Abstract
Signaling in the human growth hormone (hGH)–human GH receptor system is initiated by a controlled sequential two-step hormone-induced dimerization of two hGH receptors via their extracellular domains (ECDs). Little is currently known about the energetics governing the important regulatory step in receptor signaling (step 2) because of previously existing experimental barriers in characterizing the binding of the second receptor (ECD2). A further complication is that ECD2 binds through contacts from two spatially distinct sites: through its N-terminal domain to hGH, and to ECD1 through its C-terminal domain, which forms a pseudo-2-fold symmetrical interaction between the stems of the two receptors. We report here a detailed evaluation of the energetics of step 2 binding using a modified surface plasmon resonance method that is able to measure accurately the kinetics of the trimolecular binding process and separate the effects of the two binding sites. The binding kinetics of 23 single and 126 ECD1-ECD2 pair-wise alanine mutations was measured. Although both of the ECD2 binding interfaces were found to be important, the ECD1-ECD2 stem–stem contact is the stronger of the two. It was determined that most residues in the binding interfaces act in additive fashion, and that the six residues common in both ECDs contribute very differently to homodimerization depending on which ECD they reside in. This interface is characterized by a binding “hot-spot” consisting of a core of three residues in ECD1 and two in ECD2. There is no similar hot-spot in the N-terminal domain of ECD2 binding to Site2 of hGH. This study suggests ways to engineer ECD molecules that will bind specifically to either Site1 or Site2 of hGH, providing novel reagents for biophysical and biological studies.
The biological activities produced by the pituitary cytokine hormones, growth hormone (GH), prolactin (PRL), and placental lactogen, are initiated by a mechanism involving hormone-induced receptor homodimerization (1, 2). The formation of the active signaling complex is regulated by a set of extensive protein–protein interactions that have been highly coevolved between the cytokines and their cognate receptors (3, 4).
The pituitary hormones have an asymmetric four-α-helical-bundle structure that gives rise to two receptor-binding sites that have distinctly different topographies and electrostatic character (2, 5, 6). This feature plays an important role in the regulation of these systems by producing binding surfaces with dramatically different binding affinities to the receptor extracellular domains (ECD). As a consequence, the signaling complexes for systems that activate through receptor homodimerization are formed in a controlled sequential step-wise manner. The high-affinity site, Site1, is always occupied first by the receptor ECD (ECD1) (7). This step is followed by binding of ECD2 at Site2 of the hormone, which needs additional contacts to a patch of the C-terminal domain of ECD1 in the context of the hormone–ECD1 complex (2, 6, 8). Thus, the programmed regulatory step for triggering biological action involves a set of highly tuned interactions among binding interfaces in spatially distinct binding sites, which independently are weak but together produce a tight ECD2 association.
Previous structure–function analyses of the pituitary hormone systems have been performed primarily in the context of the 1:1 hormone/ECD1 intermediate complex (9–12) and have led to a detailed knowledge of the energetics of Site1 binding (11, 12). This 1:1 format provided a well-behaved and exceptionally productive system for a variety of protein engineering and biophysical studies (13–16). In particular, it provided the platform for the seminal studies of Wells and colleagues, using human GH (hGH) and human PRL (hPRL) systems, which demonstrated that the protein–protein interactions are characterized by binding “hot-spots.” These hot-spots efficiently focus binding energies within a cluster of relatively few residues (9–12), providing for a very efficient use of the binding interface. The concentration of the binding determinants within a relatively small area of the contact surface allows for separate adjacent areas to be used as specificity determinants without compromising the binding affinity.
In contrast to the extensive characterization of the 1:1 intermediate, there is considerably less biochemical and biophysical information about the energetics that govern the regulatory step of the receptor signaling defined by the binding of the second receptor. This is because the two consecutive binding steps cannot be synchronized in solution, and it is complicated to deconvolute signals corresponding to the second step only. Nevertheless, Cunningham et al. (1) had shown that substitution of individual hormone residues in the Site2 interface had generally little or only moderate effect on homodimerization. This observation implied that it is the ECD1-ECD2 stem–stem contact region that plays the major role in stabilizing the ternary complex. Two studies on the effects of stem–stem mutations on cell proliferation have been reported with somewhat different interpretations of which residues contribute most to homodimerization (17, 18). Although these biological assays provided a qualitative indication about which stem residues contribute most to homodimerization, it was not possible from the experimental design to discriminate whether a particular mutation produced an effect in context of its role in ECD1, ECD2, or both.
We present here a study that reveals the energetic contributions of the individual receptor contact residues that drive receptor homodimerization in the hGH-receptor (hGHR) system. Our goal was to determine the overall energetic contribution of the stem–stem interaction to homodimerization and to dissect how the individual residues in each of the two ECDs contribute to this process. Additionally, we wished to determine how the coupling of the two spatially distinct low affinity-binding sites lead to the high affinity of ECD2 measured in the ternary complex. Do the sites on ECD2 that bind exclusively to the hormone and the ones at the stem–stem act in additive or synergistic fashion? Are there binding “hot-spots” similar to the one found in the Site1 high-affinity interaction? These issues are fundamental to understanding how the cytokine systems fine tune their activities through modulating the persistence times of their signaling complexes (19).
To enable such measurements, we have modified a method based on surface plasmon resonance technology to measure the kinetics of ECD2 binding to the preassociated 1:1 hGH:ECD1 complex. This method overcomes previous problems associated with the accurate deconvolution of the kinetics for the two consecutive steps in the trimolecular binding process (hGH + ECD1, hGH:ECD1 + ECD2). We are also able to specifically target a chosen ECD mutation to act exclusively in the context of either ECD1 or ECD2, or both. This is an essential element of the procedure, especially in the case of measuring the energetics of the stem–stem interactions, because there are six residues that are common to both ECD1 and ECD2 in this contact interface. Additionally, of the six H bonds found in this interface, four involve this common set of six residues. Because the stem–stem interaction is not completely symmetrical, analogous residue positions in the two receptors have quite different structural environments.
We find that the soluble wild-type ECD2 binds to the hGH: ECD1 1:1 intermediate complex with a dissociation constant (Kd) of about 3.5 nM. Individual receptor Ala mutations both at the Site2-ECD2, as well as at the stem–stem ECD1-ECD2 interface, affect ECD2 binding over a wide dynamic range. This finding suggests that both of these two spatially distinct sites are important for forming and maintaining a stable ternary complex. However, residues in the stem–stem contact appear to play the more significant role in binding as compared with the ECD residues in the hGH-ECD2 Site2 interface. Interestingly, the binding hot-spots on each ECD together form a single complementary interface in the middle of the stem–stem contact. It is noteworthy that there are no common residues contained within the two hot-spots, emphasizing that binding energy contributions of individual ECD residues are highly dependent on their structural context. Using this information, mutations can be designed that will allow fine tuning of ECD2 binding to provide new insights into how the persistence time of homodimerization influences receptor signaling.
Materials and Methods
Expression vectors for recombinant hGH and hGHR ECD production were from Genentech. For the hGHR ECD, a truncated version was used (residues 29–238), which has unaltered activity and superior expression level compared with the 1–245 version (10). The proteins were expressed into the periplasm of Escherichia coli essentially as described (20). All protein variants were analyzed for purity by analytical HPLC, and the presence of the mutation was confirmed by electrospray mass spectrometry. Purity of the protein samples exceeded 95%. Protein concentration was determined by using molar extinction coefficients at 280 nm of 16.2 mM−1⋅cm−1 and 55 mM−1⋅cm−1 for hGH and hGHR ECD, respectively, and these values were adjusted appropriately for mutations (21).
Surface Plasmon Resonance.
All experiments were carried out on a Biacore 2000 instrument (Biacore, Piscataway, NJ) at 25°C. hGHR ECDs were coupled to a Biacore Pioneer C1 sensor chip by disulfide bond formation through an engineered cysteine on the S237C mutant following the Biacore protocol. Amounts of S237C were immobilized to achieve a level of 20–100 functional response units. The unreacted functional groups were blocked with a 30-μl injection of 50 mM reduced glutathione in 20 mM sodium acetate/1 M NaCl, pH 4.5 buffer.
The kinetic assays were carried out at a 50-μl⋅min−1 flow rate. Because varying the flow rate in a range of 25–100 μl⋅min−1 did not alter the measured association rates, we concluded that the binding of the analyte was not compromised by mass transfer effect. Lack of long spacer groups on the C1 chip limits direct interaction between the homogeneously immobilized ECD molecules. Therefore, injection of a saturating concentration of hGH creates a 1:1 rather then 1:2 surface, in which the immobilized ECD acts as ECD1 and binds to Site1 of hGH. This was confirmed by comparing the binding of wild-type hGH with a mutant (G120R) that can form only a 1:1 complex. Both the wild-type and variant hormones had essentially the same binding constant (Kd = 1.5 nM), indicating no measurable amount of 1:2 formation on the chip in the case of the wild-type hormone. These binding constants correspond with the published value (12). All data were normalized before analysis by subtracting data corresponding to blank (running buffer) injections (double blank subtraction).
Effects of mutations in the context of ECD2 were analyzed by using a single C1 sensor chip with hGHR ECD S237C immobilized. The same chip could then be reused to measure the interaction with different soluble ECD2 mutants. In contrast, multiple individual sensor chips were necessary for measuring the effect of mutations in the context of ECD1. This format required each of the eight ECD1 stem mutants to be immobilized on separate sensor chips, followed by injection of wild-type or mutant ECD2 molecules in separate measurements.
Data Analysis.
Association (kon) and dissociation (koff) rate constants were determined by using a modified trimolecular decaying surface model of the program clamp (22). Our model assumes a sequential reversible association–dissociation process, where the hGH/ECD1 intermediate is formed first followed by the binding of ECD2. ECD2 cannot bind to hGH only, and hGH cannot dissociate from the 1:2 complex before ECD2, i.e., neither a hGH:ECD2 nor an ECD1:ECD2 complex can exist. The kinetics of the standard trimolecular surface decay model in clamp allows independent dissociation of both ECD1 and ECD2 from the ternary complex and thus the model required modification for our use (see Supporting Materials and Methods, which is published as supporting information on the PNAS web site, www.pnas.org).
For each data set, the first hGH injection step was fit by using a simple 1:1 Langmuir model, and three parameters were determined for the hGH + ECD1-binding interaction; kon1, koff1, and the maximal response unit value. These values were then included in the mathematical description of the subsequent steps; formation and breakdown of the 1:2 complex on binding and dissociation of hGHR ECD2. Correction for the dissociation of the 1:1 hGH:ECD1 complex to hGH + ECD1 was necessary, because it occurs throughout the entire course of the experiment whenever the 1:1 complex is present due to nonsaturating levels of ECD2 binding. Three parameters were calculated by fitting the ECD2-binding curves by using the modified decaying surface model: the association rate constant, kon2; the dissociation rate constant koff2; and a response factor that reflects to the molecular weight relations of the three components. Equilibrium Kd values for the second ECD binding were determined by the mass action relation Kd = koff2/kon2.
Results and Discussion
Overall Description of the ECD2-Binding Interfaces.
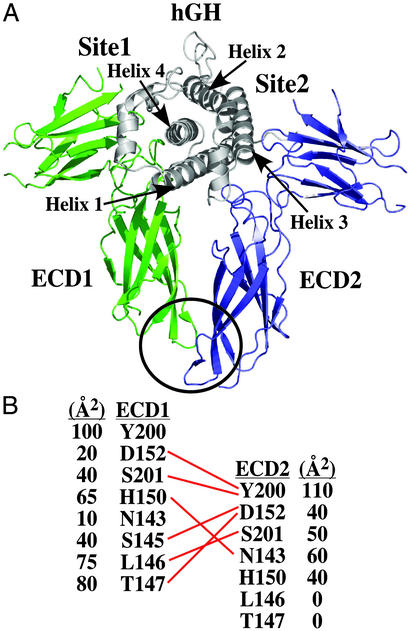
The binding of ECD2 to the 1:1 hGH:ECD1 intermediate complex involves two groups of extensive interactions with both the hormone Site2 and the C-terminal portion of ECD1 (Fig. 1). The contacts to the hormone at Site2 are primarily hydrophobic in nature, involving the burial of W104 and W169 ECD2 residues. The stem–stem ECD contact interface is comprised of eight residues from ECD1 and six residues from ECD2 (Fig. 1B). These residues are contained in the C-terminal FNIII domain (the stem region) of the hGHR ECD. In the complex, the two ECDs have a pseudosymmetric arrangement that results from an approximately 2-fold symmetry combined with an 8-Å translation along the pseudo 2-fold axis of the complex (Fig. 1). As a result, the six contact residue positions on ECD2 are also used on ECD1 and represent a large subset of the eight ECD1 contact positions. (Contact residues in ECD1 are designated by the superscript E1; those residues in ECD2, by a subscript E2).
Figure 1.
(A) Ribbon backbone representation of the (hGH:ECD1):ECD2 ternary complex. The hGH–ECD-binding sites are marked as Site1 for the high-affinity and Site2 for the low-affinity site. The ECD1/ECD2 (stem–stem) contact interface is circled. (B) H bonding pattern for residues in the stem–stem region is indicated by red lines. Buried surface areas of residues in the contact interface are given in Å2. All structure figures were generated by using PYMOL (DeLano Scientific, www.pymol.org).
The central core of the stem–stem interface consists of four residues: Ser-145E1 and Thr-147E1, D152E2 and Y200 E2, and about 20% of total buried surface area is contributed by the burial of the Y200 side chains of each ECD (Fig. 1B). There is a substantial difference in the pattern of the surface area burial comparing the two ECDs at residues L146 and T147. This difference results from the above-described 8-Å translation (Fig. 1). In ECD1, S145E1, L146E1, and T147E1 contribute an integral part of the contact interface. In ECD2, these residues are almost fully solvent exposed and are not part of the stem–stem interaction.
There are 6 H bonds between the two ECDs, with D152 and S201 contributing H bond partners in both ECDs (Fig. 1B). In ECD1, both these groups H bond to Y200E2Oη, which forms the internal core of the interface. In ECD2, their H bonding interactions are involved in the structural organization of the disordered-to-ordered transition of the 143–149 loop of ECD1.
The conformations of the polypeptide chains containing the interface groups are virtually identical in the two ECDs. Superimposing residues 140–155 and 195–201 of ECD1 with the analogous residues of ECD2 gives an rms deviation for the Cα atoms of ≈0.5 Å. It is noteworthy that the close similarities extend to the orientation of the side chains, where the positions of most atoms differ by less than 1 Å between ECDs. This finding suggests that many of the interface groups might form a somewhat rigid template to facilitate binding. It is noteworthy, however, that in the absence of the stem–stem contact, e.g., in the structure of the 1:1 hGH:ECD1 complex, the short loop containing residues 143–149 is disordered (10). It is likely that the disordered–ordered transition has evolved as part of the mechanism of association.
Energetics of the Stem–Stem Interface Residues.
A notable characteristic of the stem–stem interface is that a number of the contact residue positions contribute binding energy in the context of both ECD1 and ECD2, albeit differently. The energetic contributions of these residues in their context in both ECD1 and ECD2 were determined by alanine scanning mutagenesis. The resulting changes in binding affinities for these mutants were measured by trimolecular surface plasmon resonance (TM-SPR) (see Materials and Methods) (23).
The method is based on direct immobilization of the hGHR ECD onto a Biacore Pioneer C1 chip through an engineered Cys residue (S237C) located at the tail of its C-terminal domain. This type of coupling allows homogeneous orientation and unrestricted accessibility from the solution to the surface-immobilized ECD, whereas the very short linkers on the C1 chip minimize the possibility of direct interactions between the immobilized species. Therefore, the immobilized ECD functions as ECD1 in all of the trimolecular binding measurements. An important advantage of this format is that it controls the sequential binding steps allowing specific effects of individual contact residue mutations involving either ECD1 or ECD2 to be readily differentiated.
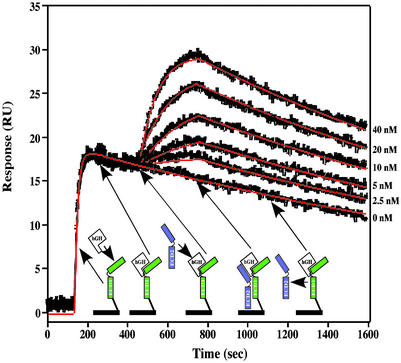
Fig. 2 shows the steps for obtaining the TM-SPR sensogram plot for the binding of wild-type ECD2 to the 1:1 hGH:ECD1 complex (hGH + ECD1, hGH:ECD1 + ECD2). Fitting of the curves by clamp (22, 23), using a modified decaying surface model (see Materials and Methods) accommodated to the special characteristics of the ternary complex formation, gave a kon = 2.5 × 105 M−1⋅s−1 association rate constant value for ECD2 binding to the 1:1 hGH:ECD1 complex. The decay of the ternary complex is described by a koff = 8.8 × 10−4⋅s−1 dissociation rate constant value. From these two values, an equilibrium Kd of 3.5 nM was calculated for the wild-type interaction. It is noteworthy that the Ala scanning mutations altered the koff values while affecting the kon values only slightly, if at all. Thus, the association constant values were quite similar to each other and to the wild-type value. Another observation is that all of the stem mutants, when used as ECD1, bound hGH with kinetic parameters identical to that of wild-type ECD1 (data not shown). This observation indicates that the stem mutants are properly folded and that Site1 binding and the stem–stem interaction are not functionally coupled.
Figure 2.
Determination of the kinetic parameters for ECD2 binding by trimolecular SPR. The surface-immobilized receptor functions as ECD1 in this format and binds to Site1 of hGH. In step 1, a 250 nM solution of wild-type hGH is injected into the flow cell and allowed to saturate the immobilized ECD, forming 1:1 complexes. Then serial dilutions (0, 5, 10, 20, and 40 nM) of soluble ECD (ECD2) are injected. The soluble ECD flows over the preformed 1:1 complex and associates with it through both receptor–hGH Site2 and receptor–receptor interactions to form a 1:2 complex. The resulting sensogram clearly shows the creation of 1:1 complexes and the subsequent formation and dissociation of stoichiometric 1:2 complexes. Data from the serial dilutions are combined and used simultaneously for fitting the kinetic parameters.
Ala Scan of the ECD1 Contact Residues.
The Ala-scanning mutagenesis on the ECD1 contact surface indicates that the binding energetics are dominated by three residues: S145E1A (≈70-fold decrease in binding), T147E1A (≈40-fold decrease), and Y200E1A (≈20-fold decrease) (Table 1, Fig. 3). Because the hydroxyl group of Tyr-200E1 forms no H bonds, the observed effect of the Y200E1A mutation is likely due to the loss of substantial van der Waals energy from the hydrophobic packing of the buried side chain (100 Å2). To test this assumption, we constructed the Y200E1F mutant, which was expected to participate in an analogous packing interaction. Indeed, the results for this mutant show only a ≈5-fold reduction in stem–stem binding affinity (Table 1).
Table 1.
Normalized Kd values for pairs of wild-type and/or single mutant ECDs in the stem–stem interface
| ECD2 | WT | N143A | S145A | L146A | T147A | H150A | D152A | Y200A | Y200F | S201A |
|---|---|---|---|---|---|---|---|---|---|---|
| WT | 1.0 | 1.4 | 71 | 2.9 | 40 | 6 | 6 | 20 | 4.6 | 0.4 |
| N143A | 0.1 | 0.3 | 2.9 | 0.4 | 7 | 8 | 0.2 | 2.6 | 0.6 | 0.4 |
| S145A | 0.9 | 2.1 | 103 | 3.7 | 91 | 5 | 7 | 48 | 69 | 2.6 |
| L146A | 0.9 | 1.3 | 57 | 2.7 | 56 | 7 | 6 | 29 | 8 | 0.7 |
| T147A | 1.1 | 2.1 | 71 | 4.0 | 60 | 10 | 9 | 63 | >285 | 1.3 |
| H150A | 1.9 | 8 | >285 | >285 | 137 | >285 | >285 | 80 | >285 | 9 |
| D152A | >285 | >285 | >285 | >285 | >285 | >285 | >285 | >285 | >285 | >285 |
| Y200A | >285 | >285 | >285 | >285 | >285 | >285 | >285 | >285 | >285 | >285 |
| S201A | 2.6 | >285 | 74 | >285 | >143 | >285 | >285 | >143 | 10 | 3.1 |
Equilibrium Kd (nM) values calculated from the ratios of the measured kon and koff rate constant values. The values are normalized to the Kd of the wild-type interaction (3.5 nM) Kdmut/Kdwt. ECD1 mutants are immobilized on the sensor chip. Mutants having values indicating a >285-fold decrease in binding generally had no detectable binding. ECD2 mutants are listed in the first column. ECD1 mutants are listed horizontally. Values shown in bold indicate nonadditive pair-wise interactions >1.5 kcal/mol.
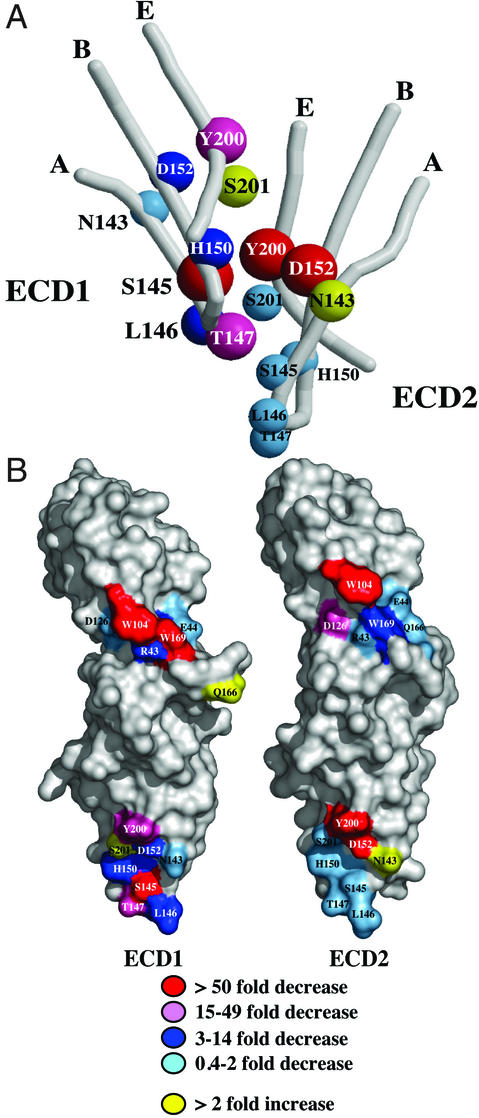
Figure 3.
(A) Structural relationship of residues in the stem–stem interface. Decreases in binding affinity due to individual wild-type-to-Ala substitutions are represented by color coding and the size of the representative spheres. (B) Space filling representation of Ala scan data for ECD1 and ECD2 binding to hGH Site1 and Site2 and the stem–stem interaction. The residues in the N-terminal domains of the ECDs (upper) represent only subsets of the corresponding contact interfaces including most, but not all, of the energetically important positions. The residues in the N-terminal domains of ECD1 and ECD2 bind to Site1 and Site2 of hGH, respectively.
The side chain hydroxyls of both S145E1 and T147E1 H bond to the D152E2 carboxylate of ECD2, and the decreased binding affinity detected for both Ala mutations is presumably partially due to the loss of their respective H bond interactions. These two hydroxyl side chains also H bond to each other that orders the sharp turn of the polypeptide chain at L146E1. The turn is further stabilized by an H bond between the peptide N of T147E1 and the hydroxyl of S145E1. There is no direct structural evidence indicating that the individual Ala mutations at residues 145E1 and 147E1 would perturb the structural organization of this region of the polypeptide chain, but the extent to which these mutations decrease stem–stem interaction suggests that some correlation may exist. The H150E1A and D152E1A show ≈5-fold reductions in binding, which might be attributed to a combination of eliminating an H bond to ECD2 and losing van der Waals contacts. The N143E1A mutant has little effect, which was anticipated because it represents only a very small buried surface area and makes no H bond to ECD2 (Fig. 1B).
Ala Scan of the ECD2 Contact Residues.
In general, the relative contributions of the eight residues that were investigated to the binding energy differ significantly in ECD2 from those determined for ECD1 (Fig. 3B). The S145E2A, L146E2A, and T147E2A mutations produced little or no decrease in binding (Table 1), which reflects the absence of direct contact with ECD1. The intra-ECD2 H bonds involving the side chains of S145E2 and T147E2 are identical to those observed in ECD1, as is the overall conformation of the tight turn involving L146E2.
Asp-152E2 and Y200E2 in ECD2 appear to play the dominant role in regulating homodimerization, based on the fact that changing either of these to an alanine results in complete loss of ECD1-ECD2 association, i.e., no second receptor binding. The side chains of both residues form two H bonds to groups in ECD1 (Fig. 1B, Table 1). The carboxylate side chain of D152E2 H bonds to the hydroxyls of S145E1 and T147E1 of ECD1. That an Ala substitution at any one of these three groups, D152E2, S145E1, and T147E1, is highly deleterious to ECD dimerization identifies this set of interactions as a binding hot-spot shared between the two ECDs. In contrast, the H bonding network involving Tyr-200E2 does not by itself appear to contribute significantly to ECD dimerization. The ECD1 D152E1A mutant eliminates an H bond to the Y200E2 hydroxyl group, which produces a 6-fold decrease in binding (see above). Surprisingly, eliminating the other H bond to the hydroxyl through the S201E1A mutation slightly increases ECD binding affinity (≈2-fold increase). These findings suggest that the total loss of binding for the Y200E2A mutant (>285-fold decrease) has more to do with the hydrophobic nature of the ring than with loss of its H bonds to D152E1 and S201E1. Removing this side chain presumably creates a substantial packing defect in the interface.
Pair-Wise Interactions Between ECD1 and ECD2 Mutants.
We applied a systematic approach to reveal inter-ECD relationships between individual ECD1 and ECD2 residues and to assess whether these act independently or in a cooperative fashion. This involved measuring the binding energy of each of the nine alanine mutants of ECD1 in conjunction with each of the eight alanine mutants of ECD2. In this format, the binding kinetics of 72 ECD1-ECD2 pair-wise combinations was evaluated (Table 1). These pair-wise ECD mutants produced a variety of effects revealing sets of energy relationships between pairs of residues in the two different ECDs.
In most cases, the sets of mutations produce reasonably additive effects (Table 1). The ECD2 alanine mutants: S145E2A, L146E2A, and T147E2A, which have binding characteristics similar to wild-type ECD2, produced little overall effects when paired with the set of ECD1 mutants. The H150E2A and S201E2A mutants in most cases produced effects somewhat larger than might be expected through strict additivity. The data suggest some cooperativity between H150E2 and ECD1 positions L146; H150; D152; Y200 and S201, as well as between S201E2 and ECD1 positions N143; L146; H150 and D152. (Pair-wise interactions showing nonadditivity relationships >1.5 kcal/mol are indicated in bold type in Table 1.) Interestingly, L146, H150, and D152 are common to both ECD sets.
The N143E2A ECD2 mutant, which showed a 14-fold increase in binding affinity to wild-type ECD1, generally enhanced binding affinity in an additive fashion when combined with the ECD1 mutants. For example, the Y200E1A single mutant binds to wild-type ECD2 with a Kd of 75 nM. The same ECD1 mutant binds with a Kd of 16 nM when paired with N143E2A, which represents a 5-fold increase in binding over the Y200E1A single mutant. A notable exception is when N143E2 is combined with H150E1A, the group that it H bonds to in the ECD interface. This combination of ECD1-ECD2 stem mutants shows no difference in affinity over the H150E1A–wild-type ECD2 pair, suggesting a nonadditive coupling relationship between the two side chains. These data could be interpreted to suggest that removing the H bond interaction between these two groups enhances affinity by allowing H150E1 to move into a more productive orientation in the interface.
Ala Scan on the Hormone-Binding Site2 of ECD2.
Evaluation of cross correlation of binding energies between ECD1 and ECD2 was extended by including mutations of ECD2 residues that interact with the hGH Site2-binding site. These ECD2-binding Site2 mutations are spatially distinct from the stem region, being located in the N-terminal FNIII domain. Six ECD2 residues that, based on the structure of the ternary complex, dominate hGH Site2 interactions were Ala scanned, and their effects on hormone Site2 binding were measured in the context of the ECD1 stem mutants (Table 2). In ECD2, W104E2 and W169E2 form extensive hydrophobic contacts with hGH Site2 interface (≈300 Å2 of total surface burial). R43E2, E44E2, and D126E2 are involved in H bonding interactions with Site2 hGH residues and provide much of the structural framework for W169E2 to contact the hormone (2). Residue Q166E2 is near the interface but does not directly participate in any Site2 interaction and is used as a control for this study.
Table 2.
Normalized Kd values for pairs of wild-type and/or single ECD1 stem mutants and ECD2 Site2-binding site mutants
| ECD2 | WT | N143A | S145A | L146A | T147A | H150A | D152A | Y200A | Y200F | S201A |
|---|---|---|---|---|---|---|---|---|---|---|
| WT | 1.0 | 1.4 | 71 | 2.9 | 40 | 6 | 6 | 20 | 4.6 | 0.4 |
| R43A | 2.6 | 0.5 | 16 | 0.7 | 40 | 1.3 | 1.3 | 9 | 1.7 | 0.5 |
| E44A | 1.0 | 1.6 | 63 | 1.9 | 37 | 3.4 | 5 | 23 | 2.9 | 0.8 |
| D126A | 40 | >100 | >285 | >100 | >285 | >100 | 24 | >285 | 29 | 3.4 |
| Q166A | 1.0 | 1.5 | 74 | 2.9 | 37 | 4.6 | 7 | 23 | 6 | 0.6 |
| W104A | >285 | >285 | >285 | >285 | >285 | >285 | >285 | >285 | >285 | >285 |
| W169A | 11 | >285 | >285 | >285 | >285 | >285 | >285 | >285 | 7 | 14 |
Values are normalized to the wild-type Kd = 3.5 nM. ECD1 mutants are immobilized on the sensor chip. ECD2 mutants are listed in the first column, whereas ECD1 mutants are listed horizontally. Values shown in bold indicate nonadditive pair-wise interactions >1.2 kcal/mol.
D126E2A shows a 40-fold decrease in binding affinity, presumably because of the disrupted structural network around the W169E2. Although the W169E2A mutation results in a 10-fold decrease in binding affinity, the W104E2A mutation results in a total loss of binding to the 1:1 hGH:ECD1 complex. This finding indicates that interactions through both tryptophans are important for effective ternary complex formation, but W104E2 plays the larger role in the Site2 interface.
In contrast to the large effect of D126E2A, the other partners in the structural network around W169E2–R43E2A and E44E2A have little or no effect on ECD2 binding (Table 2). This is surprising, because they appear to form an integral structural network between the hormone and the receptor. R43E2 makes three H bonds and provides an extensive hydrophobic surface (≈30 Å2) against which W169E2 packs. Likewise, through a salt bridge, E44E2 positions the Arg-16 side chain of the hormone to form the other side of the hydrophobic pocket for the W169E2 side chain. Interestingly, besides the fact that these mutations do not affect ECD2 binding to the wild-type 1:1 complex, there is a rather remarkable lack of effect when the measurements combine these with practically any of the ECD1 stem mutants.
The combination of D126E2A with six of the nine investigated ECD1 stem mutations suggests additive relation to these positions (Table 2). On the other hand, when this mutation is combined with either D152E1A, S201E1A, or Y200E1F, the results are nonadditive binding effects. Binding affinities corresponding to these mutant combinations are 5- to 10-fold higher than they would be in the case of an additive relation, suggesting some interdomain cooperative effects. Similar results were found for the ECD2 residue, W169E2. The dissociation constant for the W169E2A mutant binding to the wild-type 1:1 complex is 40 nM. When the W169E2A mutant is combined with the ECD1 stem mutants S145AE1, T147AE1, Y200E1A, or S201E1A, the measured effects are nearly additive. However, when this mutant is combined with either N143E1A, L146E1A, H150E1A, or D152E1A, the resulting decreases in binding affinity are somewhat greater than would be expected from an additive relation.
Conclusion
Binding energy contributions of individual residues at the hormone–receptor- and receptor–receptor-binding interfaces for ECD1 and ECD2 are color coded and summarized in Fig. 3. The information for the N-terminal domain of ECD1 was taken from ref. 9; the data used for the C-terminal domain of ECD1 and both domains of ECD2 are from this study. The figure emphasizes several important points with regard to the energetics that drive receptor homodimerization in the hGH system:
Binding Hot-Spots.
Although the stem–stem contact interfaces of ECD1 and ECD2 are nearly identical in content and conformation, significant differences exist in the size and location of their respective binding “hot-spots.” In ECD1, Ala mutations at three sites produce decreases in binding of 20-fold or greater, with two others producing 5-fold effects. In ECD2, five of the eight scanned residues caused little change in binding affinity. However, Ala mutations at two positions (D152, Y200) completely eliminate ECD2 binding. Interestingly, a mutation at one of these positions, D152, causes idiopathic Type II short stature (Laron dwarfism) (24). On the basis of our analysis and model building, we find that the D152H mutation inhibits effective homodimerization in the context of its presence in ECD2, rather than ECD1.
Conformational Adaptability of Binding Interface Residues.
The spatial organization of equivalent residues on the two ECDs involved in both the hGH Site1 binding (in ECD1) and the hGH Site2 binding (in ECD2) are very different. The pairs of equivalent residues in the two ECDs generally contribute differently to Site1 and Site2 hormone binding. One exception is W104, which plays a critical role in binding in both ECDs. On the other hand, W169 is crucial only in ECD1, whereas D126 contributes binding energy only in the context of ECD2. Such differences are not unexpected, because the Site1- and Site2-binding surfaces of the asymmetric hormone are completely unrelated. These findings will facilitate the construction of site-selective mutants that can serve only as either ECD1 or ECD2. Full-length forms of such receptor mutants would be valuable tools for in vivo experiments, allowing for ECD-selective alterations of the corresponding cytoplasmic domains.
Importance of the Stem–Stem Interaction.
Each of the domains of both receptor ECDs contributes significantly to the stability of the active ternary complex. Our results, however, suggest that the stem–stem contact interface plays a somewhat more important role compared with the hormone Site2–ECD2 contact. The stem–stem interaction seems to be rather specific, governed by multiple highly organized H bonds. The hGH Site2-ECD2 interface, on the other hand, appears to be dominated by less specific hydrophobic packing interactions. The functional significance of side-chain H bonds at this binding site was found only in case of receptor residue D126.
Binding Additivity.
With a few exceptions, there is overall additivity between the ECD1 and ECD2 stem regions. A similar situation exists between the ECD2–hormone-binding and the ECD1-ECD2 stem–stem interactions. Significant deviation from additivity in this relation was found only in case of two ECD2 positions, R43 and W169. Such long-range interdomain energy coupling could be a result of changes in the orientations of the N- and C-terminal ECD2 domains in the ternary complex due to altered stereochemistry for hormone Site2 binding in the case of the R43A and W169A mutants.
Evolutionary Considerations for Different Hot-Spot Residues in ECD1 and ECD2.
Although there is a large overlap between receptor residues used for binding to hGH Site1 or hGH Site2, only one residue, W104 was found to be crucial for both interactions. Similarly, the stem–stem contact residues also overlap, but only Y200 plays a crucial role in both ECDs. This finding may reflect on the evolutionary process that resulted in these two asymmetrical interfaces: it seems to be inherently more complicated to optimize any given residue for more than one specific function. Apparently, most of the receptor residues were optimized for binding in the context of either ECD1 or ECD2 escaping from the double requirement for specific contributions in both.
Implications of the Affinity-Enhanced N143E2A Mutant.
This mutation eliminates a stem–stem H bond yet increases binding affinity by ≈14-fold. It has been shown in cell-based assays that this effect produces a constitutively active signaling system (18). Although in solution assays this mutant does not dimerize in the absence of hormone, this finding suggests that in the environment of the membrane surface, a small enhancement in the stem–stem binding energy alone can produce a dimer with a long enough persistence time to initiate a biological response.
Supplementary Material
Acknowledgments
We acknowledge Dr. Scott Walsh for assistance in making the figures. This work was supported by National Institutes of Health Grant DK61602.
Abbreviations
- GH
growth hormone
- PRL
prolactin
- ECD
extracellular domain
- hGH
human GH
- hPRL
human PRL
- hGHR
hGH receptor
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Cunningham B C, Ultsch M, De Vos A M, Mulkerrin M G, Clauser K R, Wells J A. Science. 1991;254:821–825. doi: 10.1126/science.1948064. [DOI] [PubMed] [Google Scholar]
- 2.De Vos A M, Ultsch M, Kossiakoff A A. Science. 1992;255:306–312. doi: 10.1126/science.1549776. [DOI] [PubMed] [Google Scholar]
- 3.Yi S, Bernat B, Pal G, Kossiakoff A, Li W H. Mol Biol Evol. 2002;19:1083–1092. doi: 10.1093/oxfordjournals.molbev.a004166. [DOI] [PubMed] [Google Scholar]
- 4.Wallis M. J Mol Evol. 1994;38:619–627. doi: 10.1007/BF00175882. [DOI] [PubMed] [Google Scholar]
- 5.Somers W, Ultsch M, De Vos A M, Kossiakoff A A. Nature. 1994;372:478–481. doi: 10.1038/372478a0. [DOI] [PubMed] [Google Scholar]
- 6.Elkins P A, Christinger H W, Sandowski Y, Sakal E, Gertler A, de Vos A M, Kossiakoff A A. Nat Struct Biol. 2000;7:808–815. doi: 10.1038/79047. [DOI] [PubMed] [Google Scholar]
- 7.Fuh G, Cunningham B C, Fukunaga R, Nagata S, Goeddel D V, Wells J A. Science. 1992;256:1677–1680. doi: 10.1126/science.256.5064.1677. [DOI] [PubMed] [Google Scholar]
- 8.Kossiakoff A A, Somers W, Ultsch M, Andow K, Muller Y, de Vos A M. Protein Sci. 1994;3:1697–1705. doi: 10.1002/pro.5560031008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Clackson T, Wells J A. Science. 1995;267:383–386. doi: 10.1126/science.7529940. [DOI] [PubMed] [Google Scholar]
- 10.Clackson T, Ultsch M H, Wells J A, de Vos A M. J Mol Biol. 1998;277:1111–1128. doi: 10.1006/jmbi.1998.1669. [DOI] [PubMed] [Google Scholar]
- 11.Cunningham B C, Wells J A. Science. 1989;244:1081–1085. doi: 10.1126/science.2471267. [DOI] [PubMed] [Google Scholar]
- 12.Cunningham B C, Wells J A. J Mol Biol. 1993;234:554–563. doi: 10.1006/jmbi.1993.1611. [DOI] [PubMed] [Google Scholar]
- 13.Cunningham B C, Henner D J, Wells J A. Science. 1990;247:1461–1465. doi: 10.1126/science.247.4949.1461. [DOI] [PubMed] [Google Scholar]
- 14.Cunningham B C, Wells J A. Proc Natl Acad Sci USA. 1991;88:3407–3411. doi: 10.1073/pnas.88.8.3407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lowman H B, Bass S H, Simpson N, Wells J A. Biochemistry. 1991;30:10832–10838. doi: 10.1021/bi00109a004. [DOI] [PubMed] [Google Scholar]
- 16.Pearce K H J, Ultsch M H, Kelley R F, de Vos A M, Wells J A. Biochemistry. 1996;35:10300–10307. doi: 10.1021/bi960513b. [DOI] [PubMed] [Google Scholar]
- 17.Tsunekawa B, Wada M, Ikeda M, Banba S, Kamachi H, Tanaka E, Honjo M. J Biol Chem. 2000;275:15652–15656. doi: 10.1074/jbc.M001236200. [DOI] [PubMed] [Google Scholar]
- 18.Chen C, Brinkworth R, Waters M J. J Biol Chem. 1997;272:5133–5140. doi: 10.1074/jbc.272.8.5133. [DOI] [PubMed] [Google Scholar]
- 19.Gertler A, Grosclaude J, Strasburger C J, Nir S, Djiane J. J Biol Chem. 1996;271:24482–24491. doi: 10.1074/jbc.271.40.24482. [DOI] [PubMed] [Google Scholar]
- 20.Fuh G, Mulkerrin M G, Bass S, McFarland N, Brochier M, Bourell J H, Light D R, Wells J A. J Biol Chem. 1990;265:3111–3115. [PubMed] [Google Scholar]
- 21.Demchenko A P. Ultraviolet Spectroscopy of Proteins. Berlin: Springer; 1986. [Google Scholar]
- 22.Joss L, Morton T A, Doyle M L, Myszka D G. Anal Biochem. 1998;261:203–210. doi: 10.1006/abio.1998.2744. [DOI] [PubMed] [Google Scholar]
- 23.Morton T A, Myszka D G. Methods Enzymol. 1998;295:268–294. doi: 10.1016/s0076-6879(98)95044-3. [DOI] [PubMed] [Google Scholar]
- 24.Duquesnoy P, Sobrier M L, Duriez B, Dastot F, Buchanan C R, Savage M O, Preece M A, Craescu C T, Blouquit Y, Goossens M, et al. EMBO J. 1994;13:1386–1395. doi: 10.1002/j.1460-2075.1994.tb06392.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.