Abstract
Seven secretory mammalian kexin-like subtilases have been identified that cleave a variety of precursor proteins at monobasic and dibasic residues. The recently characterized pyrolysin-like subtilase SKI-1 cleaves proproteins at nonbasic residues. In this work we describe the properties of a proteinase K-like subtilase, neural apoptosis-regulated convertase 1 (NARC-1), representing the ninth member of the secretory subtilase family. Biosynthetic and microsequencing analyses of WT and mutant enzyme revealed that human and mouse pro-NARC-1 are autocatalytically and intramolecularly processed into NARC-1 at the (Y,I)VV(V,L)(L,M)↓ motif, a site that is representative of its enzymic specificity. In vitro peptide processing studies and/or Ala substitutions of the P1–P5 sites suggested that hydrophobic/aliphatic residues are more critical at P1, P3, and P5 than at P2 or P4. NARC-1 expression is highest in neuroepithelioma SK-N-MCIXC, hepatic BRL-3A, and in colon carcinoma LoVo-C5 cell lines. In situ hybridization and Northern blot analyses of NARC-1 expression during development in the adult and after partial hepatectomy revealed that it is expressed in cells that have the capacity to proliferate and differentiate. These include hepatocytes, kidney mesenchymal cells, intestinal ileum, and colon epithelia as well as embryonic brain telencephalon neurons. Accordingly, transfection of NARC-1 in primary cultures of embryonic day 13.5 telencephalon cells led to enhanced recruitment of undifferentiated neural progenitor cells into the neuronal lineage, suggesting that NARC-1 is implicated in the differentiation of cortical neurons.
Keywords: cleavage specificity‖hypercholesterolemia‖neurogenesis‖hepatogenesis
Proproteins are the fundamental units from which bioactive proteins and peptides are derived by limited proteolysis. Eight of the mammalian proprotein convertases (PCs) responsible for the tissue-specific processing of secretory precursors have been identified since 1990. Of these, seven belong to the yeast kexin subfamily of subtilases and exhibit cleavage- specificity for basic sites respecting the consensus (K,R)-(X)n-(K,R)↓, where n = 0, 2, 4, or 6. They are known as furin, PC1/3, PC2, PC4, PACE4, PC5/6, and PC7/LPC, respectively (1–3). Although less generalized, limited proteolysis also occurs at amino acids such as L, V, M, A, T, and S (4, 5). Recently, a subtilisin-kexin-like isozyme called SKI-1/S1P, belonging to the pyrolysin subfamily of subtilases was identified (6, 7). It cleaves at nonbasic residues in the (R,K)-X-(hydrophobic)-(L,T,K,F)↓ motif (3, 6–11).
Evidence for the existence of processing sites not recognized by the known PCs (12) (M. J. Vincent, personal communication) prompted us to mine databases. By using short, conserved segments of the SKI-1 catalytic subunit as baits and the protein blast program (www.ncbi.nlm.nih.gov/BLAST), we identified in the patented database a putative convertase called neural apoptosis-regulated convertase 1 (NARC-1; Millenium Pharmaceuticals, Cambridge, MA, patent no. WO 01/57081 A2) and LP251 (Eli Lilly, patent no. WO 02/14358 A2) recently cloned by two pharmaceutical companies. NARC-1 was identified via the cloning of cDNAs up-regulated after apoptosis induced by serum deprivation in primary cerebellar neurons, whereas LP251 was discovered via global cloning of secretory proteins. Aside from the fact that NARC-1 mRNA is expressed in liver ≫ testis > kidney and that the gene localizes to human chromosome 1p33-p34.3, no information is available on NARC-1 activity, cleavage specificity, cellular and tissue expression, and biological function.
In this article, we show that NARC-1 belongs to the proteinase K subfamily of subtilases. It is synthesized as a soluble zymogen that undergoes autocatalytic intramolecular processing in the endoplasmic reticulum (ER) at the (Y,I)VV(V,L)(L,M)↓ motif, resulting in the cleavage of its prosegment that remains associated with the secreted enzyme. Tissue distribution analysis in adulthood and during ontogeny in mouse and rat by Northern blots and in situ hybridization histochemistry (ISH) revealed that NARC-1 mRNA is expressed in a limited number of tissues including the liver, kidney, cerebellum, and small intestine. Its expression is sometimes transient, as in kidney epithelial and brain telencephalon cells. On induction of liver regeneration after partial hepatectomy (PHx) in the rat, NARC-1 mRNA levels peaked on days 2–3, whereas those of PC5 were maximal on day 1. NARC-1 overexpression in primary embryonic telencephalon cells obtained from embryonic day 13.5 (E13.5) mice results in a higher percentage of differentiated neurons.
Materials and Methods
Cloning of Mouse, Rat, and Human NARC-1 and Their Mutants.
The GenBank accession nos. of human, mouse, and rat NARC-1 cDNAs are AX127530, AX207688, and AX207690, respectively. Full-length human NARC-1 (hNARC-1) (nucleotides 233–2311) was cloned by RT-PCR from HepG2 cells, and full-length mouse (nucleotides 137–2259) and partial rat (nucleotides 1215–2284) orthologs were obtained from the liver. RT-PCRs were performed by using a Titan (Roche Diagnostics) one-tube kit. The cDNAs were transferred to a derivative of the mammalian expression vector pIRES2-EGFP (enhanced GFP) (CLONTECH) that contains a C-terminal V5 epitope, allowing protein detection with a V5 mAb (Invitrogen) (11, 13). Mutations were introduced by PCR, as described (11).
Biosynthetic Analysis.
Recombinant vectors were used to express hNARC-1 and mouse NARC-1 (mNARC-1) in HK293, Chinese hamster ovary (CHO), and Neuro2A cells by transfection with Effectene/Lipofectamine. Transfected cells were pulse- or pulse–chase-labeled with [35S]EasyTag Express Protein labeling mix (Perkin–Elmer) for various times in the presence or absence of tunicamycin (5 μg/ml) or brefeldin A (2.5 μg/ml). The media and cell lysates were immunoprecipitated with V5 mAb, and the immune complexes were resolved by SDS/PAGE on 8% polyacrylamide/tricine or glycine gels (13).
In Vitro Enzymatic Assays.
Media from HK293 cells expressing pIRES2-EGFP alone or recombinant h/mNARC-1 were concentrated 10-fold in a Centricon 30 (Millipore). Ten microliters of 10× concentrated media was incubated at 37°C for 3–18 h with 50 μM of Suc-RPFHLLVY-MCA (4-methylcoumarin-7-amide), Suc-LLVY-MCA (Enzyme Systems Products, Livermore, CA) or Boc-VVVL-MCA (A.B., unpublished work) in 25 mM Tris/Mes, pH 7.4 + 2.5 mM CaCl2 and 0.5% SDS. Fluorescence and matrix-assisted laser desorption ionization time-of-flight analysis of the products were as described (14).
Northern Blot and ISH.
Northern blot analyses (15) were performed on poly(A)+ mRNA from cell lines (16) or 3-month-old ≈250-g Sprague–Dawley male rat tissues, using an RNeasy protocol (Qiagen, Chatsworth, CA). The blots were hybridized overnight at 68°C with [32P]UTP NARC-1 cRNA probes comprising nucleotides 784–1406, 1197–2090, or 1215–2284 of human, mouse, or rat NARC-1, respectively. For ISH, mouse or rat sense and antisense cRNA probes coding for NARC-1, furin, SKI-1, PC7, PACE4, and PC5 were labeled with 35S-UTP and 35S-CTP (1,250 Ci/mmol; Amersham Pharmacia) (17), to obtain high specific activities of ≈1,000 Ci/mmol. ISH was undertaken by using 8- to 10-μm mouse tissue cryostat sections fixed for 1 h in 4% formaldehyde and hybridized overnight at 55°C. For autoradiography, the sections were dipped in photographic emulsion (NTB-2, Kodak), exposed for 6–12 days, developed in D19 solution (Kodak), and stained with hematoxylin.
PHx.
PHx was achieved by removal of two-thirds of the rat liver (18), and tissues were isolated 1, 2, 3, 6, and 10 days postsham operation or PHx for Northern blotting and ISH.
Telencephalic Neural Progenitor Cell Culture and Transient Transfections.
Primary neural progenitor cell cultures were established from dorsal telencephalic cortices dissected from E13.5 mouse embryos (19, 20). At day 0, 4 × 105 cells were seeded into 4-well chamber slides (Nalge Nunc) coated with 0.1% poly-d-lysine and 0.2% laminin (BD Biosciences, Mississauga, Canada). The cells were cultured in Neurobasal medium supplemented with 1% N2, 2% B27, 0.5 mM glutamine, 1% penicillin plus streptomycin (Invitrogen), and 40 ng/ml FGF2 (Collaborative Research). Transfections were performed at day 2, when no significant neuronal differentiation is observed (21). Plasmid DNA (0.5 μg per well) was incubated for 5 min in OptiMEM (Invitrogen). An equal volume of OptiMEM and Lipofectamine 2000 reagent (Invitrogen; 1.25 μl/μg of DNA) was added to the DNA mixture and incubated for 20 min. The DNA/Lipofectamine 2000 mix was added dropwise to each well. The cells were allowed to differentiate until days 4 and 5, fixed, and subjected to double-label immunocytochemical analysis of the expression of GFP and Ki-67 (a marker of undifferentiated neural progenitor cells) or NeuN (a marker of differentiated neurons) (22). Ki-67 and NeuN antibodies were from PharMingen and Chemicon, respectively.
Results
Protein Sequence Analysis and Phylogeny of hNARC-1, mNARC-1, and Rat NARC-1.
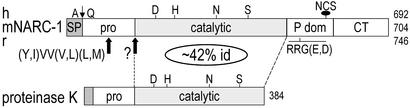
hNARC-1, mNARC-1, and rat NARC-1 (Fig. 1) catalytic domains (amino acids 143–473 in hNARC-1) share 91–92% identity. Using the pileup program (GCG), alignment of NARC-1 sequences with those of all other known subtilases (23) revealed that this enzyme is a mammalian PC member of the proteinase K subfamily of subtilases (≈42% identity in the catalytic domain of proteinase T, R, and K). NARC-1 sequences essentially differ by an N-terminal extension of the rat and mouse signal peptides (85 and 44 aa compared with 30 aa for hNARC-1) because of the presence of an upstream in-frame AUG. However, we cannot rule out that the first AUG is not the initiator, as reported for the metalloprotease nardilysin (24). Following the signal peptide, hNARC-1 exhibits a putative prosegment preceding the catalytic subunit characterized by the presence of the canonical Asp-186, His-226, and Ser-386 catalytic triad and the oxyanion hole Asn-317. The conserved subtilase active site motif Asp-X-Gly is replaced by Asp-X-Ser188 in NARC-1. The catalytic domain is followed by a putative P-domain containing a typical RRG(D,E)498 motif (5), possibly playing a role in folding (25). It is estimated to be only 100 aa long (amino acids 474–573) and ends with the sequence GCSSHWEVEDLGT573, which is followed by a β-turn (26, 27). The best pileup alignments of the catalytic and putative P-domain of hNARC-1 with those of the dibasic PCs gave ≈24% and 12% identity, respectively. The P-domain contains the only N-glycosylation site (533NCS; Fig. 1). The C-terminal domain of NARC-1 contains nine Cys residues that do not align well with the Cys-rich domain of PC5, PACE4, and furin (5).
Figure 1.
Schematic of NARC-1 and its closest family members. Their catalytic subunits share ≈42% identity (id) and their lengths in amino acids are indicated on the right. The positions of the RRG(E,D) and the single N-glycosylation site in the P domain (P dom) are emphasized along with that of the signal peptide (SP) and the primary and putative secondary (?) cleavage sites in the prosegment (pro). CT, C-terminal domain.
Biosynthesis of hNARC-1 and mNARC-1.
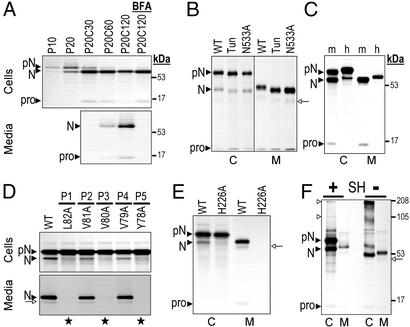
In Fig. 2A, we present a pulse–chase analysis of hNARC-1 C-terminally tagged with a V5 epitope and stably expressed in a pool of CHO cells. Initially synthesized as a 72-kDa zymogen, pro-NARC-1 is cleaved into a 63-kDa product and efficiently secreted as a 65-kDa protein. Microsequencing of the 3H-Leu-labeled 72-kDa protein revealed no Leu within the first 25 residues. Because signal peptidase cleavage was predicted to occur at the SRA30↓QEDEDGDYEELVLAL sequence, we suspected that the generated N-terminal Gln cyclized to pyroglutamine and blocked Edman degradation (28). When Gln-31 was mutated to Asn-31 we indeed obtained a Leu-11,13,15 sequence (not shown). The same sequence could be obtained for the 14-kDa protein coimmunoprecipitating with NARC-1, indicating that it is the N-terminal prosegment. The intracellular 63-kDa and secreted 65-kDa forms shared identical [3H]Thr-4 and [3H]Leu-6 sequences, indicating that they both result from processing at the YVVVL82↓ KEETHL sequence. Mutation of the single N-glycosylation site (N533A), tunicamycin treatment (Fig. 2B), or endoglycosidase F digestion (not shown) resulted in a 1.5-kDa decrease of the apparent molecular mass of pro-NARC-1 and NARC-1. However, the intracellular and secreted forms of NARC-1 still differed by ≈2 kDa, indicating that a posttranslational modification other than N-glycosylation occurred. Finally, the N533A mutation as well as tunicamycin treatment (Fig. 2B) did not affect NARC-1 processing and secretion, suggesting that different from PC1/3, PC2, and SKI-1 (6, 29), but similar to furin (30), N-glycosylation of NARC-1 is not critical for autoproteolytic processing or secretion. Biosynthetic analysis of mNARC-1 revealed a similar processing pattern (Fig. 2C). Both mouse pro-NARC-1 and mNARC-1 migrate faster than their human counterparts. This behavior seems to be a structural property of the protein, because we identified almost the same processing site, IVVLM96↓EET, shifted by only one residue (we obtained a [3H]Thr-3 sequence), and the same glycosylation pattern. Thus, the h/mNARC-1 processing site, predicted to occur at the end of a β-sheet, occurs at the consensus (Y,I)VV(V,L)(L,M)↓(K,E)E. Ala substitutions of the proposed P1, P3, or P5 residues completely abolished pro-NARC-1 processing whereas substitution of P2 or P4 residues was relatively well tolerated (Fig. 2D), suggesting that the hydrophobic residues at P1, P3, and P5 may be the most critical. A proline at P1 also inhibited hNARC-1 processing (not shown). The active site H226A NARC-1 mutant remained as pro-NARC-1 (Fig. 2E), probably in the ER because it was endoglycosidase H sensitive (not shown) and was not secreted. In addition, its coexpression with WT untagged NARC-1 did not result in significant zymogen processing (not shown). This finding indicates that NARC-1 maturation is an autocatalytic intramolecular reaction and that prosegment cleavage is necessary for its exit from the ER, in a similar fashion to the PCs (except PC2) (1, 3, 11). Like PC7 (31) and PACE4 (32), pro-NARC-1, but not NARC-1, may multimerize. Indeed, in the absence of reducing agent, the intensity of the pro-NARC-1 band decreased with a concomitant increase in higher molecular mass signals that may correspond to oligomeric species (open arrows, Fig. 2F). However, Ala substitution of the single Cys-67 in the prosegment did not affect multimerization or zymogen processing (not shown). NARC-1 oligomerization may thus depend on disulfide linkages outside the prosegment.
Figure 2.
Biosynthesis of NARC-1 and its mutants. CHO (A) or HK293 (B–F) cells stably or transiently transfected, respectively, with vectors expressing hNARC-1-V5 (A–F) or mNARC-1-V5 (C) were pulse-labeled (P) with [35S]EasyTag Express mix for the indicated time (in min), 4 h (B–E), or 2 h (F). Cell extracts (C) and media (M) were immunoprecipitated with a V5 antibody and the precipitates were resolved by SDS/PAGE on an 8% tricine (A and C–F) or glycine (B) gel. The migration positions of molecular mass standards (kDa), pro-NARC-1 (pN), NARC-1 (N), its secondary site-cleaved product (open arrow), or its pro-segment (pro) are emphasized. (A) Pulse–chase analysis in the absence or presence of brefeldin A (BFA). (B) Effect of tunicamycin (Tun) and the N-glycosylation mutant N533A on NARC-1 processing. (C) Comparison of mNARC-1 and hNARC-1 processing. (D) Processing of WT, P1, P2, P3, P4, and P5 Ala mutants; stars indicate the absence of processing. (E) Processing of the active site mutant H226A. (F) Oligomerization (open triangles) of proNARC-1 in the presence (+) or absence (−) of reducing agent (SH).
In Vitro Assays of NARC-1.
Three fluorogenic substrates were used to test the activity of NARC-1 in vitro. Boc-VVVL-MCA was based on the processing site of hNARC-1, and the commercial Suc-RPFHLLVY-MCA and shorter form Suc-LLVY-MCA substrates contain hydrophobic residues at P1 and P3. As seen in Fig. 3, analysis of the crude reaction products by matrix-assisted laser desorption ionization time-of flight showed that NARC-1 can specifically process these substrates to release 7-amino-4-methylcoumarin, which is significantly higher than in the control. The pH optimum of this reaction was found to be close to neutrality (not shown). Between 30% and 45% of the substrate Suc-RPFHLLVY-MCA was cleaved by m/hNARC-1 in 8 h whereas only 15–20% of Suc-LLVY-MCA was cleaved under identical conditions. NARC-1 is likely to be a Ca2+-dependent enzyme because its activity is inhibited by EGTA. Precise determination of the optimum Ca2+ concentration will have to await the synthesis of a sensitive fluorogenic substrate. It should be noted that incubations were done in the presence of 0.5% SDS, likely needed to release latent activity, which is significantly lower in the absence of SDS (not shown). We interpret these findings to mean that the presumed inhibitory prosegment forms a complex with NARC-1 (Fig. 2), which requires the action of SDS to dissociate it. Preincubation with carboxypeptidase Y instead of SDS did not lead to enhanced activity. We also showed that secreted h/mNARC-1 can correctly process a 19-mer peptide 72WRLPGTYVVVL↓KEETHLSQ90-amide encompassing the zymogen-processing site of hNARC-1 (not shown).
Figure 3.
In vitro fluorogenic assay of NARC-1 activity. Incubations of 10 μl of 10× concentrated media of HK293 cells expressing hNARC-1, mNARC-1, or empty vector (Ctl): (A) for 3 or 8 h with Suc-RPFHLLVY-MCA, (B) for 8 h with Suc-LLVY-MCA, or (C) for 18 h with Boc-VVVL-MCA. (Upper) The matrix-assisted laser desorption ionization time-of-flight mass spectra of the 8-h (A and B) or 18-h (C) mNARC-1 digests of each substrate. (Lower) The quantitation of released 7-amino-4-methylcoumarin fluorescence from time 0 to the indicated incubation times. Note the formation of RPHFLLVY-OH, Suc-LLVY-OH, and Boc-VVVL-OH as indicated by arrows, demonstrating cleavage N-terminal to MCA.
Ontogeny, Tissue, and Cellular Expression of NARC-1.
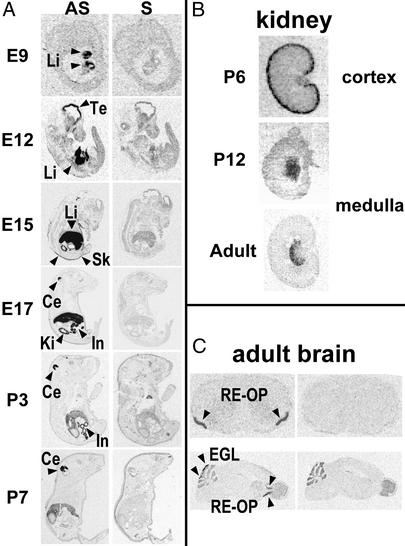
ISH in mouse embryos (Fig. 4A) revealed NARC-1 expression in the liver at E9, transient expression in the telencephalon peaking at E12, when it was also first detected in the skin, and in the kidney, small intestine and cerebellum at E15. In the kidney, labeling became undetectable in the cortex (after postpartum day 6, P6) and appeared in the medulla at P12 (Fig. 4B). Because P6 and P12 stages coincide with the differentiation of kidney mesonephros into functional metanephrons (33), a possible nephrogenic role of NARC-1 is suggested. The intensity of NARC-1 expression in the liver and small intestine decreased significantly postpartum but remained easily detectable. At E17, NARC-1 transcripts colocalized with those of albumin, indicating a major expression in hepatocytes (not shown). NARC-1 expression in the cerebellum was high up to P15 and limited to the external granular layer in adults. The other major brain expression site was the rostral extension of the olfactory peduncle (RE-OP; Fig. 4C), a location reported to contain multipotential stem cells en route to more rostral brain areas and giving rise to new olfactory neurons that are renewed throughout life (34). Northern blot analysis of 21 cell lines revealed that NARC-1 is well expressed in human neuro-epithelioma SK-N-MCIXC, colon carcinoma LoVo-C5, hepatic HepG2 and BRL-3A, pancreatic βTC3 and Rin-m5F cell lines, and Schwann cells (Fig. 5A). Notably, it is expressed only at low levels in commonly used HK293, AtT20, PC12 (Fig. 5A), CHO, COS, and 3T3 cells (not shown). Although highly expressed in 12-day embryos, NARC-1 was detected at low levels in all adult rat tissues tested, with the exception of the liver, small intestine ileum, and jejunum and to a lesser extent in the kidney, lung, spleen, and thymus (Fig. 5B). In the testis, a 2.2-kb NARC-1 mRNA was detected instead of the 2.8-kb form found in other tissues. In conclusion, the tissue and cell line distributions of NARC-1 are unique with respect to those of other convertases (6, 16) and suggest that NARC-1 may be implicated in hepatogenesis, nephrogenesis, and/or neurogenesis.
Figure 4.
ISH of NARC-1 during mouse/rat development and in adulthood. (A) ISH of mNARC-1 in mouse whole mounts at E9, E12, E15, and E17 and P3 and P7 by using antisense (AS) or control sense (S) riboprobes. (B) ISH of rNARC-1 in the rat kidney at P6 and P12 and in adulthood. (C) Coronal and sagital analysis of mNARC-1 expression in the adult brain. Li, liver; Ki, kidney; Sk, skin; In, intestine; Ce, cerebellum; Te, telencephalon; and EGL, external granular layer of the cerebellum. [Magnification: (A) E9 = ×3.8, E12 = ×2.1, E15 = ×1.2, E17 = ×0.8, P3 = ×0.7, P7 = ×0.6; (B) P6, P12 = ×2.2, adult = ×0.7; (C) ×1.3.]
Figure 5.
Cell, tissue, and post-PHx expression of NARC-1. Northern blot analysis of NARC-1 mRNA in 21 rat, mouse, and human cell lines (A) and 17 rat tissues (B) (Sm, smooth; Sk, skeletal; d, day). The open arrow points to the smaller testicular mRNA. The stars denote an independent experiment for LoVo-C5 cells. (C) mRNA expression of NARC-1, PC7, PC5, SKI-1, PACE4, and furin in intact or sham-operated rat liver compared with that in the remaining tissue after PHx, as estimated by semiquantitative ISH (17). Asterisks indicate the 4- and 2.5-fold up-regulation of PC5 and NARC-1, respectively.
Regulation of NARC-1 During Liver Regeneration After PHx.
Because NARC-1 is highly expressed in embryonic and mature livers (Figs. 4A and 5B), we tested its possible implication in rat liver regeneration that involves proliferation of hepatocytes and liver progenitor cells. After PHx (18), rapid and massive growth of the leftover lobes occurs to make up for the mass of resected lobes, a process that takes 5–10 days. We compared the expression of NARC-1 to that of furin, PACE4, PC5, PC7, and SKI-1. Semiquantitative ISH (Fig. 5C) and Northern blot (not shown) analyses revealed that NARC-1 expression was clearly up-regulated and peaked (×2.5) at 2 days post-PHx (Fig. 5C). Expression of PC5 and, to a lesser extent, PACE4 and furin peaked on day 1, whereas SKI-1 and PC7 mRNA levels did not change significantly. NARC-1 may thus play a distinct role in hepatic proliferation and/or growth.
NARC-1 Stimulates Differentiation of Cortical Neurons.
To evaluate the impact of NARC-1 in cortical neurogenesis, we examined the effects of its overexpression in a primary culture of embryonic neural progenitor cells originating from the dorsal telencephalon of E13.5 mouse embryos. Coexpression of EGFP with WT NARC-1 or its active site mutant H226A allowed the labeling of transfected cells (<5%). Western blot analysis of the transfected cells and their media (V5 immunoprecipitation followed by Western blot) shows the synthesis and processing of WT-proNARC-1, whereas the H226A mutant is neither processed nor secreted from these cells (Fig. 6A). Immunocytochemical analysis (Fig. 6B) with antibodies against the neuronal-specific marker protein NeuN (35) revealed that exogenous expression of NARC-1 led to an increase in the number of postmitotic neurons after 4 days in vitro; in parallel, there was a decrease in the number of undifferentiated neuroepithelial cells, as shown from the expression of the protein Ki-67, a marker for proliferating cells (36). In contrast, H226A-NARC-1 expression had no significant effect. These findings show that NARC-1 promotes cortical neurogenesis and suggest that this effect is the result of an increased recruitment of undifferentiated neural progenitor cells into the neuronal lineage.
Figure 6.
Stimulation of cortical neurogenesis. Primary cultures of E13.5 mouse embryonic neural progenitor cells transfected with plasmids encoding EGFP alone (CTL) or both EGFP and NARC-1 (WT) or its mutant H226A. (A) Western blot analysis before fixation of cell lysates or media previously immunoprecipitated as in Fig. 2. Note the similarity of the migration position of secreted NARC-1 from HK293 (WT-C) and primary cortical cells. pN and N are as in Fig. 2. (B) Double-labeling analysis of the expression of EGFP and either the mitotic protein Ki-67 or the neuronal NeuN cell markers. Results from four different experiments performed in duplicate were quantitated as the percentage of EGFP-positive cells that were also positive for Ki-67 or NeuN expression. Expression of NARC-1, but not its H226A mutant, caused a decrease in the number of mitotic neural progenitor cells (P < 0.005) and an increase in the number of postmitotic neurons (P < 0.01).
Discussion
Although the existence of nonbasic processing sites in precursors that transit through the secretory pathway has been recognized for many years (4, 5), the recent discovery of SKI-1 provided an example of a PC cleaving downstream of nonbasic residues (6, 7). However, because not all nonbasic sites are cleaved by SKI-1 (refs. 10 and 12; M. J. Vincent, personal communication), we mined databases for an additional PC. This process led us to the identification of an uncharacterized subtilase, NARC-1, cloned and sequenced during biopharmaceutical screenings.
In this work, we showed that hNARC-1 and mNARC-1 are synthesized as zymogens, rapidly processed in the ER. As observed for most other PCs, zymogen processing is an autocatalytic intramolecular event, and microsequence analysis has identified the cleavage site as (Y,I)VV(V,L)(L,M)↓(K,E)E, indicative of the enzymic specificity of NARC-1. Point mutations have revealed that the hydrophobic P1, P3, and P5 sites may be critical, whereas P2 and P4 would be more flexible. This site was confirmed by in vitro enzymatic assays (Fig. 3). Interestingly, database screening revealed a candidate Fugu rubripes (puffer fish) NARC-1 ortholog exhibiting a 62% sequence identity within the catalytic subunit (FuguGenscan 21252). Strikingly, the critical residues at P1, P3, and P5 for zymogen processing are completely conserved. Like SKI-1, which differs from the basic specific PCs by its P2/P4 requirement instead of P1/P4 (3), NARC-1's specificity is distinct from all known convertases. In addition, whereas SKI-1 and furin require the presence of a basic amino acid at P4, NARC-1 does not.
Data presented in Fig. 2B clearly show that the cleaved prosegment remains associated with NARC-1, even when secreted, similar to PC7 (37) and SKI-1 (9, 11). In agreement, we detected only low levels of enzyme activity in vitro, even though NARC-1 is very efficiently secreted. Activity was enhanced by exposing NARC-1 to 0.5% SDS (Fig. 3), which is thought to favor the dissociation of its putative inhibitory prosegment (3, 38, 39). The strong association of NARC-1 and its prosegment may be caused by the absence of an effective secondary maturation site, and the question of how NARC-1 becomes maximally active remains open. Even though the enzyme is secreted as a 65-kDa product, NARC-1 activity was directly measured in vitro with fluorogenic substrates (Fig. 3) and indirectly in primary cultures of neural progenitor cells (Fig. 6). Although we did not clearly identify a second processing site in either HK293, CHO, or Neuro2A cells, traces of an ≈58-kDa V5-immunoreactive protein were detected in the medium (Fig. 2 B–F). This shorter form, also observed in the absence of N-glycosylation, may represent a further N-terminally processed NARC-1. However, its scarcity has precluded its N-terminal sequencing. If autocatalytic, cleavage may occur somewhere between residues 110 and 144. Future mutagenesis should clarify this point.
Ontogeny studies (Fig. 4) showed that NARC-1 is transiently expressed in the forming telencephalon, cerebellum, and kidney cortex. In the kidney, NARC-1 expression appears within the cortex around E15 and disappears around P12, when it becomes localized to the medulla. It is tempting to speculate that NARC-1 plays a role in the development of the kidney, which is thought to originate through a series of complex interactions between epithelial and mesenchymal components of the intermediate mesoderm (40). This type of transient expression was also observed in the telencephalon, where NARC-1 expression is seen only between E12 and E15 and in the cerebellar primordium (Fig. 4A). The E12–E15 gestational period is characterized by active neurogenesis in the developing forebrain and other regions of the nervous system (41). Our results suggest that overexpression of hNARC-1 enhances the recruitment of undifferentiated neural telencephalic progenitor cells toward the neuronal lineage (Fig. 6). The precise nature of the secretory pathway substrate(s) processed by NARC-1 is not known. In the adult brain, NARC-1 expression is evident in the RE-OP (Fig. 4C), resembling the location of multipotential neurogenic progenitor cells (34). These neurons originate from the subventricular zone, migrate within the RE-OP, to eventually reach the olfactory bulb. This finding suggests that NARC-1 may participate in the differentiation and/or proliferation of these progenitor-like cells within the RE-OP.
Although NARC-1 expression is very high in developing liver hepatocytes and intestinal villi and decreases significantly in adulthood, these remain the major expressing tissues. Within the digestive system, NARC-1 expression is prominent in the ileum/jejunum between E15 and E17, just at the time when the gut tube increases in length and luminal diameter as well as in the complexity of the villous lining of this region. In the adult rat digestive system, the small intestine's ileum and jejunum are major sites of NARC-1 expression, compared with the stomach, caecum (not shown), duodenum, and colon (Fig. 5B); this tissue distribution somewhat resembles PC5 expression, which also shows maximal expression in the ileum (16). These results suggest that NARC-1 could regulate the physiology of the small intestine.
Analysis of NARC-1 expression in cell lines revealed that it is not as widely expressed as SKI-1 (6), PC7 (15), or furin (16). In fact, NARC-1 mRNA is richest in neuroepithelioma SK-N-MCIXC and in colon carcinoma LoVo-C5 (but not its derivative LoVo-D) cells. It is also found in liver-derived hepatocytes HepG2 and BRL-3A, pancreatic βTC3 and Rin-m5F, and colon HT-29 cells. Although we could detect NARC-1 by Northern blot (Fig. 5A) and quantitative RT-PCR (not shown) in many other cell lines, its expression therein is much less pronounced.
Restoration of liver mass after 2/3 PHx is controlled by the complex interplay of cytokines, growth factors, and metabolic status (42). Interestingly, the mRNA levels of PC5 and, to a lesser extent, PACE4 and furin (but not SKI-1 and PC7) seem to peak on day 1 after PHx, i.e., during the acute response phase (Fig. 5C), and likely process factors participating in the growth and/or differentiation of hepatocytes. In contrast, NARC-1 expression peaks on day 2, in a similar fashion to that of liver and intestinal apolipoprotein B (ApoB) (43). It is noteworthy that the localization of NARC-1 to human chromosome 1p33–34.3 is close to that of a major locus (FH3) for autosomal dominant hypercholesterolemia located at 1p34.1-p32 (44). This locus is associated with an increase in the hepatic secretion of low-density lipoprotein (of which ApoB is the major protein) but not high-density lipoprotein cholesterol or triglycerides (44, 45).
In conclusion, characterization of the enzymic properties and specificity of the secretory NARC-1, its ontogeny, tissue expression, and neurogenic and possibly hepatogenic properties paves the way for a molecular understanding of its biological functions. The enzymic specificity of NARC-1 suggests that its cognate substrates are likely to be very different from those of either the dibasic PCs or SKI-1. Future work on the development of specific inhibitors and gene silencing of this convertase should serve to delineate in more detail its biology and possible implication in pathologies such as familial hypercholesterolemia.
Acknowledgments
We acknowledge the technical help of A. Chen for microsequencing and PHx, M.-C. Asselin for cell cultures, A. Chamberland for Western blots and quantitative RT-PCR, and S. Basak for peptide synthesis and in vitro assays. The preparation of stable CHO transfectants by G. Siegfried, the bioinformatics assistance of J. Duhaime, and the advice of J. Cromlish and T. Reudelhuber are greatly appreciated. The secretarial work of B. Mary is greatly appreciated. This work was supported in part by Canadian Institutes of Health Research Grants GR11474 (to N.G.S. and M.C.) and MT-14766 (to N.G.S.) as well as Protein Engineering Network Centres of Excellence and Canadian Institutes of Health Research Grant MOP-42479 (to S.S.). S.S. is a Scholar of the Fonds de la Recherche en Sante du Quebec.
Abbreviations
- EGFP
enhanced GFP
- En
embryonic day
- ER
endoplasmic reticulum
- ISH
in situ hybridization histochemistry
- MCA
4-methylcoumarin-7-amide
- NARC-1
neural apoptosis-regulated convertase 1
- hNARC-1
human NARC-1
- mNARC-1
mouse NARC-1
- Pn
postpartum day
- PC
proprotein convertase
- CHO
Chinese hamster ovary
- PHx
partial hepatectomy
- RE-OP
rostral extension of the olfactory peduncle
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Steiner D F. Curr Opin Chem Biol. 1998;2:31–39. doi: 10.1016/s1367-5931(98)80033-1. [DOI] [PubMed] [Google Scholar]
- 2.Zhou A, Webb G, Zhu X, Steiner D F. J Biol Chem. 1999;274:20745–20748. doi: 10.1074/jbc.274.30.20745. [DOI] [PubMed] [Google Scholar]
- 3.Seidah N G, Chretien M. Brain Res. 1999;848:45–62. doi: 10.1016/s0006-8993(99)01909-5. [DOI] [PubMed] [Google Scholar]
- 4.Docherty K, Steiner D F. Annu Rev Physiol. 1982;44:625–638. doi: 10.1146/annurev.ph.44.030182.003205. [DOI] [PubMed] [Google Scholar]
- 5.Seidah N G, Mbikay M, Marcinkiewicz M, Chretien M. In: Proteolytic and Cellular Mechanisms in Prohormone and Neuropeptide Precursor Processing. Hook V Y, editor. Georgetown, TX: Landes; 1998. pp. 49–76. [Google Scholar]
- 6.Seidah N G, Mowla S J, Hamelin J, Mamarbachi A M, Benjannet S, Toure B B, Basak A, Munzer J S, Marcinkiewicz J, Zhong M, et al. Proc Natl Acad Sci USA. 1999;96:1321–1326. doi: 10.1073/pnas.96.4.1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sakai J, Rawson R B, Espenshade P J, Cheng D, Seegmiller A C, Goldstein J L, Brown M S. Mol Cell. 1998;2:505–514. doi: 10.1016/s1097-2765(00)80150-1. [DOI] [PubMed] [Google Scholar]
- 8.Cheng D, Espenshade P J, Slaughter C A, Jaen J C, Brown M S, Goldstein J L. J Biol Chem. 1999;274:22805–22812. doi: 10.1074/jbc.274.32.22805. [DOI] [PubMed] [Google Scholar]
- 9.Toure B B, Munzer J S, Basak A, Benjannet S, Rochemont J, Lazure C, Chretien M, Seidah N G. J Biol Chem. 2000;275:2349–2358. doi: 10.1074/jbc.275.4.2349. [DOI] [PubMed] [Google Scholar]
- 10.Ye J, Rawson R B, Komuro R, Chen X, Dave U P, Prywes R, Brown M S, Goldstein J L. Mol Cell. 2000;6:1355–1364. doi: 10.1016/s1097-2765(00)00133-7. [DOI] [PubMed] [Google Scholar]
- 11.Elagoz A, Benjannet S, Mammarbassi A, Wickham L, Seidah N G. J Biol Chem. 2002;277:11265–11275. doi: 10.1074/jbc.M109011200. [DOI] [PubMed] [Google Scholar]
- 12.Sanchez A J, Vincent M J, Nichol S T. J Virol. 2002;76:7263–7275. doi: 10.1128/JVI.76.14.7263-7275.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Benjannet S, Elagoz A, Wickham L, Mamarbachi M, Munzer J S, Basak A, Lazure C, Cromlish J A, Sisodia S, Checler F, et al. J Biol Chem. 2001;276:10879–10887. doi: 10.1074/jbc.M009899200. [DOI] [PubMed] [Google Scholar]
- 14.Basak A, Chretien M, Seidah N G. FEBS Lett. 2002;514:333–339. doi: 10.1016/s0014-5793(02)02394-3. [DOI] [PubMed] [Google Scholar]
- 15.Seidah N G, Hamelin J, Mamarbachi M, Dong W, Tardos H, Mbikay M, Chretien M, Day R. Proc Natl Acad Sci USA. 1996;93:3388–3393. doi: 10.1073/pnas.93.8.3388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Seidah N G, Chretien M, Day R. Biochimie. 1994;76:197–209. doi: 10.1016/0300-9084(94)90147-3. [DOI] [PubMed] [Google Scholar]
- 17.Marcinkiewicz M, Seidah N G. J Neurochem. 2000;75:2133–2143. doi: 10.1046/j.1471-4159.2000.0752133.x. [DOI] [PubMed] [Google Scholar]
- 18.Kountouras J, Boura P, Lygidakis N J. Hepatogastroenterology. 2001;48:556–562. [PubMed] [Google Scholar]
- 19.Ghosh A, Greenberg M E. Neuron. 1995;15:89–103. doi: 10.1016/0896-6273(95)90067-5. [DOI] [PubMed] [Google Scholar]
- 20.Slack R S, El Bizri H, Wong J, Belliveau D J, Miller F D. J Cell Biol. 1998;140:1497–1509. doi: 10.1083/jcb.140.6.1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nuthall H N, Husain J, McLarren K W, Stifani S. Mol Cell Biol. 2002;22:389–399. doi: 10.1128/MCB.22.2.389-399.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Toma J G, El Bizri H, Barnabe-Heider F, Aloyz R, Miller F D. J Neurosci. 2000;20:7648–7656. doi: 10.1523/JNEUROSCI.20-20-07648.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Siezen R J, Leunissen J A. Protein Sci. 1997;6:501–523. doi: 10.1002/pro.5560060301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hospital V, Nishi E, Klagsbrun M, Cohen P, Seidah N G, Prat A. Biochem J. 2002;367:229–238. doi: 10.1042/BJ20020822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rovere C, Luis J, Lissitzky J C, Basak A, Marvaldi J, Chretien M, Seidah N G. J Biol Chem. 1999;274:12461–12467. doi: 10.1074/jbc.274.18.12461. [DOI] [PubMed] [Google Scholar]
- 26.Lipkind G M, Zhou A, Steiner D F. Proc Natl Acad Sci USA. 1998;95:7310–7315. doi: 10.1073/pnas.95.13.7310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gluschankof P, Fuller R S. EMBO J. 1994;13:2280–2288. doi: 10.1002/j.1460-2075.1994.tb06510.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dragon N, Seidah N G, Lis M, Routhier R, Chretien M. Can J Biochem. 1977;55:666–670. doi: 10.1139/o77-096. [DOI] [PubMed] [Google Scholar]
- 29.Benjannet S, Rondeau N, Paquet L, Boudreault A, Lazure C, Chretien M, Seidah N G. Biochem J. 1993;294:735–743. doi: 10.1042/bj2940735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Creemers J W, Vey M, Schafer W, Ayoubi T A, Roebroek A J, Klenk H D, Garten W, Van de Ven W J. J Biol Chem. 1995;270:2695–2702. doi: 10.1074/jbc.270.6.2695. [DOI] [PubMed] [Google Scholar]
- 31.Creemers J W, Van de Loo J W, Plets E, Hendershot L M, Van de Ven W J. J Biol Chem. 2000;275:38842–38847. doi: 10.1074/jbc.M006758200. [DOI] [PubMed] [Google Scholar]
- 32.Nagahama M, Taniguchi T, Hashimoto E, Imamaki A, Mori K, Tsuji A, Matsuda Y. FEBS Lett. 1998;434:155–159. doi: 10.1016/s0014-5793(98)00970-3. [DOI] [PubMed] [Google Scholar]
- 33.Yan W, Wu F, Morser J, Wu Q. Proc Natl Acad Sci USA. 2000;97:8525–8529. doi: 10.1073/pnas.150149097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gritti A, Bonfanti L, Doetsch F, Cameron A, Caille I, Alvarez-Buylla A, Lim D A, Galli R, Verdugo J M, Herrera D G, et al. J Neurosci. 2002;22:437–445. doi: 10.1523/JNEUROSCI.22-02-00437.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Manitt C, Colicos M A, Thompson K M, Rousselle E, Peterson A C, Kennedy T E. J Neurosci. 2001;21:3911–3922. doi: 10.1523/JNEUROSCI.21-11-03911.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kubbutat M H, Key G, Duchrow M, Schluter C, Flad H D, Gerdes J. J Clin Pathol. 1994;47:524–528. doi: 10.1136/jcp.47.6.524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Munzer J S, Basak A, Zhong M, Mamarbachi A, Hamelin J, Savaria D, Lazure C, Hendy G N, Benjannet S, Chretien M, et al. J Biol Chem. 1997;272:19672–19681. doi: 10.1074/jbc.272.32.19672. [DOI] [PubMed] [Google Scholar]
- 38.Zhong M, Munzer J S, Basak A, Benjannet S, Mowla S J, Decroly E, Chretien M, Seidah N G. J Biol Chem. 1999;274:33913–33920. doi: 10.1074/jbc.274.48.33913. [DOI] [PubMed] [Google Scholar]
- 39.Yabuta Y, Takagi H, Inouye M, Shinde U. J Biol Chem. 2001;276:44427–44434. doi: 10.1074/jbc.M107573200. [DOI] [PubMed] [Google Scholar]
- 40.Dressler G. Trends Cell Biol. 2002;12:390–395. doi: 10.1016/s0962-8924(02)02334-6. [DOI] [PubMed] [Google Scholar]
- 41.Schuurmans C, Guillemot F. Curr Opin Neurobiol. 2002;12:26–34. doi: 10.1016/s0959-4388(02)00286-6. [DOI] [PubMed] [Google Scholar]
- 42.Fausto N. J Hepatol. 2000;32:19–31. doi: 10.1016/s0168-8278(00)80412-2. [DOI] [PubMed] [Google Scholar]
- 43.Sparks J D, Corsetti J P, Sparks C E. Metabolism. 1994;43:681–690. doi: 10.1016/0026-0495(94)90115-5. [DOI] [PubMed] [Google Scholar]
- 44.Varret M, Rabes J P, Saint-Jore B, Cenarro A, Marinoni J C, Civeira F, Devillers M, Krempf M, Coulon M, Thiart R, et al. Am J Hum Genet. 1999;64:1378–1387. doi: 10.1086/302370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hunt S C, Hopkins P N, Bulka K, McDermott M T, Thorne T L, Wardell B B, Bowen B R, Ballinger D G, Skolnick M H, Samuels M E. Arterioscler Thromb Vasc Biol. 2000;20:1089–1093. doi: 10.1161/01.atv.20.4.1089. [DOI] [PubMed] [Google Scholar]