenylated cysteine), in RhoA was cleaved off by YopT. (Upper) Scintillation counting of the chloroform-extracted YopT cleavage product of RhoA with either [14C]Gly or [35S]Cys labeling. (Lower) Levels of radiolabeled RhoA by autoradiography (Left) and total RhoA protein by Western blot (Right) present in each cleavage reaction. (B) Nanoelectrospray MS/MS analysis of the lipid YopT cleavage product from RhoA. Shown is the fragmentation pattern for the GGCM standard (Bottom) and for a single charged parent ion (m/z 408.7) from the RhoA sample after a cleavage reaction with YopT (Middle). (Top) Chemical structure of the GGCM standard. The highlighted fragments correspond to the fragment ions observed in the MS/MS spectrum of the standard and the cleavage product from RhoA.
To unambiguously identify the YopT cleavage product, MS/MS was used to determine its chemical structure. Following the in vitro cleavage assays performed as described above, the chloroform extract was concentrated and prepared for MS analysis. A molecular ion was detected at m/z 408.7 in the full MS mode. When subjected to MS/MS analysis, this parent ion produced a fragmentation pattern consistent with that of a GGCM (Fig. 2B Bottom). The fragment ion at m/z 391.3 represents loss of NH3, whereas the fragment ion at m/z 135.7 corresponds to the carboxyl-methylated cysteine immonium ion. Also, several product ions corresponding to fragmentation of the geranylgeranyl group were observed. The product ion at m/z 274.2 represents the full geranylgeranyl group, whereas the mass values at m/z 137.0 and m/z 204.3 correspond to fragments of the geranylgeranyl group. The observed fragmentation of the lipid group is very similar to the pattern previously documented for the geranylgeranyl group when analyzed by GS/MS (28). To confirm the interpretation of the fragmentation pattern, an authentic sample of the synthetic GGCM was subjected to MS/MS analysis (Fig. 2B Top). From comparing the fragmentation patterns of the standard and the chloroform-extractable cleavage product, it is clear that the YopT-induced RhoA cleavage product is identical to the GGCM standard. Collectively, our results unambiguously demonstrate that the site of cleavage in Rho GTPases by Yop T is N-terminal to the geranylgeranyl-modified cysteine residue.
YopT Cleavage Does Not Depend on Rho GTPases Conformation.
The conformation of small GTP-binding proteins is regulated by nucleotide cycling, a mechanism that switches a GTPase from an inactive conformation (GDP bound) to an active conformation (GTP bound). A number of bacterial toxins specifically target the GTP-bound form of small GTP-binding proteins (29, 30). To gain additional insights into how YopT functions, we examined whether YopT prefers the GTP-bound conformation of Rho GTPases as a substrate. Tritium-labeled geranylgeranyl-modified RhoAL63 (a constitutive GTP-bound form of RhoA) produced in HEK293T cells was assayed as an in vitro substrate in parallel with RhoAN19 (a nucleotide-free form of RhoA in an inactive conformation). As shown in Fig. 3A, both forms of RhoA were cleaved by YopT equally shown by loss of the 3H label. In addition, wild-type RhoA was loaded with either guanosine 5′-[γ-thio]triphosphate or guanosine 5′-[β-thio]diphosphate (GTP[γS] and GDP[βS], respectively; nonhydrolyzable nucleotide analogues) in vitro (31) to lock RhoA in the GTP-bound or GDP-bound conformation, respectively, and used in the YopT in vitro cleavage assay (Fig. 3B). Both RhoA-GTP and RhoA-GDP were cleaved equally by YopT. Similar results were obtained with Rac1 and Cdc42 (data not shown). Taken together, these results demonstrate that YopT does not distinguish between the conformations of its GTPase substrates, suggesting that the conformation of Rho GTPases is not important for YopT substrate recognition.
Figure 3.
YopT cleaves both GDP- and GTP-bound forms of Rho GTPases. (A) In vitro YopT cleavage assay with RhoAL63 and RhoAN19. GST-tagged RhoA variants were expressed and purified from HEK293T cells labeled with [3H]mevalonate and incubated with YopT. Cleavage products were separated by SDS/PAGE and analyzed by autoradiography. Cleavage efficiency was assessed by loss of the 3H-geranylgeranyl group from RhoA (Upper). Total RhoA proteins were measured by anti-RhoA Western blot analysis (Lower). (B) In vitro YopT cleavage assay with RhoA-GTP[γS] and RhoA-GDP[βS]. Wild-type GST-RhoA was purified from HEK293T cells labeled with [3H]mevalonate and loaded with either GTP[γS] or GDP[βS] in vitro and used in the YopT cleavage assay described in the text.
The Polybasic Sequence in RhoA Is Essential for YopT Cleavage and Recognition.
RhoA, Rac1, and Cdc42 all harbor a polybasic sequence N-terminal to the CaaX box (Fig. 1B). To further our understanding of the substrate specificity of YopT, we wanted to determine if the polybasic sequence contributes to the substrate recognition. The five basic residues in the C terminus of RhoA (two arginines and three lysines) were simultaneously mutated to glutamine, and the resulting RhoA mutant, designated RhoA[K(R)5Q], was assayed as a substrate for YopT. As observed in Fig. 4A Left, YopT failed to cleave the 3H-geranylgeranyl-labeled GST-RhoAL63[K(R)5Q] when it was coexpressed in HEK293T cells. It has been reported that the polybasic sequences could influence the cellular localization of Rho GTPases (32). For this reason, we thought that it was important to exclude the possibility that the lack of YopT cleavage of the polybasic RhoA mutant was due to mislocalization. Therefore, we performed the cleavage assay in vitro using the recombinant YopT protein and obtained the same result (Fig. 4A Right). These results support that the polybasic sequence in RhoA is an important determinant for YopT substrate recognition. We next examined whether the polybasic sequence is indeed required for substrate binding. A GST-pulldown assay using GST-RhoAL63[K(R)5Q] was carried out as previously described (24). As shown in Fig. 4B, in contrast to RhoA with an intact C terminus (RhoAL63), mutation of the polybasic sequence drastically disrupted the binding between Flag-tagged YopT (C139S) and its substrate GST-RhoA. This result suggests that the C-terminal polybasic sequence is indispensable for YopT binding of RhoA. Collectively, our results point to the importance of both the polybasic sequence and the prenyl modification in playing key roles in the substrate specificity of YopT.
Figure 4.
The C-terminal polybasic sequence in RhoA is essential for YopT recognition and cleavage. (A) Cleavage of the polybasic RhoA mutant by YopT in vivo (Left) and in vitro (Right). RhoA[K(R)5Q] represents the polybasic RhoA mutant with the five basic residues in the C terminus (two arginines and three lysines) substituted to glutamine. Cleavage assays were performed as described in the text. (B) GST-pulldown assay of the polybasic RhoA mutant. GST-RhoA variants were coexpressed with Flag-tagged YopT C139S in HEK293T cells and purified by GST affinity chromatography. The amounts of GST-RhoA on the glutathione beads and the associated YopT were determined by anti-GST (Lower) and anti-Flag (Upper) Western blot, respectively. RhoAΔCaaX refers to the CaaX deletion mutant of RhoA and was used as a negative control. (C) YopT cleavage assay with chimeric GST fusion proteins. GST-CLVL and GST-ARRGKKKSGCLVL correspond to fusion proteins consisting of RhoA CaaX box (CLVL) or the last 13 residues in the C terminus of RhoA (ARRGKKKSGCLVL) and the N-terminal GST tag. GST-H-Ras (1–166)-ARRGKKKSGCLVL is a chimeric construct generated by replacing the carboxyl 23-residue sequence in H-Ras with the 13-residue sequence from the C terminus of RhoA. These GST fusion proteins were expressed in HEK293T cells labeled with [3H]mevalonate. The resulting prenylated GST fusion proteins with 3H-geranylgeranyl labeling were then subjected to YopT cleavage assay as described in the text.
The two YopT recognition elements (polybasic sequence and prenylation) identified reside within the unstructured carboxyl terminus of RhoA. We wanted to determine if these two elements are sufficient for substrate recognition and cleavage by YopT. To examine this question, a GST fusion protein containing either the C-terminal 13 residues (ARRGKKKSGCLVL) or the CLVL sequence of RhoA was produced in HEK293T cells labeled with [3H]mevalonate. It has been established that the C-terminal CaaX motif itself is sufficient for proteins to undergo CaaX-dependent modifications (23, 33). We observed that both GST-CLVL and GST-ARRGKKKSGCLVL are prenylated in cells as demonstrated by [3H]mevalonate incorporation into the GST fusion proteins (Fig. 4C). These prenylated GST fusion proteins were then assayed in vitro for cleavage by YopT. As shown in Fig. 4C, CaaX alone did not render the prenylated GST fusion protein susceptible to YopT cleavage, whereas the CaaX-modified GST-ARRGKKKSGCLVL was cleaved by YopT. To further confirm the importance of the polybasic sequence, we replaced the C-terminal unstructured sequence of H-Ras (23 residues) with the RhoA C-terminal 13 residues described above and carried out a YopT cleavage reaction with this chimeric Ras protein. As expected, this modified H-Ras protein was efficiently cleaved by YopT (Fig. 4C), although YopT fails to cleave the CaaX-modified wild-type H-Ras protein (F.S. and J.E.D., unpublished data). Our results suggest that the C-terminal 13 residues in RhoA are sufficient for YopT recognition and cleavage.
We have previously demonstrated that YopT selectively binds to the prenylated form of Rho GTPases but not the unmodified Rho protein in a GST-pulldown assay (24). However, under physiological conditions, the prenyl group is buried in the lipid bilayer functioning as the membrane anchor, leading us to question how YopT recognizes the buried lipid anchor. The polybasic sequence recognition provides a plausible explanation. We propose that YopT binding to the polybasic sequence of RhoA could be the initial substrate recognition signal. The binding of the polybasic sequence to the active site region of YopT may allow the protease to shift the position of the lipid anchor from the membrane to a presumed hydrophobic binding pocket at the active site of YopT. The details of the recognition between YopT and its substrate will be more clearly resolved by structural studies. Some of the same features that contribute to the binding of RhoA to YopT seem to be used in the cycling of RhoA on and off the membrane with Rho GDI (34).
Recognition of Prenylated Cysteine Methyl Ester by YopT.
Prenylation can encompass two forms of lipid modification: farnesylation (C15 unit) and geranylgeranylation (C20 unit). The X residue in the CaaX box specifies farnesylation or geranylgeranylation. When the X residue is Leu or Phe, as is the case for Rac, RhoA, and Cdc42, geranylgeranylation takes place (35, 36). It has been reported that substitution of Leu-193 (the X in RhoA CaaX box) to Met switches RhoA prenylation from geranylgeranylation to farnesylation (32). We took advantage of this well characterized mutant (L193M) to examine whether YopT could use the farnesyl-modified RhoA as a substrate. As observed in Fig. 5A, YopT was able to cleave farnesylated RhoA expressed from RhoA L193M as efficiently as geranylgeranylated RhoA when YopT was coexpressed in the cells. Similar results were obtained when the cleavage took place in vitro using recombinant YopT protein (data not shown). Our results suggest that YopT does not distinguish between farnesylation and geranylgeranylation of its substrates.
Figure 5.
The role of the prenylated cysteine methyl ester in YopT recognition. (A) YopT cleavage assay with farnesylated RhoA. The cleavage assay was carried out in vivo by coexpressing YopT and RhoA L193M (a RhoA mutant that undergoes farnesylation instead of geranylgeranylation) in HEK293T cells labeled with [3H]mevalonate. Cleavage was assessed by loss of the 3H-geranylgeranyl group from RhoA (Upper). Expression of RhoA was detected by anti-RhoA Western blot (Lower). (B) YopT cleavage assay with recombinant RhoA that was geranylgeranylated in vitro (no aaX proteolysis and methylation). Recombinant GST-RhoA was geranylgeranyl-modified in vitro using geranylgeranyl transferase and 3H-geranylgeranyl pyrophosphate as described in the text. The resulting labeled GST-RhoA (RhoA-CLVL) was immobilized onto glutathione beads and incubated with recombinant YopT. Fully modified GST-RhoA produced from HEK293T cells was used as a positive control (RhoA-CoMe). (C) In vivo YopT cleavage assay with coexpressed YopT and RhoA V192Y, a RhoA mutant that blocks the aaX proteolysis as well as subsequent methylation of the geranylgeranylated cysteine. (D) GST-pulldown assay of the RhoA V192Y mutant. The experiment was performed as described in the text. The binding affinity between the mutant RhoA and YopT C139S was assessed by anti-Flag Western blot of the GST pulldowns.
The steps involved in RhoA posttranslational modification include conjugation of the prenyl group to the cysteine followed by a proteolytical removal of the aaX residues (CaaX protease) and methyl esterification to form a cysteine methyl ester. To test whether partially modified RhoA containing the prenylated cysteine, but an intact aaX sequence, would serve as a substrate for YopT, we modified the bacteria-expressed GST-RhoA protein by incubation of this protein with 3H-labeled geranylgeranyl pyrophosphate and geranylgeranyl transferase I. The resulting 3H-labeled GST-RhoA protein with an intact aaX sequence at the C terminus was then assayed for YopT cleavage along with the fully modified 3H-labeled GST-RhoA protein purified from HEK293T cells. As shown in Fig. 5B, the in vitro prenylated RhoA with the additional aaX residues was resistant to YopT cleavage, whereas RhoA containing the cysteine methyl ester was efficiently cleaved by YopT. We also carried out a similar experiment in vivo using a RhoA V192Y mutant. It has been shown that mutation of Val-192 in RhoA to a bulky Tyr residue blocks the removal of the aaX residue by CaaX protease (37, 38). Therefore, RhoA V192Y functions as an in vivo equivalent to the in vitro modified RhoA noted above. Consistent with the result observed in vitro, the lipid-modified RhoA with an intact aaX sequence resulting from the V192Y mutation was not cleaved by YopT in the cells (Fig. 5C). We further examined whether the untrimmed aaX sequence actually disrupts substrate binding, thus resulting in the inability of YopT to cleave the unmodified form of RhoA. We carried out a GST-pulldown assay with RhoA V192Y, along with wild-type RhoA and RhoAΔCaaX, which were used as the positive and negative control, respectively. The results shown in Fig. 5D demonstrated an intermediate binding affinity between YopT (C139S) and RhoA with the aaX sequence (V192Y). We interpret this result to suggest that the aaX sequence following the prenylated cysteine impairs the correct binding between YopT and its GTPase substrate, although it does not completely abolish the substrate binding. Taken together, our results demonstrate that YopT recognizes and cleaves both farnesylated and geranylgeranylated RhoA. The methyl ester, or at least the proteolytic removal of the aaX residues, also seems to be important for substrate recognition by YopT. Failure to remove the aaX sequence by the CaaX protease results in a lower binding affinity of this form of RhoA to YopT and an inefficient cleavage of RhoA.
In this study, we have demonstrated that the Yersinia YopT effector functions as a cysteine protease to cleave Rho family GTPases including RhoA, Rac1, and Cdc42. The cleavage occurs at a specific site N-terminal to the carboxyl-prenylated cysteine in Rho GTPases. We have also shown that YopT recognizes both GTP- and GDP-bound forms of Rho GTPases, and cleavage by YopT is not dictated by the prenylation forms of GTPases. In addition, the polybasic sequence in the C terminus of RhoA seems to be essential for YopT recognition. Our data also suggest that a fully modified RhoA containing a cysteine methyl ester is required for highly efficient recognition and cleavage of the RhoA substrate by YopT.
Acknowledgments
We are grateful for the critical review of this manuscript by Dr. Christine McDonald and Robert Kruger. We thank Dr. Bruce J. Mayer (University of Connecticut Health Science Center) for kindly providing the pEBB vector. We also thank Dr. Zhaohui Xu and members of Dixon laboratory for helpful discussions and technical assistance. F.S. was supported by the Anthony and Lillian Lu graduate student fellowship, and K.E.B. was supported by a postdoctoral fellowship from National Institutes of Health. This work was funded by National Institutes of Health Grants 18024 (to J.E.D.) and GM40602 (to C.A.F.), the Walther Cancer Institute (to J.E.D.), and the Ellison Medical Foundation (to J.E.D.).
Abbreviations
- GGCM
geranylgeranyl cysteine methyl ester
- GTP[γS]
guanosine 5′-[γ-thio]triphosphate
- GDP[βS]
guanosine 5′-[β-thio]diphosphate
- MS/MS
tandem MS
References
- 1.Hueck C J. Microbiol Mol Biol Rev. 1998;62:379–433. doi: 10.1128/mmbr.62.2.379-433.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cornelis G R, Van Gijsegem F. Annu Rev Microbiol. 2000;54:735–774. doi: 10.1146/annurev.micro.54.1.735. [DOI] [PubMed] [Google Scholar]
- 3.Cornelis G R, Boland A, Boyd A P, Geuijen C, Iriarte M, Neyt C, Sory M P, Stainier I. Microbiol Mol Biol Rev. 1998;62:1315–1352. doi: 10.1128/mmbr.62.4.1315-1352.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cornelis G R. J Cell Biol. 2002;158:401–408. doi: 10.1083/jcb.200205077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cornelis G R. Nat Rev Mol Cell Biol. 2002;3:742–754. doi: 10.1038/nrm932. [DOI] [PubMed] [Google Scholar]
- 6.Juris S J, Shao F, Dixon J E. Cell Microbiol. 2002;4:201–211. doi: 10.1046/j.1462-5822.2002.00182.x. [DOI] [PubMed] [Google Scholar]
- 7.Guan K L, Dixon J E. Science. 1990;249:553–556. doi: 10.1126/science.2166336. [DOI] [PubMed] [Google Scholar]
- 8.Andersson K, Carballeira N, Magnusson K E, Persson C, Stendahl O, Wolf-Watz H, Fallman M. Mol Microbiol. 1996;20:1057–1069. doi: 10.1111/j.1365-2958.1996.tb02546.x. [DOI] [PubMed] [Google Scholar]
- 9.Black D S, Bliska J B. EMBO J. 1997;16:2730–2744. doi: 10.1093/emboj/16.10.2730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Persson C, Carballeira N, Wolf-Watz H, Fallman M. EMBO J. 1997;16:2307–2318. doi: 10.1093/emboj/16.9.2307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Galyov E E, Hakansson S, Forsberg A, Wolf-Watz H. Nature. 1993;361:730–732. doi: 10.1038/361730a0. [DOI] [PubMed] [Google Scholar]
- 12.Juris S J, Rudolph A E, Huddler D, Orth K, Dixon J E. Proc Natl Acad Sci USA. 2000;97:9431–9436. doi: 10.1073/pnas.170281997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Von Pawel-Rammingen U, Telepnev M V, Schmidt G, Aktories K, Wolf-Watz H, Rosqvist R. Mol Microbiol. 2000;36:737–748. doi: 10.1046/j.1365-2958.2000.01898.x. [DOI] [PubMed] [Google Scholar]
- 14.Black D S, Bliska J B. Mol Microbiol. 2000;37:515–527. doi: 10.1046/j.1365-2958.2000.02021.x. [DOI] [PubMed] [Google Scholar]
- 15.Palmer L E, Hobbie S, Galan J E, Bliska J B. Mol Microbiol. 1998;27:953–965. doi: 10.1046/j.1365-2958.1998.00740.x. [DOI] [PubMed] [Google Scholar]
- 16.Orth K. Curr Opin Microbiol. 2002;5:38–43. doi: 10.1016/s1369-5274(02)00283-7. [DOI] [PubMed] [Google Scholar]
- 17.Orth K, Xu Z, Mudgett M B, Bao Z Q, Palmer L E, Bliska J B, Mangel W F, Staskawicz B, Dixon J E. Science. 2000;290:1594–1597. doi: 10.1126/science.290.5496.1594. [DOI] [PubMed] [Google Scholar]
- 18.Iriarte M, Cornelis G R. Mol Microbiol. 1998;29:915–929. doi: 10.1046/j.1365-2958.1998.00992.x. [DOI] [PubMed] [Google Scholar]
- 19.Grosdent N, Maridonneau-Parini I, Sory M P, Cornelis G R. Infect Immun. 2002;70:4165–4176. doi: 10.1128/IAI.70.8.4165-4176.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zumbihl R, Aepfelbacher M, Andor A, Jacobi C A, Ruckdeschel K, Rouot B, Heesemann J. J Biol Chem. 1999;274:29289–29293. doi: 10.1074/jbc.274.41.29289. [DOI] [PubMed] [Google Scholar]
- 21.Hall A. Science. 1998;279:509–514. doi: 10.1126/science.279.5350.509. [DOI] [PubMed] [Google Scholar]
- 22.Sorg I, Goehring U M, Aktories K, Schmidt G. Infect Immun. 2001;69:7535–7543. doi: 10.1128/IAI.69.12.7535-7543.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang F L, Casey P J. Annu Rev Biochem. 1996;65:241–269. doi: 10.1146/annurev.bi.65.070196.001325. [DOI] [PubMed] [Google Scholar]
- 24.Shao F, Merritt P M, Bao Z, Innes R W, Dixon J E. Cell. 2002;109:575–588. doi: 10.1016/s0092-8674(02)00766-3. [DOI] [PubMed] [Google Scholar]
- 25.Guan K L, Dixon J E. Anal Biochem. 1991;192:262–267. doi: 10.1016/0003-2697(91)90534-z. [DOI] [PubMed] [Google Scholar]
- 26.Hightower K E, Casey P J, Fierke C A. Biochemistry. 2001;40:1002–1010. doi: 10.1021/bi002237d. [DOI] [PubMed] [Google Scholar]
- 27.Zhang L, Tschantz W R, Casey P J. J Biol Chem. 1997;272:23354–23359. doi: 10.1074/jbc.272.37.23354. [DOI] [PubMed] [Google Scholar]
- 28.Yamane H K, Farnsworth C C, Xie H Y, Evans T, Howald W N, Gelb M H, Glomset J A, Clarke S, Fung B K. Proc Natl Acad Sci USA. 1991;88:286–290. doi: 10.1073/pnas.88.1.286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Barbieri J T, Riese M J, Aktories K. Annu Rev Cell Dev Biol. 2002;18:315–344. doi: 10.1146/annurev.cellbio.18.012502.134748. [DOI] [PubMed] [Google Scholar]
- 30.Boquet P. Microbes Infect. 2000;2:837–843. doi: 10.1016/s1286-4579(00)90369-1. [DOI] [PubMed] [Google Scholar]
- 31.Vikis H G, Li W, He Z, Guan K L. Proc Natl Acad Sci USA. 2000;97:12457–12462. doi: 10.1073/pnas.220421797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Michaelson D, Silletti J, Murphy G, D'Eustachio P, Rush M, Philips M R. J Cell Biol. 2001;152:111–126. doi: 10.1083/jcb.152.1.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Goldstein J L, Brown M S, Stradley S J, Reiss Y, Gierasch L M. J Biol Chem. 1991;266:15575–15578. [PubMed] [Google Scholar]
- 34.Hoffman G R, Nassar N, Cerione R A. Cell. 2000;100:345–356. doi: 10.1016/s0092-8674(00)80670-4. [DOI] [PubMed] [Google Scholar]
- 35.Baron R, Fourcade E, Lajoie-Mazenc I, Allal C, Couderc B, Barbaras R, Favre G, Faye J C, Pradines A. Proc Natl Acad Sci USA. 2000;97:11626–11631. doi: 10.1073/pnas.97.21.11626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yokoyama K, Goodwin G W, Ghomashchi F, Glomset J A, Gelb M H. Proc Natl Acad Sci USA. 1991;88:5302–5306. doi: 10.1073/pnas.88.12.5302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kato K, Cox A D, Hisaka M M, Graham S M, Buss J E, Der C J. Proc Natl Acad Sci USA. 1992;89:6403–6407. doi: 10.1073/pnas.89.14.6403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Figueroa C, Taylor J, Vojtek A B. J Biol Chem. 2001;276:28219–28225. doi: 10.1074/jbc.M101763200. [DOI] [PubMed] [Google Scholar]