Abstract
The nuclear pore complex (NPC) is a protein assembly that contains several distinct subcomplexes. The mammalian nucleoporin (Nup)-107 is part of a hetero-oligomeric complex, that also contains Nup160, Nup133, Nup96, and the mammalian homolog of yeast Sec13p. We used transfection of HeLa cells with small interfering RNAs to specifically deplete mRNA for Nup107. In a domino effect, Nup107 depletion caused codepletion of a subset of other Nups on their protein but not on their mRNA level. Among the affected Nups was a member of the Nup107 subcomplex, Nup133, whereas two other tested members of this complex, Nup96 and Sec13, were unaffected and assembled into Nup107/Nup133-deficient NPCs. We also tested several phenylalanine-glycine repeat-containing Nups that serve as docking sites for karyopherins. Some of these, such as Nup358, Nup214 on the cytoplasmic, and Nup153 on the nucleoplasmic side of the NPC, failed to assemble into Nup107/Nup133-depleted NPCs, whereas p62, a Nup at the center of the NPC, was unaffected. Interestingly, the filamentous, NPC-associated protein Tpr also failed to assemble into the NPCs of Nup107-depleted cells. These data indicate that Nup107 functions as a keystone Nup that is required for the assembly of a subset of Nups into the NPC. Despite the depletion of Nup107 and the accompanying effects on other Nups, there was no significant effect on the growth rate of these cells and only a partial inhibition of mRNA export. These data indicate redundancy of Nups in the function of the mammalian NPC.
Keywords: nucleoporin stability‖cell viability‖mRNA export
Nuclear pore complexes (NPCs), the mediators of nucleocytoplasmic transport, are the largest protein assemblies in eukaryotic cells (60 MDa in yeast and 125 MDa in vertebrates; refs. 1–5). They are embedded in circular openings of the double nuclear envelope membrane and exhibit eightfold rotational symmetry along a nucleo-cytoplasmic axis and a pseudo twofold axis of symmetry along a perpendicular axis in the nuclear envelope membrane. Each NPC is a modular structure consisting of a central cylinder, a spoke-ring structure that anchors the cylinder to the pore membrane domain of the nuclear envelope, a cytoplasmic and nucleoplasmic ring and extensions that emanate from these rings into the cytoplasm and the nucleoplasm. On the nucleoplasmic side, these extensions are organized into the pore basket that, in turn, is connected to a filamentous intranuclear network (6–9).
A comprehensive analysis of purified yeast NPCs revealed the existence of 30 nucleoporins (Nups) that are present in multiple copies per NPC (10). With a few exceptions, most Nups show a symmetrical distribution around the twofold axis of symmetry in the plane of the nuclear envelope (10). A similar analysis with mammalian NPCs has identified 47 Nups or Nup-associated proteins (11). As in the yeast NPC, some of the mammalian Nups are localized asymmetrically at the NPC. For instance, Nup358 and Nup214 are located on the cytoplasmic side (12–14), whereas Nup153 and the NPC-associated, filamentous protein Tpr occur at or near the nucleoplasmic pore basket (15–17). By contrast, other Nups, such as p62, Nup107 and Nup133, were localized to both sides of the NPC (18–20). Nups have also been grouped into phenylalanine-glycine (FG) repeat-containing and non-FG repeat-containing Nups (2). The FG repeats serve as docking sites for karyopherins and other transport factors (21–24).
Despite these advances, it is still not possible to unequivocally assign Nups to the distinct modular substructures of the NPC. Several subcomplexes have been isolated after partial disassembly of either yeast or vertebrate NPCs under various harsh dissociating conditions (18–20, 25–28). It is not clear, however, whether these subcomplexes are physiologically relevant, because the dissociating conditions could have resulted in a redistribution of Nups. The resulting subcomplexes, therefore, may not represent intermediates in the assembly of the NPC.
A very promising approach has recently been reported by Lutzmann et al. (29), who expressed recombinant Nups in Escherichia coli with the goal to assemble subcomplexes. These data revealed that the yeast hetero-oligomeric Nup84p subcomplex that was previously obtained via dissociation of NPCs and consisted of Nup84p, Nup85p, Nup145p-C, Nup120p, Sec13p, and Seh1p (27, 28) could be assembled from recombinant proteins that were coexpressed in E. coli as dimers, trimers, and even pentamers. Nup133p was reported as another potential member of the yeast Nup84p subcomplex (20, 27). Coexpression of Nup84p and Nup133p in E. coli yielded a stable heterodimer (29). The assembled Nup84p subcomplex exhibited a Y-shaped, triskelion-like morphology (28, 29), but it is not clear how these triskelions form any of the known NPC modules.
Deletion of NUP84 is not lethal but causes clustering of NPCs and inhibition of mRNA export (27). The yeast cells, in which genes coding for individual members of the Nup84p subcomplex were deleted, showed defects in mRNA export but not in protein import (27, 30–32). However, the yeast genetic studies did not determine whether the deletion of a single member of the Nup84p subcomplex affects the integration and stability of other members of the Nup84p subcomplex.
In mammalian cells, Nup107 is the homolog of yeast Nup84p. A hetero-oligomeric Nup107 complex, whose members (Nup107, Nup133, Nup96, Nup160, and Sec13) are homologous to the members of the yeast Nup84p subcomplex, has been obtained by dissociation of NPCs (20, 25, 26). Nup107 contains a leucine zipper motif in its carboxyl-terminal region and numerous kinase consensus sites, but does not contain FG repeats (33). Fluorescence recovery after photobleaching experiments revealed that GFP-tagged Nup107 or Nup133 are tightly attached to NPCs during interphase and are exchanged only once per cell cycle (20). Moreover, immunoprecipitation and immunofluorescence analysis of GFP-tagged Nup107 and Nup133 showed that both remain associated during mitosis and are targeted at early stages to the reforming nuclear envelope (20).
Here, we show that depletion of mammalian Nup107 by RNA interference (RNAi; refs. 34–37) resulted in the failure of a subset of Nups to assemble into NPCs followed by degradation of these proteins. Although the incompletely assembled NPCs were also partially defective in mRNA export, they did not affect the growth rate of cells, indicating the existence of considerable redundancy in the function of individual Nups.
Materials and Methods
Small Interfering RNA (siRNA) Preparation and Transfection.
Specific siRNAs were designed as described by Elbashir et al. (34). We used the 21-nt sense strand (5′-GCUGCAAAAGAAGUAUUUGdTdT, coding region 2119–2138 relative to the start codon) and the 21-nt antisense strand (5′-CAAAUACUUCUUUUGCAGCdTdT) of Nup107 mRNA (GenBank accession no. NM 020401.1). The mock siRNA sequences used as control (34) were the reverse sequences of the human lamin A/C coding region 608–630 relative to the start codon (GenBank accession no. X03444). siRNA duplexes were prepared as described (34). HeLa and HeLa S3 cells were grown at 37°C/5% CO2 in DMEM (Invitrogen) supplemented with 10% (vol/vol) FBS, penicillin, and streptomycin. The day before transfection, cells at 50–80% confluency were trypsinized and diluted 1:5 with fresh medium without antibiotics. Transient transfection with siRNAs was performed by using oligofectamine (Invitrogen), as described by the manufacturer. siRNA duplexes were used at a concentration of 100 nM.
RT-PCR.
Total RNAs were extracted from HeLa cells by using an RNeasy RNA-preparation Kit (Qiagen, Chatsworth, CA). Reverse transcription and PCR were carried out by using a OneStep RT-PCR Kit (Qiagen). One microgram of template RNAs and 30 pmol of gene-specific primers were used in a 50-μl reaction. The PCR amplifications were in the linear range, as established by preliminary tests with increasing numbers of cycles, and were analyzed by agarose gel electrophoresis.
Immunoblotting.
HeLa cells were washed twice and harvested in ice-cold PBS containing protease inhibitors (Complete Mini EDTA-free, Roche Molecular Biochemicals). Cells were lysed in RIPA buffer (150 mM NaCl/50 mM Tris/HCl, pH 8.0/0.5 mM EDTA/0.5% deoxycholate/0.1% SDS/0.5% Nonidet P-40/protease inhibitors). The lysates were sonicated twice for 5 sec (Micro Ultrasonic Cell Disruptor, Kontes) and incubated for 10 min at 65°C. After centrifugation at 14,000 rpm for 15 min at 4°C (yielding no visible pellet), the supernatants were diluted with SDS/PAGE sample buffer. After determining the protein content by Bradford assay (Bio-Rad), equal amounts of proteins were separated by SDS/PAGE using 8%, 4–12%, or 4–20% Tris-glycine gels (NOVEX, San Diego). Proteins were transferred to nitrocellulose membranes (Bio-Rad), and specific proteins were detected by immunoblotting with indicated primary antibodies and peroxidase-conjugated secondary antibodies (Amersham Pharmacia) by using an enhanced chemiluminescence kit (Pierce).
Immunofluorescence Microscopy.
HeLa S3 cells, grown on coverslips, were washed twice with PBS, fixed in 2% (vol/vol) formaldehyde/PBS for 30 min at room temperature, and permeabilized with 0.2% Triton X-100/PBS for 5 min or with 0.5% saponin/PBS for 10 min. After blocking with 2% BSA/PBS for 1 h, the cells were incubated with specific antibodies diluted in 1% BSA/PBS for 1 h. Cells were washed three times with PBS and were then incubated with FITC-conjugated goat α-rabbit, FITC-conjugated donkey α-goat and CY3-conjugated donkey α-mouse IgG antibodies (Jackson ImmunoResearch) in 1% BSA/PBS for 30 min. Cells were washed three times with PBS, mounted in ProLong antifade reagent (Molecular Probes) and analyzed with a Leica TCS SP spectral confocal microscope.
In Situ Hybridization of poly(A)+ RNAs.
HeLa cells, grown on coverslips, were fixed for 8 min at room temperature in 4% (vol/vol) formaldehyde/PBS. After three washes with PBS for 15 min each, the cells were permeabilized in PBS containing 0.5% Triton X-100 for 5 min. They were then labeled for 30 min with α-Nup107 antibodies diluted in PBS containing 0.2% Triton X-100, 1 mM DTT, and 200 units per ml of RNAsin (Promega). Cells were washed three times in PBS containing 0.2% Triton X-100 for 15 min each, followed by fixation as described above. Then, cells were equilibrated in 2× SSC for 10 min at 42°C, and the coverslips were inverted onto 100 μl of the hybridization mix and incubated overnight at 42°C in a humidified chamber. The hybridization mix consisted of 2× SSC containing 1 mg/ml tRNA, 10% (wt/vol) dextran sulfate, 25% (wt/vol) formamide, and 50 μg/ml biotinylated oligo-dT(45) (synthesized by The Rockefeller University Protein and DNA Resource Center). After hybridization, the cells were washed twice with 2× SSC and once with 0.5× SSC for 15 min each at 42°C. Cells were then refixed as described above. After three washes with PBS for 15 min, cells were incubated for 30 min with CY5-streptavidine and FITC-conjugated goat α-rabbit IgG antibodies (Jackson ImmunoResearch) in 0.2% Triton X-100/PBS. After three washes in 0.2% Triton X-100/PBS and two washes in PBS, coverslips were mounted and analyzed as described above.
Antibodies.
Several antibodies were kindly provided by V. Doye of Institut Curie, Paris (α-Nup107-N and α-Nup133; ref. 20), B. Fontoura of the University of Miami, Miami (α-Sec13; personal communication), L. Gerace of The Scripps Research Institute, La Jolla, CA (α-Tpr-N; ref. 38), and D. Kraemer of the University of Würzburg, Würzburg, Germany (α-Nup214; ref. 14). The antibodies α-Nup96 (39) and α-Nup358 (12) were described. Commercial antibodies were from BAbCO (mAb414) and Santa Cruz Biotechnology (α-p62-N).
Results
Nup107 Was Efficiently Depleted by RNAi.
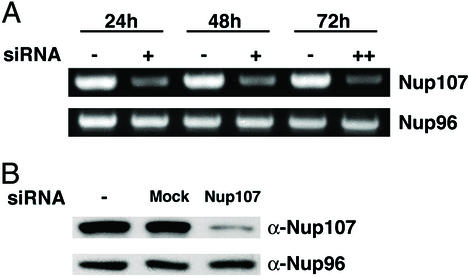
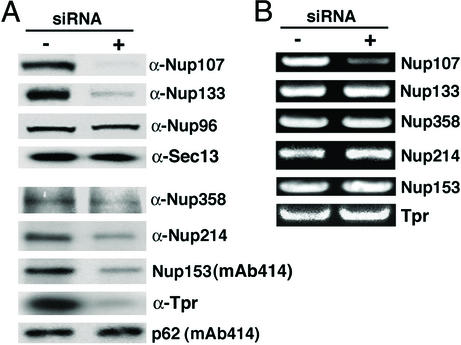
We transfected HeLa cells either with Nup107-specific siRNA duplexes or with nonspecific mock siRNAs (see Materials and Methods). The level of mRNA for Nup107 and Nup96 was monitored by RT-PCR 24, 48, and 72 h after the initial transfection. In case of the 72 h sample, cells were transfected a second time, 36 h after the first transfection. Nup107 mRNA was significantly diminished 24 and 48 h after transfection and was further reduced in the 72 h sample, whereas Nup96 mRNA was not affected, confirming the specificity of the Nup107 siRNAs (Fig. 1A).
Figure 1.
Effects of Nup107-specific RNAi on mRNA and protein levels of Nup107 and Nup96. (A) mRNA levels of Nup107 and Nup96 were determined by RT-PCR in nontransfected HeLa cells (−) or in HeLa cells 24 or 48 h after a single transfection (+) with Nup107-specific siRNAs. For the 72-h time point, a second transfection was applied 36 h after the initial transfection (++). The Nup107 mRNA level in the cells transfected with Nup107 siRNAs was significantly reduced at each time point, whereas the mRNA level of Nup96 was not affected at any time point. (B) Lysates of HeLa cells, either nontransfected (Left) or 48 h after transfection with mock (Middle) or Nup107-specific siRNAs (Right), were immunoblotted with α-Nup107 (Upper) and α-Nup96 antibodies (Lower). Cells transfected with Nup107 siRNAs showed a significant reduction of Nup107, whereas Nup96 was not affected.
To monitor the effects of RNAi on the cellular protein level, HeLa cells were lysed 48 h after transfection. Proteins of the lysate were separated by SDS/PAGE, transferred to nitrocellulose, and detected by specific antibodies. As expected, Nup107 was decreased drastically by Nup107-specific siRNAs, but was unaffected in either mock-transfected or nontransfected cells (Fig. 1B). In contrast, the level of Nup96, which is a member of the Nup107 hetero-oligomer, was not affected.
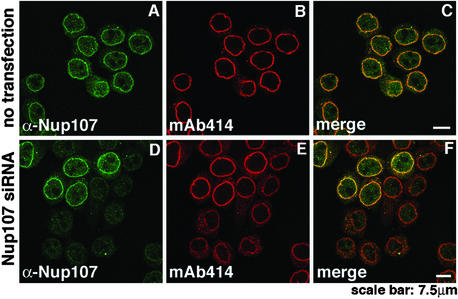
As previously shown (33), immunofluorescence microscopy of nontransfected HeLa cells with α-Nup107 antibodies exhibited nuclear rim staining that is characteristic for Nups (Fig. 2A). In agreement with the data shown in Fig. 1, the nuclear rim staining in those cells that were successfully transfected with Nup107-specific siRNAs was essentially eliminated (Fig. 2D). From images like those in Fig. 2D (and see Figs. 4–6), it was determined that 70–90% of the HeLa cells were successfully transfected by Nup107-specific siRNAs. It is not clear whether the faint, primarily intranuclear signal in the transfected cells (Fig. 2D) reflects some remaining Nup107, or whether it represents nonspecific binding of the Nup107 antibodies. It is clear, however, that most of the remaining Nup107 mRNA and protein seen in Fig. 1 is largely attributable to the 10–30% of cells that were not transfected by the Nup107-specific siRNAs. Taken together, the data of Figs. 1 and 2 strongly suggest that Nup107 was virtually eliminated in the Nup107 siRNAs-transfected cells.
Figure 2.
Immunofluorescence analysis of Nup107 depletion. HeLa S3 cells, double-labeled with α-Nup107 antibodies and mAb414, were analyzed by immunofluorescence microscopy. Nontransfected cells displayed nuclear rim staining with both antibodies (A–C). Nup107 was dramatically reduced at the nuclear rim of cells 48 h after transfection with Nup107-specific siRNAs (D and F). The five cells in the upper part of D and F displaying nuclear rim staining with α-Nup107 represented nontransfected cells. Note that the Nup107-depleted cells also exhibited reduced labeling intensity with mAb414 (E and F).
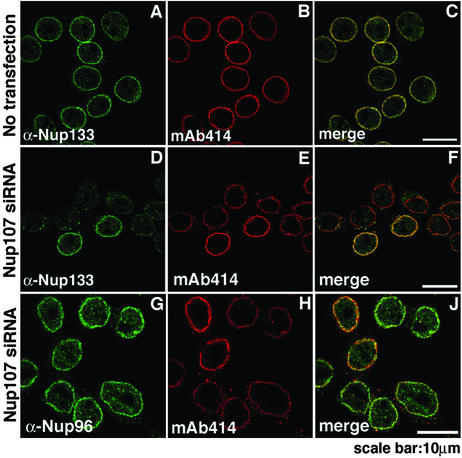
Figure 4.
Immunofluorescence analysis of other members of the Nup107 subcomplex after Nup107 depletion. HeLa S3 cells, double-labeled with indicated antibodies, were analyzed 48 h after transfection. Nontransfected cells displayed nuclear rim staining with either α-Nup133 antibodies or mAb414 (A–C). Whereas the signal of Nup133 was greatly reduced at the nuclear rim of Nup107-depleted cells (D and F), there was no change in Nup96 labeling in Nup107-depleted cells (G and J) when compared with the two nontransfected cells in H. Nup107-depleted cells showed reduced labeling with mAb414 (E and H).
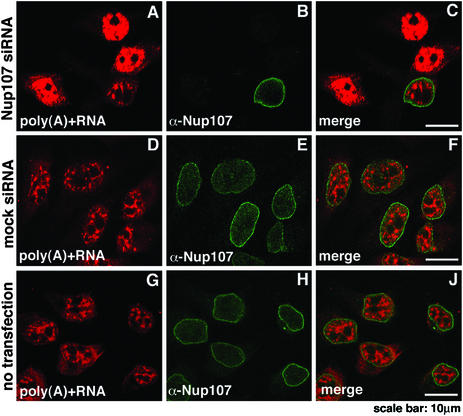
Figure 6.
Nuclear accumulation of poly(A)+ RNA in Nup107-depleted cells. Localization of poly(A)+ RNA was determined by in situ hybridization with biotinylated oligo-dT and indirect fluorescence microscopy. Nontransfected HeLa cells (G–J) and cells 72 h after double transfections with Nup107-specific (A–C) or mock siRNAs (D–F) were analyzed. The cells were immunolabeled with α-Nup107 antibodies. In comparison to mock-transfected (D–F) or nontransfected cells (G–J), the Nup107-depleted cells exhibited nuclear accumulation of poly(A)+ RNA (A–C).
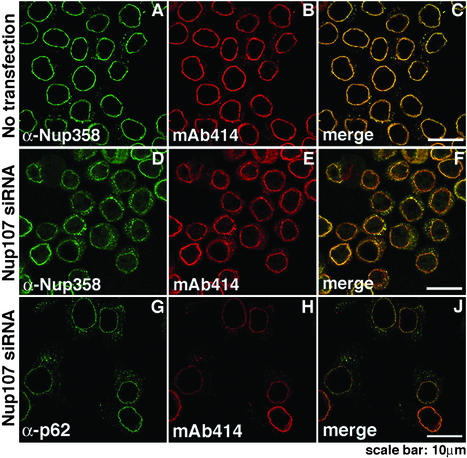
Unexpectedly, the cells transfected with Nup107–specific siRNAs exhibited reduced mAb414-labeling intensity at the nuclear rim and a shift of labeling to the cytoplasm (compare Fig. 2 B and E and the merged images in Fig. 2 C and F). mAb414 is known to interact primarily with Nup p62, but also with other FG-containing Nups, such as Nup358, Nup214, and Nup153 (40). This finding suggested that either one or several of these mAb414-reactive Nups and perhaps other Nups failed to assemble into Nup107-deficient NPCs and, therefore, were mislocalized and eliminated by degradation.
Nup107 Depletion Caused Failure of a Subset of Nups to Assemble into NPCs.
The choice of Nups that we tested as possible candidates for failure to assemble into Nup107-deficient NPCs and their subsequent elimination was largely dictated by the availability of monospecific anti-Nup antibodies. Immunoblotting of total cell lysates and immunofluorescence microscopy were carried out as described in Figs. 1B and 2, respectively.
Among the proteins that interact with Nup107 to form a hetero-oligomeric complex, Nup96 was clearly not depleted from total lysates of cells after transfection with Nup107-specific siRNAs (Figs. 1B and 3A) nor did Nup96 fail to assemble into Nup107-deficient NPCs (Fig. 4). Identical results were obtained for the mammalian homolog of yeast Sec13p (Fig. 3A, immunofluorescence data not shown). However, Nup133 was clearly codepleted with Nup107, as shown by immunoblotting of total cell lysates (Fig. 3A) and by immunofluorescence microscopy (Fig. 4). Because there was a normal level of Nup133 mRNA (Fig. 3B), these data suggested that Nup133 was synthesized but failed to assemble into Nup107-deficient NPCs and, therefore, was degraded.
Figure 3.
Effects of Nup107 depletion on protein and mRNA levels of other NPC-associated proteins. (A) Lysates of nontransfected HeLa cells (−) and of HeLa cells 48 h after transfection with Nup107 siRNAs (+) were analyzed by immunoblotting with the indicated antibodies. Nup133, Nup358, Nup214, Nup153, and Tpr were significantly reduced in the Nup107-depleted samples, whereas Nup96, Sec13, and p62 were not affected. (B) The corresponding total RNA extracts were analyzed by RT-PCR. Only mRNA for Nup107 was depleted.
Among the mAb414-reactive Nups that we tested, we found that the FG-containing Nups, Nup358, Nup214, and Nup153, were clearly codepleted, whereas the FG-containing Nup p62 was not affected (Fig. 3A). Moreover, we tested antibodies that have recently been reported to react with the NPC-anchored filamentous protein Tpr (38). Also, this protein was codepleted with Nup107 (Fig. 3A). Concomitant RT-PCR showed no reduction of mRNA levels for the proteins that were codepleted with Nup107 (Fig. 3B). The immunoblotting data of Fig. 3A were borne out by the immunofluorescence microscopy (Fig. 5). We conclude that the assembly of a subset of FG-containing Nups and of Tpr into the NPC depends on prior NPC assembly of the Nup107/Nup133 subcomplex into the NPC, and that failure to assemble into the NPC results in their subsequent degradation
Figure 5.
Immunofluorescence analysis of FG-containing Nups after Nup107 depletion. HeLa S3 cells, double-labeled with indicated antibodies, were analyzed 48 h after transfection. Compared with nontransfected cells (A–C), Nup358 labeling was reduced at the nuclear rim and increased in the cytoplasm in Nup107-depleted cells (D and F), whereas labeling with α-p62 exhibited no significant change (G and J). Nup107-depleted cells showed reduced labeling with mAb414 (E and H).
Nup107 Depletion Caused Inhibition of mRNA Export.
Nup133, Nup214, Nup153, and Tpr are known to be involved in mRNA export (26, 41–43). As these proteins were codepleted with Nup107, it was likely that mRNA export was inhibited. To test this supposition, we determined the intracellular localization of poly(A)+ RNA by in situ hybridization with biotinylated oligo-dT followed by immunofluorescence. There was an obvious retention of poly(A)+ RNA in those nuclei in which rim-staining of the Nup107 protein was significantly decreased (Fig. 6 A–C). Neither mock (Fig. 6 D–F) nor nontransfected cells (Fig. 6 G–J) exhibited an altered intracellular distribution of poly(A)+ RNA. Thus, depletion of Nup107 resulted in an inhibition of mRNA egress from the nucleus. This inhibition was clearly partial as the growth rate of cells (data not shown) was not significantly diminished.
Discussion
In this study, we successfully used Nup107-specific siRNAs to deplete Nup107 on the mRNA and protein level. In a domino effect, depletion of Nup107 interfered with the assembly of several distinct Nups into the NPC and led to their codepletion by proteolysis. mRNA export was diminished in these cells, but their growth rate was not significantly affected, suggesting considerable functional redundancy of Nups in vertebrate NPCs.
From the transfection efficiency (70–90%) and the degree of Nup107 depletion in immunoblots of total cell lysates (Figs. 1B, 3A), we estimate that Nup107 depletion was virtually complete in transfected cells 48 h after transfection with Nup107 siRNAs. This rapid and virtual complete depletion of the Nup107 pool cannot be solely attributed to dilution of the preexisting pool of Nup107 during the two to three cell doublings after siRNA transfection; it is likely accelerated by an increased rate of degradation of the protein. It is conceivable that substoichiometric quantities of Nup107 yield substoichiometric assembly of Nup107 into NPCs, which is inherently unstable and, therefore, followed by an increased rate of degradation.
The most interesting result of our studies here is the rapid codepletion of a subset of distinct Nups which accompanies the depletion of Nup107 (Figs. 3–5). We found that mRNA levels for these Nups were not affected by the Nup107-specific siRNAs, and it is likely that their rate of translation was not affected either. Hence, the codepletion of these Nups caused by the disappearance of Nup107 could only be caused by an increased rate of proteolysis of these Nups.
Among the codepleted Nups was Nup133. The yeast homologs of Nup107 and Nup133 have been shown to form a stable heterodimer when expressed together in E. coli (29); in mammalian cells, both Nup133 and Nup107 remained associated after mitotic disassembly of the NPC (20). In contrast to Nup133, two other members of the Nup107 hetero-oligomer that we tested, Nup96 and the homolog of yeast Sec13p, were not affected by Nup107 depletion. Hence, these Nups can be assembled into the NPC in the absence of the Nup107/Nup133 heterodimer preventing their degradation. This result is consistent with data showing that a partial yeast Nup84p complex, without Nup84p and Nup133p, could be assembled in E. coli by coexpression of heterodimers or heterotrimers or by in vitro assembly of separately expressed heterodimers and heterotrimers (29). Nup133p could be incorporated into this complex through its interaction with Nup84p (29).
We also tested for several peripheral Nups that contain FG repeats and that are known to serve as docking sites for karyopherins and other nuclear transport factors. We found that Nup358 and Nup214 located on the cytoplasmic site of the NPC and Nup153 on the nucleoplasmic side of the NPC were codepleted with Nup107. Also codepleted was the filamentous protein Tpr that is anchored to the NPC. These data suggest that prior stoichiometric assembly of the Nup107/Nup133 heterodimer into the NPC is required for the assembly of these peripheral proteins. Interestingly, other FG-containing Nups, such as p62 and Nup98 (data not shown), were not affected and were assembled into NPCs.
The NPCs that were assembled in the absence of Nup107 and other codepleted Nups were clearly functional, as the growth rate of these cells was not significantly affected. Hence, the observed inhibition of mRNA export (Fig. 6) must have only been partially effective.
Consistent with our results on Nup107 depletion, the deletion of the yeast homolog of Nup107, NUP84, was not lethal (27). It has not been examined whether genetic deletion of the yeast Nup84p also causes a codepletion, on the protein level, of other yeast Nups. Strikingly, individual deletion of yeast NUP84 or NUP133 resulted in clustering of NPCs, as detected by electron microscopy (27, 44). In contrast, NPCs in Nup107-depleted cells were not clustered (data not shown). The only abnormality that we could detect in these cells on the electron microscopic level was the absence of the characteristic cytoplasmic extensions of the NPC (data not shown), consistent with the depletion of Nup358 and Nup214.
The observed codepletion of a subset of distinct Nups on the protein level after degradation of mRNA for Nup107 by Nup107-specific siRNAs supports the idea that the NPC is assembled from protein subcomplexes, and that formation of these complexes and their subsequent stoichiometric assembly into the NPC protects the individual proteins from degradation. siRNA-mediated depletion of Nups and its likely domino effects on other distinct Nups will continue to serve as a valuable tool for discovering other protein–protein interactions within the NPC.
Acknowledgments
We thank Valérie Doye for Nup107 and Nup133 antibodies, Beatriz M. A. Fontoura for Nup96 and Sec13 antibodies, Larry Gerace for Tpr antibodies, Doris Kraemer for Nup214 antibodies, and Jian Wu and Elias Coutavas for Nup358 antibodies. We also thank Elias Coutavas, Beatriz M. A. Fontoura, and Aurelian Radu for helpful discussions and critical reading of the manuscript.
Abbreviations
- NPC
nuclear pore complex
- Nup
nucleoporin
- FG
phenylalanine-glycine
- RNAi
RNA interference
- siRNA
small interfering RNA
References
- 1.Allen T D, Cronshaw J M, Bagley S, Kiselewa E, Goldberg M W. J Cell Sci. 2000;113:1651–1659. doi: 10.1242/jcs.113.10.1651. [DOI] [PubMed] [Google Scholar]
- 2.Ryan K J, Wente S R. Curr Opin Cell Biol. 2000;12:361–371. doi: 10.1016/s0955-0674(00)00101-0. [DOI] [PubMed] [Google Scholar]
- 3.Rout M P, Aitchison J D. J Biol Chem. 2001;276:16593–16596. doi: 10.1074/jbc.R100015200. [DOI] [PubMed] [Google Scholar]
- 4.Vasu S K, Forbes D J. Curr Opin Cell Biol. 2001;13:363–375. doi: 10.1016/s0955-0674(00)00221-0. [DOI] [PubMed] [Google Scholar]
- 5.Yang Q, Rout M P, Akey C W. Mol Cell. 1998;1:223–234. doi: 10.1016/s1097-2765(00)80023-4. [DOI] [PubMed] [Google Scholar]
- 6.Goldberg M W, Allen T D. J Mol Biol. 1996;257:848–865. doi: 10.1006/jmbi.1996.0206. [DOI] [PubMed] [Google Scholar]
- 7.Goldberg M W, Wiese C, Allen T D, Wilson K L. J Cell Sci. 1997;110:409–420. doi: 10.1242/jcs.110.4.409. [DOI] [PubMed] [Google Scholar]
- 8.Ris H. Scanning. 1997;19:368–375. doi: 10.1002/sca.4950190504. [DOI] [PubMed] [Google Scholar]
- 9.Stoffler D, Fahrenkrog B, Aebi U. Curr Opin Cell Biol. 1999;11:391–401. doi: 10.1016/S0955-0674(99)80055-6. [DOI] [PubMed] [Google Scholar]
- 10.Rout M P, Aitchison J D, Suprapto A, Hjertaas K, Zhao Y, Chait B T. J Cell Biol. 2000;148:635–651. doi: 10.1083/jcb.148.4.635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cronshaw J M, Krutchinsky A N, Zhang W, Chait B T, Matunis M J. J Cell Biol. 2002;158:915–927. doi: 10.1083/jcb.200206106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wu J, Matunis M J, Kraemer D, Blobel G, Coutavas E. J Biol Chem. 1995;270:14209–14213. doi: 10.1074/jbc.270.23.14209. [DOI] [PubMed] [Google Scholar]
- 13.Yokoyama N, Hayashi N, Seki T, Pante N, Ohba T, Nishii K, Kuma K, Hayashida T, Miyata T, Aebi U. Nature. 1995;376:184–188. doi: 10.1038/376184a0. [DOI] [PubMed] [Google Scholar]
- 14.Kraemer D, Wozniak R W, Blobel G, Radu A. Proc Natl Acad Sci USA. 1994;91:1519–1523. doi: 10.1073/pnas.91.4.1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sukegawa J, Blobel G. Cell. 1993;72:29–38. doi: 10.1016/0092-8674(93)90047-t. [DOI] [PubMed] [Google Scholar]
- 16.Cordes V C, Reidenbach S, Kohler A, Stuurman N, van Driel R, Franke W W. J Cell Biol. 1993;123:1333–1344. doi: 10.1083/jcb.123.6.1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cordes V C, Reidenbach S, Rackwitz H R, Franke W W. J Cell Biol. 1997;136:515–529. doi: 10.1083/jcb.136.3.515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guan T, Muller S, Klier G, Pante N, Blevitt J M, Haner M, Paschal B, Aebi U, Gerace L. Mol Biol Cell. 1995;6:1591–1603. doi: 10.1091/mbc.6.11.1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hu T, Guan T, Gerace L. J Cell Biol. 1996;134:589–601. doi: 10.1083/jcb.134.3.589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Belgareh N, Rabut G, Wei Bai S, van Overbeek M, Beaudouin J, Daigle N, Zatsepina O V, Pasteau F, Labas V, Fromont-Racine M, et al. J Cell Biol. 2001;154:1147–1160. doi: 10.1083/jcb.200101081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nakielny S, Dreyfuss G. Cell. 1999;99:677–690. doi: 10.1016/s0092-8674(00)81666-9. [DOI] [PubMed] [Google Scholar]
- 22.Zolotukhin A S, Felber B K. J Virol. 1999;73:120–127. doi: 10.1128/jvi.73.1.120-127.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fontoura B M, Blobel G, Yasseen N R. J Biol Chem. 2000;275:31289–31296. doi: 10.1074/jbc.M004651200. [DOI] [PubMed] [Google Scholar]
- 24.Ben-Efraim I, Gerace L. J Cell Biol. 2001;152:411–418. doi: 10.1083/jcb.152.2.411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fontoura B M, Blobel G, Matunis M J. J Cell Biol. 1999;144:1097–1112. doi: 10.1083/jcb.144.6.1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vasu S K, Shah S, Orjalo A, Park M, Fischer W H, Forbes D J. J Cell Biol. 2001;155:339–354. doi: 10.1083/jcb.200108007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Siniossuglou S, Wimmer C, Rieger M, Doye V, Tekotte H, Weise C, Enig S, Segref A, Hurt E. Cell. 1996;84:265–275. doi: 10.1016/s0092-8674(00)80981-2. [DOI] [PubMed] [Google Scholar]
- 28.Siniossoglou S, Lutzmann M, Santos-Rosa H, Leonard K, Mueller S, Aebi U, Hurt E. J Cell Biol. 2000;149:41–54. doi: 10.1083/jcb.149.1.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lutzmann M, Kunze R, Buerer A, Aebi U, Hurt E. EMBO J. 2002;21:387–397. doi: 10.1093/emboj/21.3.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Teixeira M T, Siniossoglu S, Podtelejnikov S, Bénichou J-C, Mann M, Dujon B, Hurt E, Fabre E. EMBO J. 1997;16:5086–5097. doi: 10.1093/emboj/16.16.5086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Aitchison J D, Blobel G, Rout M P. J Cell Biol. 1995;131:1659–1675. doi: 10.1083/jcb.131.6.1659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Goldstein A L, Snay C A, Heath C V, Cole C N. Mol Biol Cell. 1996;7:917–934. doi: 10.1091/mbc.7.6.917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Radu A, Blobel G, Wozniak R W. J Cell Biol. 1994;269:17600–17605. [PubMed] [Google Scholar]
- 34.Elbashir S M, Harborth J, Lendeckel W, Yalcin A, Weber K, Tuschl T. Nature. 2001;411:494–498. doi: 10.1038/35078107. [DOI] [PubMed] [Google Scholar]
- 35.Harborth J, Elbashir S M, Bechert K, Tuschl T, Weber K. J Cell Sci. 2001;114:4557–4565. doi: 10.1242/jcs.114.24.4557. [DOI] [PubMed] [Google Scholar]
- 36.Sharp P A. Genes Dev. 2001;15:485–490. doi: 10.1101/gad.880001. [DOI] [PubMed] [Google Scholar]
- 37.Zamore P D. Nat Struct Biol. 2001;8:746–750. doi: 10.1038/nsb0901-746. [DOI] [PubMed] [Google Scholar]
- 38.Frosst P, Guan T, Subauste C, Hahn K, Gerace L. J Cell Biol. 2002;156:617–630. doi: 10.1083/jcb.200106046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Enninga J, Levy D E, Blobel G, Fontoura B M. Science. 2002;295:1523–1525. doi: 10.1126/science.1067861. [DOI] [PubMed] [Google Scholar]
- 40.Davis L I, Blobel G. Proc Natl Acad Sci USA. 1987;84:7552–7556. doi: 10.1073/pnas.84.21.7552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.van Deursen J, Boer J, Kasper L, Grosveld G. EMBO J. 1996;15:5574–5583. [PMC free article] [PubMed] [Google Scholar]
- 42.Bastos R, Lin A, Enarson M, Burke B. J Cell Biol. 1996;134:1141–1156. doi: 10.1083/jcb.134.5.1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shibata S, Matsuoka Y, Yoneda Y. Genes Cells. 2002;7:421–434. doi: 10.1046/j.1365-2443.2002.00525.x. [DOI] [PubMed] [Google Scholar]
- 44.Pemberton L F, Rout M P, Blobel G. Proc Natl Acad Sci USA. 1995;92:1187–1191. doi: 10.1073/pnas.92.4.1187. [DOI] [PMC free article] [PubMed] [Google Scholar]