Abstract
Hereditary Motor and Sensory Neuropathy – Russe (HMSNR) is a severe autosomal recessive disorder, identified in the Gypsy population. Our previous studies mapped the gene to 10q22-q23 and refined the gene region to ∼70 kb. Here we report the comprehensive sequencing analysis and fine mapping of this region, reducing it to ∼26 kb of fully characterised sequence spanning the upstream exons of Hexokinase 1 (HK1). We identified two sequence variants in complete linkage disequilibrium, a G>C in a novel alternative untranslated exon (AltT2) and a G>A in the adjacent intron, segregating with the disease in affected families and present in the heterozygote state in only 5/790 population controls. Sequence conservation of the AltT2 exon in 16 species with invariable preservation of the G allele at the mutated site, strongly favour the exonic change as the pathogenic mutation. Analysis of the Hk1 upstream region in mouse mRNA from testis and neural tissues showed an abundance of AltT2-containing transcripts generated by extensive, developmentally regulated alternative splicing. Expression is very low compared with ubiquitous Hk1 and all transcripts skip exon1, which encodes the protein domain responsible for binding to the outer mitochondrial membrane, and regulation of energy production and apoptosis. Hexokinase activity measurement and immunohistochemistry of the peripheral nerve showed no difference between patients and controls. The mutational mechanism and functional effects remain unknown and could involve disrupted translational regulation leading to increased anti-apoptotic activity (suggested by the profuse regenerative activity in affected nerves), or impairment of an unknown HK1 function in the peripheral nervous system (PNS).
Keywords: CMT disease; HMSNR, Hexokinase 1; Gypsy founder mutations
Introduction
Recessively inherited hereditary motor and sensory neuropathies (AR HMSN) are a group of individually rare, clinically and genetically heterogeneous conditions within the broad category of Charcot–Marie–Tooth (CMT) disease. Compared with autosomal dominant forms, they are characterised by earlier onset, increased severity and poorer prognosis.1 The causative molecular defects identified to-date show an astonishing diversity of possible pathogenetic mechanisms and pathways, contributing greatly to the understanding of the physiology and pathology of the peripheral nervous system (PNS).2
Hereditary Motor and Sensory Neuropathy – Russe (HMSNR), named after the Bulgarian town on the Danube River where it was first discovered, is one of at least four autosomal recessive peripheral neuropathies common in the European Gypsy population. Our previous studies have identified the ancestral Gypsy mutations causing HMSN Lom,3 the congenital cataracts facial dysmorphism neuropathy syndrome4 and CMT4C.5 With carrier rates for those mutations ranging between 4 and 20% for different population sub-isolates,6 the resulting disorders represent a serious burden to local communities and the health care system.
Hereditary Motor and Sensory Neuropathy – Russe (OMIM #605285) is characterised by delayed early motor development, distal lower limb weakness apparent at age 5–15 years, upper limb involvement at age 10–40 years, and steady progression to total muscle paralysis below the knees and often below the elbows by the fourth to fifth decade of life.7, 8, 9 Sensory loss is prominent and affects all modalities. Motor nerve conduction velocities (MNCV) are reduced in the intermediate range already in the second half of the first decade of life. The amplitudes of compound muscle action potentials (CMAPs) decrease in parallel with the progression of muscle weakness and atrophy (Table 1). A peculiar electrophysiological feature is an increase in the threshold for electrical nerve stimulation, that could be due to muscle re-innervation by abundant small-calibre, hypomyelinated fibres.8, 9 Sensory action potentials are absent. Neuropathological studies show a marked reduction in the large myelinated fibre population, but no evidence of active demyelination or fibre degeneration and lack of hypertrophic changes. The most characteristic feature of HMSNR neuropathology is the profuse regenerative activity, with numerous clusters of thinly myelinated regenerating fibres leading to an abnormally increased density of non-myelinated axons.8, 9 The findings do not provide a straightforward indication of the localisation of the primary defect – in the Schwann cell(SC) or the axon.
Table 1. Neurophysiological findings in HMSNR patients.
| MNCVs (m/sec) | CMAP amplitude (mV) | |||
|---|---|---|---|---|
| Age group | Peroneal | Median | Ulnar | Peroneal |
| Children (5–12 years)a | 26.0–33.8* | 26.7–38.7 | 26.6–36.4 | ≤2 |
| Adultsb | Unobtainable | 32.0±6.8 | 31.9±7.05 | Unobtainable |
CMAP, compound muscle action potential; HMSNR, Hereditary Motor and Sensory Neuropathies – Russe; MNCV, motor nerve conduction velocity, recorded from the flexor carpi ulnaris or the flexor digitorum profundus in adults and from the distal muscles in children.
*Unobtainable in the patient aged 12 years.
Our previous investigations placed the HMSNR gene on chromosome 10q23, telomere to EGR2, with closely related haplotypes pointing to a shared ancestral mutation.10 Further analysis reduced the region to ∼70 kb.8, 9, 11 Here we report on the detailed study of this interval and the identification of a G>C substitution in a highly conserved novel alternative untranslated exon of Hexokinase 1 (HK1), implicating this unexpected candidate gene in the pathogenesis of PNS disorders.
Subjects and methods
Subjects
The study included 34 HMSNR patients, of whom 25 were described previously8, 9, 10, 11 and 28 unaffected relatives. Mutation screening was carried out on a sample of 790 Gypsy and 233 Bulgarian population controls. Informed consent was obtained from all participating individuals. The study complied with the ethical guidelines of the institutions involved.
Methods
Novel expressed sequences in the ∼70 kb HMSNR region were identified through searching public EST and mRNA databases. Physical and genetic maps were compiled using http://www.ncbi.nlm.nih.gov, http://www.genome.ucsc.edu and http://www.pantherdb.org.
Sequencing was carried out using standard PCR-based techniques and BigDye (Applied Biosystems, Scoresby, Victoria, Australia) chemistry (primers available on request).11 A total of 66 kb, spanning all known and newly identified exons, non-overlapping ESTs, and most introns and intergenic regions, were analysed in at least one patient (carrying a recombinant haplotype of interest) and at least one carrier.
Identified sequence variants were checked for listing as polymorphisms in dbSNP and those not found were tested for co-segregation with the disease phenotype in an extended panel of samples. The putative HK1 mutations in the 5′ untranslated region of HK1 were analysed by PCR-based RFLP assays of the AluI restriction site abolished by the nucleotide substitution in alternative exon T2 (AltT2), and the Tsp509I site created by the change in the adjacent intron (Supplementary Figure 1).
Evolutionary conservation of the AltT2 exon and the flanking intron was assessed with BLAST and BLAT searches and the Vertebrate Multiz Alignment and Conservation in the UCSC Genome Browser. Pairwise alignments were carried out using EMBOSS Pairwise Alignment -Needle, and multiple sequence alignments were done with ClustalW.
Total RNA was extracted from mouse C57BL6 tissues: weeks 1 and 10, the sciatic nerve and the brain; and week 10, the testis. The tissues were ground under liquid nitrogen, suspended in Trizol (Invitrogen, Carlsbad, CA, USA) and stored at −80°C. RNA extraction was done following the Trizol protocol. RNA concentration was determined spectrophotometrically (Perkin Elmer MBA2000, Glen Waverley, Victoria, Australia) and integrity was tested on 1% agarose gels.
Reverse transcription was carried out on 2 μg of RNA with random hexamers or oligo-dT primers and MMLV-RT minus RNase H (Promega, Sydney, Australia) following standard protocols. Subsequent PCR reactions were done using 0.5–4 μl cDNA as template. Alternative transcripts in the HK1 upstream region were characterised with forward primers in exons T1 or AltT2, and reverse primers in T3 and invariable coding exon 2 (Supplementary Table 1). A fragment of the ubiquitous HK1 sequence (coding exons 3–7) was amplified in parallel. The products were size-separated by agarose gel electrophoresis, excised and purified with Qiaquick gel extraction kits (Qiagen, Hilden, Germany), and sequenced.
Hexokinase 1 expression was analysed in HMSNR and control Scs cultured as described.12 Quantitative PCR was carried out in triplicate, with primers spanning exons 12 and 13 (For 5′-CTGTGGTGAATGACACAGTG-3′, Reverse 5′-TGATCCATCGTCTCCAAAGG-3′). Each assay included a standard curve of four to five serial dilution points of a recDNA plasmid containing the Q-PCR amplicon and a no-template control. The Q-gene Excel-based analysis package was used to calculate the mean normalised gene expression by first averaging the replicate CT values and the standard error of the mean normalised gene expression in each of the samples in relation to the 28 s reference gene.
Immunohistochemical studies of HMSNR and control sural nerve biopsies were carried out on 7 μ cryosections, using goat anti-HK1 polyclonal antibody targeting the porin-binding domain of HK1 (sc-6517, Santa Cruz Biotechnology Inc., Santa Cruz, CA, USA) at a 1:5 dilution. Secondary antibody was rat anti-goat horseradish peroxidase (1:50 dilution), developed with 3,3-diaminobenzidine (DAB) (Sigma-Aldrich, Dorset, UK) as a substrate.
Total hexokinase activity was measured in cultured HMSNR and control Scs.12 Enzyme activity was measured in a kinetic G6PDH-coupled assay. Activity was expressed in nmol/min*mg protein. The assay was carried out in triplicate on each sample – undiluted, and 1:2 and 1:5 dilutions.
Bioinformatics tools were used to predict potential effects of the mutations on gene function. Effects on splicing were analysed using NNSplice 0.9 (http://biologyhelp.awardspace.com/), NetGene2 (http://www.cbs.dtu.dk/services/NetGene2/), ESEfinder 3.0 (http://rulai.cshl.edu/cgi-bin/tools/ESE3/) and RescueESE (http://genes.mit.edu/burgelab/rescue-ese/). Changes in transcription factor binding sites (TFBS) were examined with Promo (http://alggen.lsi.upc.es/). These analyses were done on 50–100 bp stretches of the human sequence, centred on the putative mutation. Analysis of the 5′UTR for functional elements was done with UTRscan on the longest human AltT2 sequence and on the full predicted testis-specific mRNA including exons T1, T2 and parts of T3 (represented by EST BG719874), until the ATG start codon. Searches for open reading frames and translation of mRNA sequences were carried out using the Translate tool at http://expasy.org/.
Results
Genetic analyses
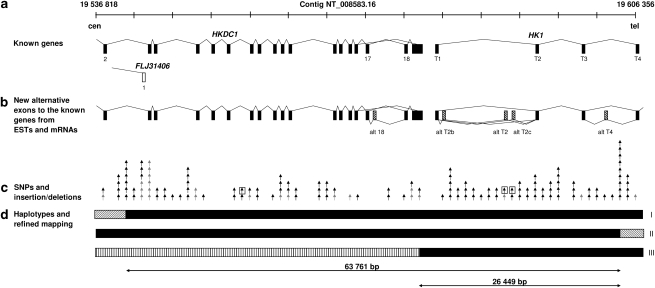
The previously reported ∼70 kb HMSNR gene region on 10q was defined by SNPs rs5030944 (HGVbase: SNP001494322) on the centromeric side and rs5030917 (HGVbase: SNP001494284) on the telomeric side.11 This interval contained three positional candidate genes: the first exon of FLJ31406 (AK055968), Hexokinase domain containing 1 (HKDC1, alias FLJ22761) and four upstream exons of Hexokinase 1 (HK1) (Figure 1a). A full representation of expressed sequences in this region was obtained by exploring EST and mRNA databases. Over 40 ESTs were found to map to the interval, of which 23 did not overlap the known genes. Spliced ESTs connecting to known gene sequences suggested the existence of five alternative exons: one between exons 17 and 18 of HKDC1 and four within the 5′ UTR of HK1. Following the HK1 exon nomenclature, based on Andreoni et al 2000,13 where the testis upstream exons are named T1–T6, the reticulocyte first exon is 1R and ubiquitous exons are numbered 1–18, we refer to our putative alternative HK1 exons as AltT2, AltT2b and AltT2c, and AltT4 (Figure 1b).
Figure 1.
Fine mapping of the 70 kb Hereditary Motor and Sensory Neuropathies – Russe (HMSNR) region on 10q (nt 19 536 818–19 606 356 in reference contig NT_008583.16). (a) Known genes: Hexokinase domain containing 1 (HKDC1) , Hexokinase 1 (HK1) and FLJ31406. (b) Alternative exons supported by mRNAs and ESTs. (c) Positions of SNPs and insertion/deletions: grey – previously published;11 black – identified in this study; and boxed – variants not excluded as possible HMSNR mutations by dbSNP and segregation analysis. (d) Recombinant haplotypes used in fine mapping: haplotypes I and II identified in patients K3 and Romanian Gypsy family;11 haplotype III in newly identified patient reduced the critical interval to 26 449 kb.
Sequencing of 66 kb of the region added 119 variants (100 nucleotide substitutions and 19 insertion/deletions) to the previously integrated physical and genetic map,11 with a final density of 1 variant/∼500 bp (Figure 1c). All but three could be classified as polymorphisms based on listing in dbSNP or lack of co-segregation with the disease. The changes, which could not be ruled out, included a G>A transition in intron 7 of HKDC1 (nt 19 556 225 in reference contig NT_008583.16), a G>C substitution in the putative AltT2 exon of HK1, and an intronic G → A 1315 bp downstream of the AltT2 change (nt 19 589 622 and 19 590 937, respectively, in NT_008583.16) (Figure 1c, boxed).
Genotyping the entire HMSNR sample showed that all affected individuals were homozygous for all three changes with one exception: a newly recruited patient (distantly related to the original HMSNR family) was heterozygous for the HKDC1 but homozygous for the HK1 variants, suggesting the presence of a new critical recombination. Using the newly identified polymorphisms in the region, the recombination breakpoint was mapped between SNP rs2611 centromeric and a novel T → C polymorphism (nt 19 578 386 in NT_008583.16) (Figure 1d, haplotype III). The complete homozygosity interval was thus reduced to 26 449 bp of fully characterised sequence, leaving the HK1 upstream region as the only possible location of the HMSNR mutation, and the changes in exon Alt2 and the flanking intron as the only candidates.
To examine the molecular epidemiology of the two variants, we screened 790 control subjects, representing a cross-section of the genetic structure of the Gypsy population.6 This analysis identified five heterozygous carriers (overall carrier rate 1:158), four of whom belonged to the same Gypsy sub-isolate as the affected families (the Kalderas), and one was a member of the genetically closely related group of the Rudari. Homozygotes were not detected among 790 Gypsy controls and the mutations were altogether absent in 233 Bulgarian controls. The results mirrored precisely the epidemiology of HMSNR in the comprehensively investigated Gypsy population of Bulgaria: a rare disorder with 24 known HMSNR cases in a population of ∼700–800 000, confined exclusively to the Kalderas – a very young Vlax sub-isolate with a high rate of consanguinity.6 These data provided strong support for the involvement of the HK1 nucleotide substitutions in the pathogenesis of HMSNR, but did not allow a distinction between a pathogenic mutation and a linked polymorphism: the two invariably occurred together in patients, carrier relatives, and unrelated heterozygous controls.
Characterisation of the 5′UTR of Hexokinase 1
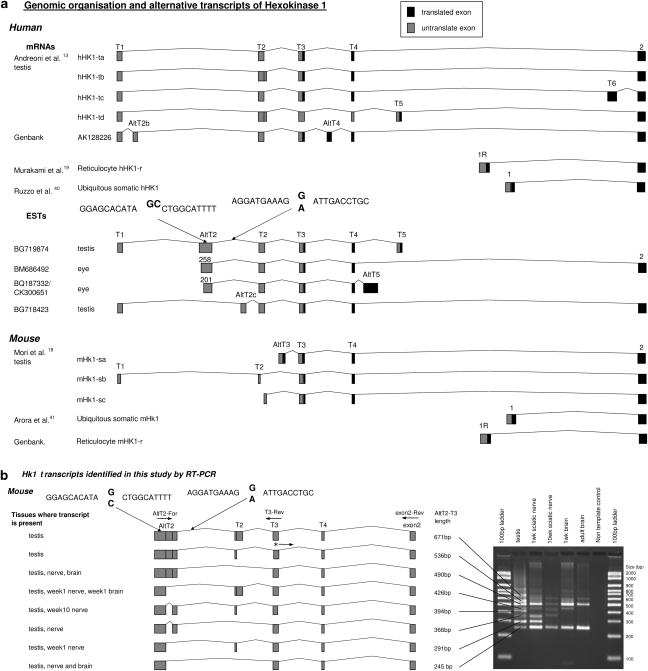
Hexokinase 1 (EC 2.7.1.1) is one of the four mammalian enzymes, catalysing the phosphorylation of glucose to glucose-6-phosphate and serving as a major point of regulation of the energy-producing glycolytic pathway. The C-terminus is conserved between the different isozymes and involved in the catalytic activity, while evolutionary divergence of the N-terminal half is thought to be related to regulatory functions.14, 15 HK1 is ubiquitously expressed and particularly abundant in the brain, the testis and erythrocytes. It contains 18 translated exons, with the last 17 present in all known transcripts. Alternative splicing in the 5′ end of the gene results in isoforms (Figure 2a), with different tissue specificity, developmental regulation, biochemical properties and cellular localisation.13, 14, 15, 16, 17, 18, 19 These include somatic, reticulocyte (R) and several testis (T) HK1 isoforms (Figure 2a), with a different first translated exon. Somatic exon 1 encodes the ‘porin-binding domain' (PBD), indispensable for HK1 binding to the outer mitochondrial membrane (OMM), and for the protein's special role in energy metabolism and cell survival.20
Figure 2.
Upstream region of the Hexokinase1 (HK1) gene. (a) Structure of the 5′UTR of human and mouse HK1, summarising published information. The positions of the two nucleotide substitutions in Hereditary Motor and Sensory Neuropathies – Russe (HMSNR) chromosomes are marked with arrows. (b) HK1 transcripts identified in this study in mouse tissues using RT-PCR with forward primer in AltT2 and reverse primers in T3 and invariable exon 2 (See Supplementary Table 1 for primer sequences). The translation start site in exon T318 is marked with an asterisk. The diagram illustrates the exon composition of alternative transcripts, with the corresponding fragments and sizes in bp shown in the agarose gel picture. Please refer to Supplementary Material online for the sequences of the different fragments.
The AltT2 exon (Figure 2a), which is not found in published HK1 mRNAs, is supported by four human ESTs: BQ187332/CK300651 from eye tissues, BG719874 from the testis and BM686492 from the optic nerve. The longest EST sequence (262 bp) spans nt 19 589 496–19 589 696 in NT_008583.16, with the G → C mutation 75 bp away from the end of the exon, (nt 19 589 622). BLAST and BLAT searches showed a high degree of evolutionary conservation, with nucleotide identity between human AltT2 and 15 other species ranging from 76 to 99%, and the guanine residue at the AltT2 mutation site conserved in all species (Supplementary Figure 2a; Supplementary Table 2). By contrast, the sequence around the intronic mutation was much less conserved, with homology detected in only 8/15 species, of which three presented with the mutant human allele (Supplementary Figure 2b).
We characterised AltT2-containing transcripts in mouse newborn and adult neural tissues in comparison with the testis, which is particularly enriched in alternative splicing,21 and where the HK1 5′upstream region has been studied in detail.13, 15, 18 RT-PCR with different combinations of primers in exons T1 and AltT2 (forward), and T3 and ubiquitous 2 (reverse) revealed a total of 13 alternative transcripts. In both the testis and neural cells, the overall level of AltT2-containing transcripts was markedly lower than the invariable coding region of HK1 (not shown). None of the five amplification products obtained with primers T1-F and S2-R included AltT2 (not shown). The remaining eight AltT2-containing transcripts resulted from a variety of internal splicing events involving the AltT2 and T2 exons (Figure 2b; sequences shown as Supplementary Material on-line). The analysis of neural tissue-derived mRNA identified six of the eight testis AltT2-containing isoforms, with expression patterns differing between the peripheral nerve and the brain and between newborn and adult tissues (Figure 2b). An interesting and functionally important feature of AltT2-containing transcripts, shared with the testis-specific isoforms described by Mori et al,18 was the skipping of the PDB-encoding exon of somatic HK1. All AltT2-containing transcripts included the exon T3 ATG initiation codon and its surrounding sequence, referred to as the ‘spermatogenic cell-specific region (SSR)'.18 New HK1 transcripts are deposited in Genbank (accession numbers EU430519–EU430533).
Our Q-PCR analysis showed no difference in HK1 mRNA levels in cultured HMSNR and control Scs (not shown).
Searching for potential functional effects of the HK1 mutation(s)
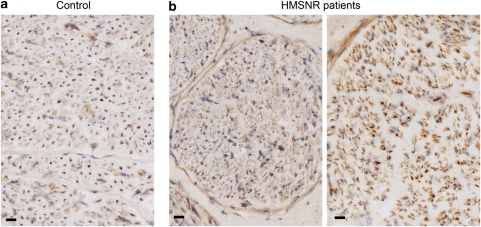
To check whether the HMSNR-associated variants affected HK1 expression and distribution in the peripheral nerve, we examined HMSNR and control sural nerve biopsies by immunohistochemistry using an anti-Hexokinase 1 antibody, raised against a peptide containing the PBD and parts of the ubiquitous exon 2 (Figure 3). In the normal nerve (Figure 3a), we obtained strong signal from myelinated axons, whereas unmyelinated axons and Scs were weakly stained. The same distribution pattern and staining intensity were found in HMSNR (Figure 3b), with the difference in appearance due to the disease-related paucity of large fibres.8, 9
Figure 3.
Hexokinase 1 (HK1) immunohistochemistry in human peripheral nerve. HK1 antibody (Santa Cruz Biotechnologies) was against the porin binding domain (PBD) and part of the downstream sequence, encoded by exon 2. The staining was done on 7 μm cryogenic sections of sural nerve biopsies; photographs were taken with a × 40 objective. Scale bar 20 μm. (a) Normal human nerve and (b) Hereditary Motor and Sensory Neuropathies – Russe (HMSNR) nerves from two independent patients.
Next, we compared total Hexokinase catalytic activity in cultured Scs from an HMSNR patient and a normal control. We obtained no evidence of enzyme deficiency – the results were 55.3 nmol/min*mg protein in HMSNR (average of the three dilutions: 53.7, 66.7 and 45.5 nmol/min*mg) and 42.5 nmol/min*mg protein in control cells (average of the three dilutions: 42.2, 48.2 and 37.3 nmol/min*mg protein).
Other putative effects on HK1 gene function were examined using bioinformatics tools. Possible changes in TFBS were analysed in a 25 bp stretch surrounding each of the two mutations (Supplementary Table 3). The mutant C allele of AltT2 was predicted to create a binding site for the Glucocorticoid Receptor α (GR). However, with a dissimilarity of 8.1% and an expectation value of 0.098, this is likely to be a false positive result. The intronic change was predicted to abolish the putative binding sites for GR α and general transcription factor II-I, again with expectation values of 0.098 and 0.037 suggesting a false positive.
Prediction of donor and acceptor splice sites at the positions of the two mutations showed no effects on splicing. ESEfinder (but not RescueESE) predicted an exonic splice enhancer upstream of the AltT2 mutation, with the mutation site itself not involved.
Similarity to an internal ribosome entry sites (IRES) motif was predicted by the UTRscan program for the end of AltT2 (nt170–262, mutation at position 188), but was not affected by the mutation, and was not found in the full EST BG719874-predicted 5′UTR.
Translation of the 556 bp human 5′UTR with the ExPASy Translate tool (Supplementary Figure 3) showed four upstream AUG codons, which produce short upstream ORFs. As translation can in rare cases also be initiated at CUG, and to a lesser extent GUG, UUG, ACG and AUA codons,22 we inspected the sequence for such codons and found 20 CUG, 11 GUG and one AUA in three ORFs. ORF1 predicted a Stop codon (UAG) at the AltT2 mutation site changed into Tyrosine (UAC) by the G>C mutation. This reading frame contains two CUG and two GUG codons, which could be potential initiation codons upstream of the mutated Stop codon. A similar situation was also found in 14 of the other species analysed.
Discussion
In this study, we have conducted comprehensive fine mapping and sequencing analysis of the HMSNR gene region, resulting in reduction of the critical interval to ∼26.4 kb and complete characterisation of the sequence within that interval. The data unambiguously point to HK1 as the gene responsible for Hereditary Motor and Sensory Neuropathy – Russe. This unexpected finding is supported by the lack of other genes in the refined critical region, the detailed analysis and exclusion of all other sequence changes as possible disease-causing mutations, and the results of the population screening, which correspond precisely to the frequency of carriers and sub-isolate distribution predicted by the disease epidemiology. We discovered two closely spaced nucleotide substitutions in complete linkage disequilibrium, which invariably occur together in HMSNR chromosomes. Our attempt to differentiate between the two was not successful – none of the 1580 chromosomes tested carried only one of the variants, separated from the other. In interpreting this, one should take into account the population history of the Gypsies,6 where a recent origin of the mutation on a rare haplotype, carrying the polymorphism, with subsequent amplification by inbreeding and drift effects, could easily explain the current observations. Our data favour the AltT2 G>C substitution as the pathogenic mutation on the basis of its exonic location, evolutionary conservation of the surrounding sequence and across-species preservation of the normal human G allele.
AltT2-containing transcripts in the peripheral nerve are rare compared with the ubiquitous somatic isoform, and show complex alternative splicing patterns with differences between newborn and adult nerve pointing to developmental regulation. The early onset of the HMSNR neuropathy suggests a pathological mechanism operating from a very early age, possibly prenatally, that could involve specific AltT2-containing isoform(s). The length of the 5′ UTR of AltT2-containing transcripts and the diversity of alternatively spliced forms point to a possible role in translational regulation.23 The existence of such regulation in HK1 expression is supported by the lack of the protein in rat large sensory neurons despite abundant HK1 mRNA,24 and our own expression array data (not shown), which revealed high HK2 mRNA levels in mouse peripheral nerve, where the protein is not expressed.24 Translation regulation by upstream ORFs (uORF) occurs in a number of genes, such as Fli-1, where it is mediated by two short uORFs, one using a GUG start codon,25 and HIAP2, whose translation is controlled by a uORF with a CUG start codon.26 One can thus speculate that a disruption in HK1 translation regulation, caused by the tyrosine for a Stop substitution in a uORF with a non-AUG start codon, could be the mutational mechanism in HMSNR.
The functional consequences are not evident. Hexokinase 1 is the major regulator of the cell's energy metabolism, whose primary role is generation of ATP.20 This function is facilitated by binding to the OMM, which provides ‘privileged' access to the intramitochondrial pool of ATP, coupling glycolysis to oxidative phosphorylation.20 Mitochondrially-associated HK1 plays a role in growth factor- and Akt-mediated cell survival, through intervening in the classical apoptotic cascade and restricting the accumulation and oligomerization of pro-apoptotic proteins at the OMM.27 Mutations in the HK1 coding sequence lead to enzyme deficiency and nonspherocytic haemolytic anaemia of varying severity and associated CNS symptoms.28, 29, 30 HK1 is highly expressed in the nervous system, localised mainly to the mitochondrial fraction in the brain and the dorsal root ganglia.20, 31 Inhibition of its activity and/or mitochondrial binding blocked growth factors NGF- and GDNF-induced neurite outgrowth in cultured adult sensory neurons.24 Our findings do not support a reduction in HK1 metabolic activity. The phenotype is limited to the PNS, nerve biopsy immunohistochemistry showed no abnormalities in the expression levels or distribution of PBD-containing isoforms, and total Hexokinase activity measurement in cultured Scs showed no deficit in HMSNR samples (one should note however the possibility that neurons, rather than Scs, may be the primary target of the disease process). An intriguing possibility of increased anti-apoptotic activity is raised by the unusual profuse regenerative activity in HMSNR nerve,8, 9 contrary to the inhibited outgrowth expected under reduced HK1 activity or OMM binding.24 Impaired apoptosis is increasingly recognised as a mechanism in CMT disease, and a role in its regulation has been proposed for a number of proteins mutated in different CMT forms, such as PMP22 (CMT1A),32 EGR2/Krox20 (CMT1D/CMT4E),33 MFN2 (CMT2A)34 and HSP27 (CMT2F).35
Relatively little is known about the functions of non-bound HK1. PBD-lacking transcripts are expressed in mouse spermatogenic cells, with alternative splicing of the upstream exons subject to developmental regulation,15, 18 similar to our observations on AltT2-containing transcripts in the peripheral nerve. The translated protein is localised in the sperm tail and not with the mitochondria.15 In the brain axons, HK1 was shown to be transported independent of the mitochondria and to associate with presynaptic terminals.36 Microarray analysis of the peripheral nerve has shown that HK1 is developmentally regulated with the highest expression observed prenatally;37 however, the data do not allow any conclusions on the differential expression of isoforms. Proposed (but not confirmed) roles of non-bound HK1 include autophosphorylation and phosphorylation of histone H2A in the brain and cell surface signal transduction in the testis.38, 39 The molecular pathogenesis of HMSNR is likely to tell a new tale about this old enzyme. The identification of the mutation has already made genetic testing and counselling possible.
Acknowledgments
We thank the HMSNR patients and families for participating in this project. The study was funded by a grant of the US Muscular Dystrophy Association to LK and of the Netherlands Organization for Scientific Research grant 050-10-010 to FB. JH was supported by a WAIMR student fellowship.
Footnotes
Supplementary Information accompanies the paper on European Journal of Human Genetics website (http://www.nature.com/ejhg)
Conflict of interest
The authors declare no conflict of interest.
Supplementary Material
References
- Harding AE, Thomas PK. Autosomal recessive forms of hereditary motor and sensory neuropathy. J Neurol Neurosurg Psychiatry. 1980;43:669–678. doi: 10.1136/jnnp.43.8.669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barisic N, Claeys KG, Sirotkovic-Skerlev M, et al. Charcot-Marie-Tooth disease: a clinico-genetic confrontation. Ann Hum Genet. 2008;72:416–441. doi: 10.1111/j.1469-1809.2007.00412.x. [DOI] [PubMed] [Google Scholar]
- Kalaydjieva L, Gresham D, Gooding R, et al. N-myc downstream-regulated gene 1 is mutated in hereditary motor and sensory neuropathy-Lom. Am J Hum Genet. 2000;67:47–58. doi: 10.1086/302978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varon R, Gooding R, Steglich C, et al. Partial deficiency of the C-terminal-domain phosphatase of RNA polymerase II is associated with congenital cataracts facial dysmorphism neuropathy syndrome. Nat Genet. 2003;35:185–189. doi: 10.1038/ng1243. [DOI] [PubMed] [Google Scholar]
- Gooding R, Colomer J, King R, et al. A novel Gypsy founder mutation, p.Arg1109X in the CMT4C gene, causes variable peripheral neuropathy phenotypes. J Med Genet. 2005;42:e69. doi: 10.1136/jmg.2005.034132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalaydjieva L, Morar B, Chaix R, Tang H. A newly discovered founder population: the Roma/Gypsies. Bioessays. 2005;27:1084–1094. doi: 10.1002/bies.20287. [DOI] [PubMed] [Google Scholar]
- Guergueltcheva V, Tournev I, Bojinova V, et al. Early clinical and electrophysiologic features of the two most common autosomal recessive forms of Charcot-Marie-Tooth disease in the Roma (Gypsies) J Child Neurol. 2006;21:20–25. doi: 10.1177/08830738060210010401. [DOI] [PubMed] [Google Scholar]
- Thomas PK, Kalaydjieva L, Youl B, et al. Hereditary motor and sensory neuropathy-russe: new autosomal recessive neuropathy in Balkan Gypsies. Ann Neurol. 2001;50:452–457. doi: 10.1002/ana.1137. [DOI] [PubMed] [Google Scholar]
- Colomer J, Iturriaga C, Kalaydjieva L, et al. HMSN-Russe in two Spanish patients: distinctive features of the disease and current genetic findings. Acta Myologica. 2001;XX:202–209. [Google Scholar]
- Rogers T, Chandler D, Angelicheva D, et al. A novel locus for autosomal recessive peripheral neuropathy in the EGR2 region on 10q23. Am J Hum Genet. 2000;67:664–671. doi: 10.1086/303053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hantke J, Rogers T, French L, et al. Refined mapping of the HMSNR critical gene region–construction of a high-density integrated genetic and physical map. Neuromuscul Disord. 2003;13:729–736. doi: 10.1016/s0960-8966(03)00098-1. [DOI] [PubMed] [Google Scholar]
- Hanemann CO, Rosenbaum C, Kupfer S, Wosch S, Stoegbauer F, Muller HW. Improved culture methods to expand Schwann cells with altered growth behaviour from CMT1A patients. Glia. 1998;23:89–98. doi: 10.1002/(sici)1098-1136(199806)23:2<89::aid-glia1>3.0.co;2-z. [DOI] [PubMed] [Google Scholar]
- Andreoni F, Ruzzo A, Magnani M. Structure of the 5′ region of the human hexokinase type I (HKI) gene and identification of an additional testis-specific HKI mRNA. Biochim Biophys Acta. 2000;1493:19–26. doi: 10.1016/s0167-4781(00)00147-0. [DOI] [PubMed] [Google Scholar]
- Griffin LD, Gelb BD, Wheeler DA, Davison D, Adams V, McCabe ER. Mammalian hexokinase 1: evolutionary conservation and structure to function analysis. Genomics. 1991;11:1014–1024. doi: 10.1016/0888-7543(91)90027-c. [DOI] [PubMed] [Google Scholar]
- Mori C, Nakamura N, Welch JE, et al. Mouse spermatogenic cell-specific type 1 hexokinase (mHk1-s) transcripts are expressed by alternative splicing from the mHk1 gene and the HK1-S protein is localized mainly in the sperm tail. Mol Reprod Dev. 1998;49:374–385. doi: 10.1002/(SICI)1098-2795(199804)49:4<374::AID-MRD4>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- Pal N, Bessman SP. Insulin effect on [14C]-valine incorporation and its relation to hexokinase activity in developing brain. Biochem Biophys Res Commun. 1988;154:450–454. doi: 10.1016/0006-291x(88)90707-3. [DOI] [PubMed] [Google Scholar]
- Gelb BD, Adams V, Jones SN, Griffin LD, MacGregor GR, McCabe ER. Targeting of hexokinase 1 to liver and hepatoma mitochondria. Proc Natl Acad Sci USA. 1992;89:202–206. doi: 10.1073/pnas.89.1.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori C, Welch JE, Fulcher KD, O'Brien DA, Eddy EM. Unique hexokinase messenger ribonucleic acids lacking the porin-binding domain are developmentally expressed in mouse spermatogenic cells. Biol Reprod. 1993;49:191–203. doi: 10.1095/biolreprod49.2.191. [DOI] [PubMed] [Google Scholar]
- Murakami K, Piomelli S. Identification of the cDNA for human red blood cell-specific hexokinase isozyme. Blood. 1997;89:762–766. [PubMed] [Google Scholar]
- Wilson JE. Isozymes of mammalian hexokinase: structure, subcellular localization and metabolic function. J Exp Biol. 2003;206:2049–2057. doi: 10.1242/jeb.00241. [DOI] [PubMed] [Google Scholar]
- Gupta S, Zink D, Korn B, Vingron M, Haas SA. Strengths and weaknesses of EST-based prediction of tissue-specific alternative splicing. BMC Genomics. 2004;5:72. doi: 10.1186/1471-2164-5-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wegrzyn JL, Drudge TM, Valafar F, Hook V. Bioinformatic analyses of mammalian 5′-UTR sequence properties of mRNAs predicts alternative translation initiation sites. BMC Bioinformatics. 2008;9:232. doi: 10.1186/1471-2105-9-232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kochetov AV, Ischenko IV, Vorobiev DG, et al. Eukaryotic mRNAs encoding abundant and scarce proteins are statistically dissimilar in many structural features. FEBS Lett. 1998;440:351–355. doi: 10.1016/s0014-5793(98)01482-3. [DOI] [PubMed] [Google Scholar]
- Wang Z, Gardiner NJ, Fernyhough P. Blockade of hexokinase activity and binding to mitochondria inhibits neurite outgrowth in cultured adult rat sensory neurons. Neurosci Lett. 2008;434:6–11. doi: 10.1016/j.neulet.2008.01.057. [DOI] [PubMed] [Google Scholar]
- Sarrazin S, Starck J, Gonnet C, Doubeikovski A, Melet F, Morle F. Negative and translation termination-dependent positive control of FLI-1 protein synthesis by conserved overlapping 5′ upstream open reading frames in Fli-1 mRNA. Mol Cell Biol. 2000;20:2959–2969. doi: 10.1128/mcb.20.9.2959-2969.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warnakulasuriyarachchi D, Ungureanu NH, Holcik M. The translation of an antiapoptotic protein HIAP2 is regulated by an upstream open reading frame. Cell Death Differ. 2003;10:899–904. doi: 10.1038/sj.cdd.4401256. [DOI] [PubMed] [Google Scholar]
- Robey RB, Hay N. Mitochondrial hexokinases, novel mediators of the antiapoptotic effects of growth factors and Akt. Oncogene. 2006;25:4683–4696. doi: 10.1038/sj.onc.1209595. [DOI] [PubMed] [Google Scholar]
- Bianchi M, Magnani M. Hexokinase mutations that produce nonspherocytic hemolytic anemia. Blood Cells Mol Dis. 1995;21:2–8. doi: 10.1006/bcmd.1995.0002. [DOI] [PubMed] [Google Scholar]
- Kanno H. Hexokinase: gene structure and mutations. Baillieres Best Pract Res Clin Haematol. 2000;13:83–88. doi: 10.1053/beha.1999.0058. [DOI] [PubMed] [Google Scholar]
- van Wijk R, Rijksen G, Huizinga EG, Nieuwenhuis HK, van Solinge WW. HK Utrecht: missense mutation in the active site of human hexokinase associated with hexokinase deficiency and severe nonspherocytic hemolytic anemia. Blood. 2003;101:345–347. doi: 10.1182/blood-2002-06-1851. [DOI] [PubMed] [Google Scholar]
- Gardiner NJ, Wang Z, Luke C, Gott A, Price SA, Fernyhough P. Expression of hexokinase isoforms in the dorsal root ganglion of the adult rat and effect of experimental diabetes. Brain Res. 2007;1175:143–154. doi: 10.1016/j.brainres.2007.08.015. [DOI] [PubMed] [Google Scholar]
- Sancho S, Young P, Suter U. Regulation of Schwann cell proliferation and apoptosis in PMP22-deficient mice and mouse models of Charcot-Marie-Tooth disease type 1A. Brain. 2001;124:2177–2187. doi: 10.1093/brain/124.11.2177. [DOI] [PubMed] [Google Scholar]
- Parkinson DB, Bhaskaran A, Droggiti A, et al. Krox-20 inhibits Jun-NH2-terminal kinase/c-Jun to control Schwann cell proliferation and death. J Cell Biol. 2004;164:385–394. doi: 10.1083/jcb.200307132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuspiel M, Zunino R, Gangaraju S, Rippstein P, McBride H. Activated mitofusin 2 signals mitochondrial fusion, interferes with Bax activation, and reduces susceptibility to radical induced depolarization. J Biol Chem. 2005;280:25060–25070. doi: 10.1074/jbc.M501599200. [DOI] [PubMed] [Google Scholar]
- Bruey JM, Ducasse C, Bonniaud P, et al. Hsp27 negatively regulates cell death by interacting with cytochrome c. Nat Cell Biol. 2000;2:645–652. doi: 10.1038/35023595. [DOI] [PubMed] [Google Scholar]
- Garner JA, Linse KD, Polk RK. Type I brain hexokinase: axonal transport and membrane associations within central nervous system presynaptic terminals. J Neurochem. 1996;67:845–856. doi: 10.1046/j.1471-4159.1996.67020845.x. [DOI] [PubMed] [Google Scholar]
- Verheijen MH, Chrast R, Burrola P, Lemke G. Local regulation of fat metabolism in peripheral nerves. Genes Dev. 2003;17:2450–2464. doi: 10.1101/gad.1116203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams V, Griffin L, Towbin J, Gelb B, Worley K, McCabe ER. Porin interaction with hexokinase and glycerol kinase: metabolic microcompartmentation at the outer mitochondrial membrane. Biochem Med Metab Biol. 1991;45:271–291. doi: 10.1016/0885-4505(91)90032-g. [DOI] [PubMed] [Google Scholar]
- Kalab P, Visconti P, Leclerc P, Kopf GS. p95, the major phosphotyrosine-containing protein in mouse spermatozoa, is a hexokinase with unique properties. J Biol Chem. 1994;269:3810–3817. [PubMed] [Google Scholar]
- Ruzzo A, Andreoni F, Magnani M. Structure of the human hexokinase type I gene and nucleotide sequence of the 5′ flanking region. Biochem J. 1998;331 Pt 2):607–613. doi: 10.1042/bj3310607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arora KK, Fanciulli M, Pedersen PL. Glucose phosphorylation in tumor cells. Cloning, sequencing, and overexpression in active form of a full-length cDNA encoding a mitochondrial bindable form of hexokinase. J Biol Chem. 1990;265:6481–6488. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.