Abstract
Algorithms for inferring population structure from genetic data (ie, population assignment methods) have shown to effectively recognize genetic clusters in human populations. However, their performance in identifying groups of genealogically related individuals, especially in scanty-differentiated populations, has not been tested empirically thus far. For this study, we had access to both genealogical and genetic data from two closely related, isolated villages in southern Italy. We found that nearly all living individuals were included in a single pedigree, with multiple inbreeding loops. Despite Fst between villages being a low 0.008, genetic clustering analysis identified two clusters roughly corresponding to the two villages. Average kinship between individuals (estimated from genealogies) increased at increasing values of group membership (estimated from the genetic data), showing that the observed genetic clusters represent individuals who are more closely related to each other than to random members of the population. Further, average kinship within clusters and Fst between clusters increases with increasingly stringent membership threshold requirements. We conclude that a limited number of genetic markers is sufficient to detect structuring, and that the results of genetic analyses faithfully mirror the structuring inferred from detailed analyses of population genealogies, even when Fst values are low, as in the case of the two villages. We then estimate the impact of observed levels of population structure on association studies using simulated data.
Keywords: population structure, genetic isolate, genetic clustering methods, Fst, kinship
Introduction
Geographic isolates represent valuable resources for the dissection of complex genetic traits.1, 2, 3, 4 In principle, geographical isolation implies that genetic determinants and environmental factors contributing to complex traits are homogeneous across individuals. Unfortunately, undetected structuring within populations may bias association studies, and concerns about population stratification exist even in apparently homogeneous communities,5 essentially because their long-term demographic histories are generally unknown. The only way to ensure that isolates are not cryptically structured is through geneaological reconstruction.6, 7 In general, however, reconstructed pedigrees tend to span very few generations, and hence one has to resort to indirect evidence about structuring, typically obtained through analyses of genetic variation. To our knowledge, there has been no empirical comparison of genealogically and genetically inferred relationships in isolated populations.
A population is structured when it departs from panmixia because it is divided into sub-populations between which there is a certain degree of reproductive isolation. A number of Bayesian clustering algorithms have been developed in recent years that have proven effective in identifying genetic clusters of individuals in analyses of human populations.8, 9, 10, 11, 12, 13 Such populations were often distributed worldwide,14, 15, 16 but sometimes geographically close and isolated.17 Nonetheless, all populations investigated so far were well differentiated; values of Wright's genetic variance between sub-populations, Fst, were always >0.01. It is unknown whether and to what extent these methods can efficiently describe the structure of scanty-differentiated populations, such as those inhabiting small geographical regions. To date, this issue has only been addressed in simulated populations.18
Detection of population structure in genetic isolates is crucial because population stratification, ie, the existence of clusters of genetically non-independent individuals, is considered to be the main source of bias in association studies.19, 20, 21 Devlin and Roeder22 suggest that a common framework, termed genomic control, may be used to control for the effect of both population stratification and inter-individual relatedness on association tests using ‘null' markers. Where genealogical information is available, and inter-individual relatedness is considered the sole source of bias, this information is used directly in association tests without the need for genomic data.19, 23, 24
In this study, we use genealogical and genetic data from Gioi25 and Cardile, two isolated villages from southern Italy, close to a previously studied isolated village.26, 27 We investigated the extent of overlap between population structure inferred from genetic analyses and from detailed studies of genealogical relationships. We then develop a computer-simulation model incorporating observed levels of kinship to quantify the potential bias of observed levels of population structure on association studies.
Materials and methods
Study sample, genetic and genealogical data
The study sample comprises 1356 individuals from the villages of Gioi (n=882) and Cardile (n=474), corresponding almost completely to current residents. According to historical sources, the village of Gioi was settled in the ninth century BC by Greeks, and in the tenth century AD founders from Gioi settled Cardile 6 km away. High levels of reproductive isolation are reported for the two villages until the middle of the twentieth century.
We collected 20 383 birth records spanning the last four centuries from registry office and parish archives. These data were used to construct pedigrees spanning 350 years (15–17 generations). Kinship coefficients (Φij) between individuals i and j were calculated as described in Karigl et al28 and implemented in the KinInbCoeff module of the CC-QLS package.23
A genome-wide scan of 1122 microsatellites (average marker spacing of 3.6 cM and mean marker heterozygosity of 0.70) was performed by the deCODE genotyping service on DNA extracted from peripheral blood from all study samples.
Genetic clustering analysis
Genetic clusters were inferred by the software Structure, under assumptions of admixture, correlated allele frequencies, and no prior population information.9, 29 For each number of clusters (K) from 1 to 8, 50 runs were performed using a burnin length of 20 000 iterations followed by 10 000 iterations. For each K, the posterior probability of clustering was estimated from the average logarithmic probability of data across runs. The second order rate of change of logarithmic probability of data between subsequent K values was estimated according to Evanno et al30 to identify the optimal number of clusters in the data. Resulting membership coefficients generated by Structure were input into CLUMPP31 and analyzed using the LargeKGreedy algorithm. No genuine multimodality was found among runs with average similarity (G' values) of 0.99, 0.79, and 0.89% for K equals 2, 3 and 4, respectively. Graphical display of membership coefficients was obtained by Distruct.32
Structure was run twice under the conditions described above. First, genotypes at 239 loci, a subset of the1122 loci available in our data, chosen to minimize the probability of linkage disequilibrium between adjacent markers on the chromosomes, were analyzed in all 1356 individuals. Then, we compared the 36 markers common to this study and to the HGDP-CEPH Human Genome Diversity Panel, using random subsamples of 37 and 22 individuals from Gioi and Cardile, respectively, and 161 European individuals available in the HGDP panel. Sizes of subsamples from the two villages were chosen to approximate sample sizes of European populations.
Spearman correlation coefficients were estimated using SPSS 13.0 (SPSS Inc., Chicago, IL, USA). Fst values were computed using the Arlequin 3.1 software.33
Assessment of bias in association tests
Both population structure and relatedness among individuals contribute to non-independence of genotypes, which, in turn, inflates the variance of tests for allelic association, by a factor λ. For quantitative traits, Bacanu et al34 proposed a method to quantify inflation of the variance of t2 using null markers (ie, λGC). An alternative, generalized least squares approach, suitable for large, inbred pedigrees with high consanguinity and no population stratification, was proposed by Abney et al.19 Here, the genealogy-based variance inflation factor, λGB, is computed exactly while computing the t2 statistic, and it corresponds to the ratio between non-corrected and corrected t2 statistics at a given marker.
Simulations for comparing λGB and λGC were carried out using the Genedrop program from the MORGAN 2.6 package.35 A quantitative trait was simulated for the 446 individuals in the largest pedigree of Cardile, and for a random sample of the same size in the largest pedigrees of Gioi and Gioi–Cardile (see Table 1) with heritability=0.3 and total phenotypic variance=0.0027. We then estimated λGC by simulating genotypes at 1122 null markers covering the whole genome for all individuals in the sample, as described in Ciullo et al.27 To mimic a realistic situation, allele frequencies and inter-marker distances matched those of the 1122 microsatellite markers genotyped in the two villages.26 A biallelic locus with a minor allele frequency of 0.3 was further simulated to estimate λGB. We considered four simulation schemes in which this locus had no effect on the trait (null model), additive, dominant, or recessive effects. For each model, the median values of λGB and λGC (expected values of λGB=1 and λGC=1) over 1000 simulations were estimated.
Table 1. Features of pedigrees reconstructed from sampled individuals using genealogical data.
| All pedigrees | Gioi–Cardile | Gioi | Cardile |
|---|---|---|---|
| Sampled individuals included | 1356 | 882 | 474 |
| No. of pedigrees | 63 | 45 | 19 |
| No. of generations | Up to 15 | Up to 15 | Up to 15 |
| Total members in pedigrees | 5272 | 4190 | 2384 |
| Largest pedigree | |||
| Sampled individuals included | 1274 | 828 | 446 |
| No. of pedigrees | 1 | 1 | 1 |
| No. of generations | 15 | 15 | 15 |
| Total members in the pedigree | 5165 | 4113 | 2354 |
Results
Genetic clustering
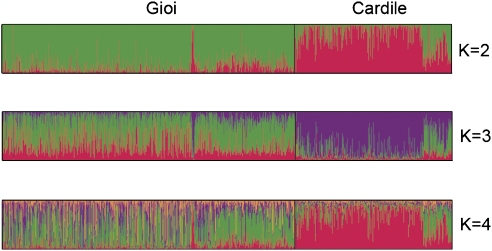
We analyzed population structure in Gioi and Cardile considering up to eight possible genetic clusters (K). Graphical representation of membership to clusters for K=2–4 is shown in Figure 1. The distribution of the logarithmic probability of the data between successive values of K showed no obvious peaks (Supplementary Figure 1); therefore, we inferred the number of clusters by Evanno's rate of change method30 rather than computing the posterior probability of the data.36 The most likely number of clusters was two, with clusters roughly corresponding to villages, despite the limited geographical distance. Individuals were clearly assigned to one of the two clusters, with 78% showing membership coefficients ≥0.75, 55%, ≥0.90 and 37%, ≥0.95 (Supplementary Table 1).
Figure 1.
Cluster membership according to analyses of genotypes at 239 markers in all individuals in the study sample, for K=2–4. Each inferred cluster is represented by a different color.
In comparison with other European populations, no population structure between the villages is apparent, as expected given the greater geographical scope of the analysis (Supplementary Figure 2). In fact, there was essentially no structure at all in the European plus Cilento data set, most likely because the limited degree of differentiation known to exist among European populations14, 37, 38 is likely to be undetectable with the low number of shared markers available for consideration. To better clarify this point, we analyzed the subsamples from Gioi and Cardile with the same 36 markers used for comparison with Europe and, again, we were unable to detect the same structure identified using 239 markers (data not shown). This suggests that comparisons across European populations are hardly informative when the number of markers is so small.
Validation of genetic clustering analysis by means of genealogical data
Kinship calculation
Using genealogical data, we backward reconstructed pedigrees starting from all contemporary individuals sampled in Gioi and Cardile (Table 1). In the same table, we report features of the largest reconstructed pedigree, comprising 5165 members and spanning 15 generations. As can be seen, for both individual villages and for the combined population a single pedigree includes nearly all individuals. This proved the presence of multiple relatedness links among individuals and confirmed, in the combined analysis, the common origin of the two villages.
Relatedness was quantified by pairwise kinship coefficients inferred from pedigree data. Summary kinship statistics are reported in Table 2, together with data on other isolated populations from the literature.7, 39, 40 Average kinship between individuals of the current generation is 0.004 in Gioi (equal to that between third cousins) and 0.009 in Cardile (approaching that between second cousins, ie, Φij=0.015), showing a high degree of inbreeding in both villages.
Table 2. Kinship (Φij) summary statistics of Gioi and Cardile compared with those of other isolated populations.
| Sample size | Average±SD | Median | 25–75% percentiles | |
|---|---|---|---|---|
| Gioi–Cardile | 1356 | 0.003±0.015 | 0.001 | 0.000–0.002 |
| Gioi | 882 | 0.004±0.018 | 0.001 | 0.000–0.002 |
| Cardile | 474 | 0.009±0.024 | 0.004 | 0.001–0.008 |
| Perdasdefogua | 821 | NA | 0.007 | 0.004–0.011 |
| Talanaa | 875 | NA | 0.014 | 0.009–0.021 |
| S-leut Hutteritesb | 806 | 0.042±0.031 | NA | NA |
| Icelandc | 37762 | 0.008±NA | NA | 0.000–0.001 |
| 1925–1949 cohort | ||||
| Icelandc | 38336 | 0.005±NA | NA | 0.000–0.001 |
| 1950–1965 cohort |
Membership and kinship
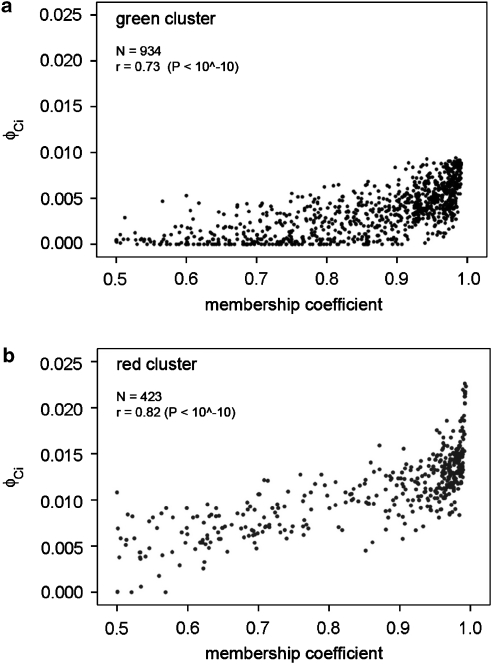
In determining the optimum clustering of the data, Structure estimates membership coefficients corresponding to the probability of an individual's genome belonging to each cluster. To investigate patterns of kinship as a function of group membership, we grouped individuals into clusters to which they had 50% or greater probability of belonging regardless of their village of origin. We compared membership coefficients for each individual with their estimated average kinship with all other members of the cluster, namely ΦCi, where C represents the cluster (C=1, 2, representing the green and red clusters in Figure 1) and i the individual considered (934 in the green and 423 in the red clusters). We found highly significant correlations between cluster membership coefficients and ΦCi, namely r=0.73 (P<10−10, N=934) and r=0.082 (P<10−10, N=423), respectively, for the green and red clusters in Figure 2.
Figure 2.
Relationship between average kinship of individuals with other cluster members (ΦCij; Y axis) and membership coefficients (X axis) for (a) green and (b) red clusters for K=2. Rank correlation coefficients (r) and number of observations (N) are also shown.
Fst and kinship
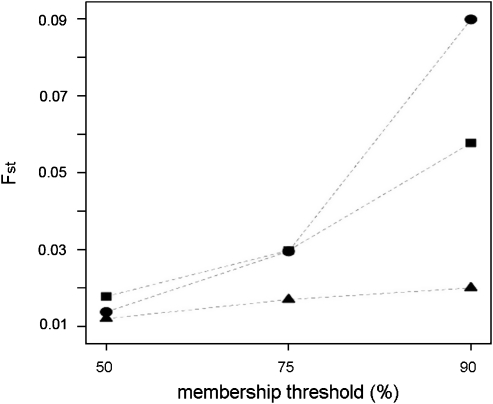
Pairwise Fst between samples from the two villages evaluated using genotypes at the 239 unlinked loci from all sampled individuals is a low 0.008. We investigated whether Fst calculations would be affected with subsamples of individuals with increasing relatedness. To this end, for K=2–4, individuals were clustered with increasing stringency of membership threshold requirements (threshold coefficient levels of 50, 75, 90, 95 and 99 percent; individuals in higher threshold clusters are also found in lower clusters). Regardless of the value of K considered, average kinship within clusters and Fst between clusters increases with increasing membership threshold (Supplementary Table 1, Figure 3). This is true especially for high levels of kinship; with 99% threshold and K=2, kinship in the two groups is between half-siblings (0.125) and first cousins (0.06) and Fst is 0.113.
Figure 3.
Genetic distances (overall Fst) among clusters for varying K values (triangles: K=2; circles: K=3; squares: K=4) and threshold levels of cluster membership. Individuals in higher threshold clusters are also found in lower clusters. Within each K, Fst increases with increasingly stringent threshold levels required for cluster membership (and thus kinship).
Substructure effect on association studies
The variance correction factor of the test for quantitative association was estimated by simulation, in Gioi and Cardile separately and in the combined Gioi–Cardile sample, using both genomic (λGC) and genomic/genealogical (λGB) approaches.
Genomic- and genealogy-based corrections performed similarly in our data with similar median values of λGC and λGB (Table 3), regardless of the model used for λ computation, in Gioi and Cardile, where no population structure is described. When the two populations were combined, λGC and λGB values remained close to those estimated in the two, unstructured, individual populations, and hence close to 1. This result suggests that existing population substructure does not substantially impact the simple association test for quantitative traits when Gioi and Cardile are analyzed together. Once inter-individual relatedness is correctly accounted for using either genomic or genealogical data, the impact of these levels of population substructure appears negligible.
Table 3. Comparison of λGC and λGB in Gioi–Cardile, Gioi, and Cardile samples.
| Median λGC (95% CI) | Gioi—Cardile | Gioi | Cardile |
|---|---|---|---|
| Null | 1.126 (1.015–1.262) | 1.161 (1.035–1.322) | 1.276 (1.103–1.487) |
| Additive | 1.158 (1.043–1.301) | 1.196 (1.060–1.363) | 1.317 (1.133–1.567) |
| Dominant | 1.149 (1.027–1.278) | 1.193 (1.057–1.359) | 1.310 (1.127–1.554) |
| Recessive | 1.142 (1.021–1.274) | 1.186 (1.054–1.353) | 1.300 (1.121–1.549) |
| Median λGB (95% CI) | |||
| Null | 1.123 (1.043–1.227) | 1.168 (1.063–1.307) | 1.275 (1.126–1.461) |
| Additive | 1.136 (1.053–1.242) | 1.183 (1.077–1.333) | 1.304 (1.141–1.511) |
| Dominant | 1.127 (1.043–1.241) | 1.180 (1.073–1.327) | 1.298 (1.134–1.507) |
| Recessive | 1.126 (1.044–1.245) | 1.177 (1.071–1.333) | 1.295 (1.129–1.504) |
Median values obtained from null distributions of the tests, considering different genetic effects, are reported.
Discussion
According to available historical data, the populations of Gioi and Cardile share a largely common origin, separating approximately 1000 years ago. Whether or not such a recent separation, combined with the close geographical distance and small population sizes (approximately 1000 for Gioi and 500 for Cardile), could result in significant differentiation was not obvious from the start. We found that genetic differentiation measured by Fst is an apparently low 0.8%. However, Rosenberg et al14 found that Fst between random (ie, non-isolated) European populations is even lower (namely 0.7%). Therefore, it is safe to conclude that small population sizes and even limited degrees of geographical isolation may rapidly lead to what can be considered a relatively sharp genetic divergence on a European scale. The high level of observed consanguinity confirms that these villages do represent genetic isolates, and hence are potentially useful for the study of complex traits.
Availability of genealogical records since the seventeenth century allowed us to calculate pairwise kinship as an estimator of relatedness between individuals. Strictly speaking, although kinship estimates based on genealogy path counting may not be as accurate as estimates based on genomic data, especially in populations with histories of consanguinity,41, 42 kinship inferred from genealogies allowed us to quantify relationships between individuals independently from data used to infer genetic clusters.
The average kinship in Gioi and Cardile is slightly lower than in other Italian genetic isolates40 and one order of magnitude lower than in the highly inbred S-leut Hutterites.39 A high correlation emerged between the two independent descriptors of population structure. This study confirms early findings in populations from New Guinea43, 44 that limited population size and nonrandom mate choice (resulting in genealogical structure in the population, and ultimately in inbreeding) are indeed reflected in distributions of allele frequencies. To our knowledge, our study represents the first empirical demonstration of that finding based on a thorough comparison of DNA and genealogical data.
Latch et al18 compared simulated data at 10 co-dominant loci in 100 individuals from five populations, assuming Fst values in the range of 1–10%. They concluded that available methods assign genotypes to clusters with accuracy >97% only if Fst is greater than 5%. Conversely, in this study we were able to identify two clusters and assign individuals almost consistently to their geographic origin, despite Fst being only 0.8%, probably due to the fact that Latch and colleagues considered lower numbers of markers and individuals.9, 45 We corroborated our result showing that: (a) individuals belonging to clusters are also genealogically related to other cluster members (by correlating membership coefficients inferred from genomic analysis and average kinship with other cluster members; Figure 2); and (b) the higher the average membership coefficient for a cluster, the greater the kinship between cluster members (as shown by the significant increase in average kinship with increasing membership stringency; Supplementary Table 1).
Population-based studies of complex traits are known to be sensitive to undetected structuring. When genealogical data are not available, genomic-based corrections represent the only viable alternative. By simulation, we estimated to what extent structuring may inflate measures of phenotype–genotype association. We expected that when Gioi and Cardile are treated as a single population, the differences between them, albeit limited, could lead to a poorer performance of genomic- versus genealogy-based corrections. To the contrary, the results show that the effects of the levels of structuring observed in Gioi and Cardile are unlikely to affect these measures to any substantial degree. Once inter-individual relatedness within each population is correctly handled, the effects of subtle differentiation between populations seem limited and indeed not large enough to significantly bias results of association studies.
In short, this study shows that populations may be structured even in geographically close localities whose inhabitants shared ancestors in the recent past. Furthermore, a limited number of neutral genetic markers (eg, 239 in our study) is sufficient to detect these low levels of structuring, and the results of genetic analyses reproduce faithfully the structuring inferred from detailed analyses of population genealogies. Therefore, when the complete genealogy of a study population cannot be reconstructed, which is more often the rule than the exception: (a) typing a few hundred polymorphisms allows one to recognize the effects of kinship and (b) the effects of kinship can be incorporated into models that predict possible biases in association studies. A still open question, which we plan to address soon, is the minimum number of polymorphisms necessary to recognize the population structure at this level of differentiation.
Acknowledgments
We thank the populations of Gioi and Cardile for their kind cooperation. We thank Don Guglielmo Manna and Dr Leopoldo Errico for helping in the interaction with the populations; Raffaella Romano and Teresa Rizzo for the organization of the study in the villages; Valerio Di Vico for providing us with historical information; Agnar Helgason, Giorgio Bertorelle, and Catherine Bourgain for several important suggestions and comments. This study was supported by funds of the Italian Ministry of Universities (PRIN 2006 and FIRB 2008) to G.B, (FIRB -RBIN064YAT) and the Fondazione Banco di Napoli to MC.
Footnotes
Supplementary Information accompanies the paper on European Journal of Human Genetics website (http://www.nature.com/ejhg)
Supplementary Material
References
- Peltonen L. Positional cloning of disease genes: advantages of genetic isolates. Hum Hered. 2000;50:66–75. doi: 10.1159/000022892. [DOI] [PubMed] [Google Scholar]
- Shifman S, Darvasi A. The value of isolated populations. Nat Genet. 2001;28:309–310. doi: 10.1038/91060. [DOI] [PubMed] [Google Scholar]
- Wright AF, Carothers AD, Pirastu M. Population choice in mapping genes for complex diseases. Nat Genet. 1999;23:397–404. doi: 10.1038/70501. [DOI] [PubMed] [Google Scholar]
- Kristiansson K, Naukkarinen J, Peltonen L. Isolated populations and complex disease gene identification. Genome Biol. 2008;9:109. doi: 10.1186/gb-2008-9-8-109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helgason A, Yngvadottir B, Hrafnkelsson B, Gulcher J, Stefansson K. An Icelandic example of the impact of population structure on association studies. Nat Genet. 2005;37:90–95. doi: 10.1038/ng1492. [DOI] [PubMed] [Google Scholar]
- Madrigal L, Melendez-Obando M. Grandmothers' longevity negatively affects daughters' fertility. Am J Phys Anthropol. 2008;136:223–229. doi: 10.1002/ajpa.20798. [DOI] [PubMed] [Google Scholar]
- Helgason A, Palsson S, Gudbjartsson DF, Kristjansson T, Stefansson K. An association between the kinship and fertility of human couples. Science. 2008;319:813–816. doi: 10.1126/science.1150232. [DOI] [PubMed] [Google Scholar]
- Chen C, Durands E, Forbes F, Francois O. Bayesian clustering algorithms ascertaining spatial population structure: a new computer program and a comparison study. Mol Ecol Notes. 2007;7:747–756. [Google Scholar]
- Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corander J, Waldmann P, Marttinen P, Sillanpaa MJ. BAPS 2: enhanced possibilities for the analysis of genetic population structure. Bioinformatics. 2004;20:2363–2369. doi: 10.1093/bioinformatics/bth250. [DOI] [PubMed] [Google Scholar]
- Dawson KJ, Belkhir K. A Bayesian approach to the identification of panmictic populations and the assignment of individuals. Genet Res. 2001;78:59–77. doi: 10.1017/s001667230100502x. [DOI] [PubMed] [Google Scholar]
- Guillot G, Estoup A, Mortier F, Cosson JF. A spatial statistical model for landscape genetics. Genetics. 2005;170:1261–1280. doi: 10.1534/genetics.104.033803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huelsenbeck JP, Andolfatto P. Inference of population structure under a Dirichlet process model. Genetics. 2007;175:1787–1802. doi: 10.1534/genetics.106.061317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg NA, Pritchard JK, Weber JL, et al. Genetic structure of human populations. Science. 2002;298:2381–2385. doi: 10.1126/science.1078311. [DOI] [PubMed] [Google Scholar]
- Li JZ, Absher DM, Tang H, et al. Worldwide human relationships inferred from genome-wide patterns of variation. Science. 2008;319:1100–1104. doi: 10.1126/science.1153717. [DOI] [PubMed] [Google Scholar]
- Jakobsson M, Scholz SW, Scheet P, et al. Genotype, haplotype and copy-number variation in worldwide human populations. Nature. 2008;451:998–1003. doi: 10.1038/nature06742. [DOI] [PubMed] [Google Scholar]
- Vitart V, Biloglav Z, Hayward C, et al. 3000 years of solitude: extreme differentiation in the island isolates of Dalmatia, Croatia. Eur J Hum Genet. 2006;14:478–487. doi: 10.1038/sj.ejhg.5201589. [DOI] [PubMed] [Google Scholar]
- Latch EK, Dharmarajan G, Glaubitz JC, Rhodes OE. Relative performance of Bayesian clustering software for inferring population substructure and individual assignment at low levels of population differentiation. Conservation Genetics. 2006;7:295–302. [Google Scholar]
- Abney M, Ober C, McPeek MS. Quantitative-trait homozygosity and association mapping and empirical genomewide significance in large, complex pedigrees: fasting serum-insulin level in the Hutterites. Am J Hum Genet. 2002;70:920–934. doi: 10.1086/339705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourgain C, Genin E. Complex trait mapping in isolated populations: are specific statistical methods required. Eur J Hum Genet. 2005;13:698–706. doi: 10.1038/sj.ejhg.5201400. [DOI] [PubMed] [Google Scholar]
- Newman DL, Abney M, McPeek MS, Ober C, Cox NJ. The importance of genealogy in determining genetic associations with complex traits. Am J Hum Genet. 2001;69:1146–1148. doi: 10.1086/323659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devlin B, Roeder K. Genomic control for association studies. Biometrics. 1999;55:997–1004. doi: 10.1111/j.0006-341x.1999.00997.x. [DOI] [PubMed] [Google Scholar]
- Bourgain C, Hoffjan S, Nicolae R, et al. Novel case-control test in a founder population identifies P-selectin as an atopy-susceptibility locus. Am J Hum Genet. 2003;73:612–626. doi: 10.1086/378208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thornton T, McPeek MS. Case-control association testing with related individuals: a more powerful quasi-likelihood score test. Am J Hum Genet. 2007;81:321–337. doi: 10.1086/519497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciullo M, Nutile T, Dalmasso C, et al. Identification and replication of a novel obesity locus on chromosome 1q24 in isolated populations of Cilento. Diabetes. 2008;57:783–790. doi: 10.2337/db07-0970. [DOI] [PubMed] [Google Scholar]
- Colonna V, Nutile T, Astore M, et al. Campora: a young genetic isolate in South Italy. Hum Hered. 2007;64:123–135. doi: 10.1159/000101964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciullo M, Bellenguez C, Colonna V, et al. New susceptibility locus for hypertension on chromosome 8q by efficient pedigree-breaking in an Italian isolate. Hum Mol Genet. 2006;15:1735–1743. doi: 10.1093/hmg/ddl097. [DOI] [PubMed] [Google Scholar]
- Karigl G. A recursive algorithm for the calculation of identity coefficients. Ann Hum Genet. 1981;45:299–305. doi: 10.1111/j.1469-1809.1981.tb00341.x. [DOI] [PubMed] [Google Scholar]
- Falush D, Stephens M, Pritchard JK. Inference of population structure using multilocus genotype data: linked loci and correlated allele frequencies. Genetics. 2003;164:1567–1587. doi: 10.1093/genetics/164.4.1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evanno G, Regnaut S, Goudet J. Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol. 2005;14:2611–2620. doi: 10.1111/j.1365-294X.2005.02553.x. [DOI] [PubMed] [Google Scholar]
- Jakobsson M, Rosenberg NA. CLUMPP: a cluster matching and permutation program for dealing with label switching and multimodality in analysis of population structure. Bioinformatics. 2007;23:1801–1806. doi: 10.1093/bioinformatics/btm233. [DOI] [PubMed] [Google Scholar]
- Rosenberg NA. DISTRUCT: a program for the graphical display of population structure. Mol Ecol Notes. 2004;4:137–138. [Google Scholar]
- Excoffier L, Laval LG, Schneider S. Arlequin ver. 3.0: An integrated software package for population genetics data analysis. Evol Bioinformatics Online. 2005;1:47–50. [PMC free article] [PubMed] [Google Scholar]
- Bacanu SA, Devlin B, Roeder K. Association studies for quantitative traits in structured populations. Genet Epidemiol. 2002;22:78–93. doi: 10.1002/gepi.1045. [DOI] [PubMed] [Google Scholar]
- Wijsman EM, Rothstein JH, Thompson EA. Multipoint linkage analysis with many multiallelic or dense diallelic markers: Markov chain-Monte Carlo provides practical approaches for genome scans on general pedigrees. Am J Hum Genet. 2006;79:846–858. doi: 10.1086/508472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritchard JK.WW: Documentation for structure software:version 2 available on line at http://pritch.bsd.uchicago.edu/software/readme_2_1/readme.html , 2004
- Lao O, Lu TT, Nothnagel M, et al. Correlation between genetic and geographic structure in Europe. Curr Biol. 2008;18:1241–1248. doi: 10.1016/j.cub.2008.07.049. [DOI] [PubMed] [Google Scholar]
- Novembre J, Johnson T, Bryc K, et al. Genes mirror geography within Europe. Nature. 2008;456:98–101. doi: 10.1038/nature07331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abney M, McPeek MS, Ober C. Estimation of variance components of quantitative traits in inbred populations. Am J Hum Genet. 2000;66:629–650. doi: 10.1086/302759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falchi M, Forabosco P, Mocci E, et al. A genomewide search using an original pairwise sampling approach for large genealogies identifies a new locus for total and low-density lipoprotein cholesterol in two genetically differentiated isolates of Sardinia. Am J Hum Genet. 2004;75:1015–1031. doi: 10.1086/426155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leutenegger AL, Prum B, Genin E, et al. Estimation of the inbreeding coefficient through use of genomic data. Am J Hum Genet. 2003;73:516–523. doi: 10.1086/378207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu F, Elefante S, van Duijn CM, Aulchenko YS. Ignoring distant genealogic loops leads to false-positives in homozygosity mapping. Ann Hum Genet. 2006;70:965–970. doi: 10.1111/j.1469-1809.2006.00279.x. [DOI] [PubMed] [Google Scholar]
- Long JC. The allelic correlation structure of Gainj- and Kalam-speaking people. I. The estimation and interpretation of Wright's F-statistics. Genetics. 1986;112:629–647. doi: 10.1093/genetics/112.3.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood JW, Johnson PL, Kirk RL, McLoughlin K, Blake NM, Matheson FA. The genetic demography of the Gainj of Papua New Guinea. I. Local differentiation of blood group, red cell enzyme, and serum protein allele frequencies. Am J Phys Anthropol. 1982;57:15–25. doi: 10.1002/ajpa.1330570105. [DOI] [PubMed] [Google Scholar]
- Rosenberg NA, Mahajan S, Ramachandran S, Zhao C, Pritchard JK, Feldman MW. Clines, clusters, and the effect of study design on the inference of human population structure. PLoS Genet. 2005;1:e70. doi: 10.1371/journal.pgen.0010070. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.