Abstract
Primary cutaneous amyloidosis (PCA) is an itchy skin disorder associated with amyloid deposits in the superficial dermis. The disease is relatively common in Southeast Asia and South America. Autosomal dominant PCA has been mapped earlier to 5p13.1–q11.2 and two pathogenic missense mutations in the OSMR gene, which encodes the interleukin-6 family cytokine receptor oncostatin M receptor beta (OSMRβ), were reported. Here, we investigated 29 Taiwanese pedigrees with PCA and found that 10 had heterozygous missense mutations in OSMR: p.D647V (one family), p.P694L (six families), and p.K697T (three families). The mutation p.P694L was associated with the same haplotype in five of six families and also detected in two sporadic cases of PCA. Of the other 19 pedigrees that lacked OSMR pathology, 8 mapped to the same locus on chromosome 5, which also contains the genes for 3 other interleukin-6 family cytokine receptors, including interleukin-31 receptor A (IL31RA), which can form a heterodimeric receptor with OSMRβ through interleukin-31 signaling. In one family, we identified a point mutation in the IL31RA gene, c.1562C>T that results in a missense mutation, p.S521F, which is also sited within a fibronectin type III-like repeat domain as observed in the OSMR mutations. PCA is a genetically heterogeneous disorder but our study shows that it can be caused by mutations in two biologically associated cytokine receptor genes located on chromosome 5. The identification of OSMR and IL31RA gene pathology provides an explanation of the high prevalence of PCA in Taiwan as well as new insight into disease pathophysiology.
Keywords: primary cutaneous amyloidosis, OSMR, IL31RA, genetic heterogeneity, haplotype
Introduction
Primary cutaneous amyloidosis (PCA) is a relatively common itchy skin disorder in South America and Southeast Asia.1, 2, 3 The disease is characterized histologically by focal deposition of amyloid in the dermal papillae of lesional skin. The precise pathogenesis of PCA is unclear, but it is considered to be multifactorial, involving both genetic and environmental contributions. Earlier reports have implicated frictional epidermal damage,4 apoptosis,5 viral infection,6 and other triggers in the disease etiology.
Most PCA cases are sporadic, although a significant proportion of patients from certain geographical areas may have an autosomal dominant pattern of inheritance.7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18 Notably, Ollague et al2 reported that about one-third of the cases of PCA in South America have a positive family history. In Southeast Asia, familial PCA (FPCA) is also prevalent, especially in Chinese families, more so than in Malays and Indians.3 Thus, familial aggregation and differing racial susceptibilities suggest that genetic factors may have a significant role in the pathogenesis of FPCA.
Our group has reported 56 cases of PCA with a positive family history.19 Using a candidate gene approach, we reported earlier possible linkage to chromosome 1q23.20 Subsequently, however, we showed significant linkage of FPCA to a different locus on chromosome 5p13.1–q11.2 using a genome-wide scan with microsatellite markers.21 Further analysis, focusing on two large Taiwanese families with FPCA, identified a common haplotype shared by all affected individuals located between D5S1490 and D5S623.
Within this critical region, Arita et al22 have replicated the linkage data for chromosome 5p13.1–q11.2 in a large Brazilian family. Moreover, they identified a missense mutation (c.2072T>C; p.I691T) in all affected individuals in the OSMR gene, encoding oncostatin M-specific receptor β (OSMRβ). Additional studies in two other white families from the United Kingdom and South Africa discovered another heterozygous missense mutation (c.1853G>C; p.G618A) common to the affected individuals in both pedigrees, thus establishing OSMR as a disease gene for some cases of FPCA.22
The OSMR gene belongs to the interleukin-6 (IL-6) cytokine receptor gene family. Cytokines of the IL-6 family include IL-6, IL-11, IL-27, and IL-31, ciliary neurotrophic factor, cardiotrophin-1, cardiotrophin-like cytokine, leukemia inhibitory factor, neuropoietin, and OSM.23 IL-6-type cytokines exert their action through the signal transduction leading to the activation of the JAK/STAT, MAPK, and PI3K/Akt cascades.24 Several cytokine receptors have been reported to be structurally similar to but functionally distinct from the prototype IL-6 receptor, which consists of two gp130 and one IL-6R molecule. In the OSM type I receptor, leukemia inhibitory factor receptor (LIFR) heterodimerizes with gp130, whereas in the OSM type II receptor, OSMRβ with gp130. Furthermore, OSMRβ couples with the IL-31 receptor A subunit to constitute the IL-31 receptor. Notably, four genes encoding the IL-6-type receptor components are located in chromosome 5: LIFR (38.510–38.592 Mb), OSMR (38.881–38.970 Mb), IL31RA (55.183–55.248 Mb), and IL6ST (55.272–55.326 Mb), encoding LIFR, OSMRβ, IL-31 receptor A, and gp130, respectively.
To investigate the genetic basis of FPCA in Taiwan, we sequenced OSMR in families that mapped to 5p13.1–q11.2 and defined the genetic backgrounds in these cases that contribute to the high prevalence of PCA in Taiwan. To further characterize the possible genetic heterogeneity observed in our collection of FPCA pedigrees,16 we also used the Affymetrix GeneChip Human Mapping 10K Array to conduct a whole-genome scan on nine pedigrees that showed earlier weak positive linkage. On the basis of linkage analysis, we further investigated gene mutations in the chromosome 5 critical region and identified mutations in two genes related to the IL-6 cytokine pathway. Moreover, we have identified specific chromosome 5 haplotypes that are associated with new and recurrent mutations in the OSMR gene.
Materials and methods
Study subjects
Genomic DNA samples from 29 families showing autosomal dominant form of PCA were available for this study. Fifteen of these pedigrees were analyzed earlier in a published genome-wide linkage analysis, and a significant linkage region was mapped to chromosome 5p13.1–q12.1 in some, but not all, pedigrees.21 Of the other 14 families, 7 have at least two subjects affected with PCA and these were included for linkage replication. Taken together, the 29 multiplex families contained 73 affected individuals and 69 phenotypically normal subjects. In addition, we also collected DNA from 91 sporadic PCA subjects, including 23 singleton trio families and 68 affected individuals (Figure 1). As controls, 142 clinically unaffected subjects were recruited for this study. Each participant was examined by a dermatologist to determine the disease status of PCA. The diagnosis of PCA was made based on the typical clinical pictures of the skin lesions. For those cases whose clinical pictures were ambiguous, skin biopsies were performed to confirm the diagnosis.
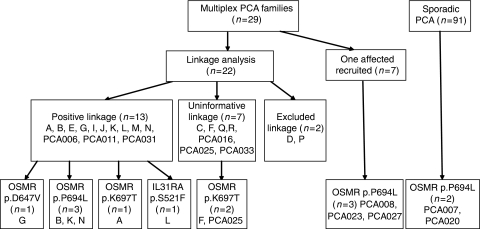
Figure 1.
Outline of subject recruitment, linkage results, and mutation findings. DNA samples were collected from affected and unaffected individuals to investigate the genetic causes of PCA.
Genotyping and linkage replication study
A 10-ml whole blood sample was taken from each individual for DNA analysis. Genomic DNA was extracted using Puregen kit (Gentra Systems, Minneapolis, MN, USA) according to the manufacturer's instructions. Seven microsatllites (D5S1993, D5S426, D5S1490, D5S418, D5S623, D5S1969, and D5S407), spanning the significant linkage region on chromosome 5, were selected for genotyping to replicate our previous linkage findings. PCR amplification, electrophoresis, and data analysis were carried out as reported earlier.21
Genotyping with the Affymetrix GeneChip Human Mapping 10K Array
As microsatellite-based genome scan set may not provide adequate marker density for certain chromosomal regions, the Affymetrix GeneChip Human Mapping 10K Array with an average density between SNPs of 0.32 c (210 kb) was also performed on nine FPCA families (B, E, G, I, J, K, L, N, P). DNA labeling, hybridization, washing, and staining of 10K SNP arrays were performed according to the manufacturer's protocol (Affymetrix Inc., Santa Clara, CA, USA). The 10 K SNP mapping data were analyzed with the Affymetrix GeneChip DNA Analysis Software (GDAS) to generate genotype calls for each of the SNP probes on the array.
Sequencing of the OSMR, IL31RA, and IL6ST genes
We sequenced the OSMR gene in 306 individuals, including 73 individuals from PCA families, 91 cases of sporadic PCA, and 142 control subjects. Direct PCR sequencing was used to examine the promoter region and all 18 exons and flanking introns of the OSMR gene, using 20 pairs of primers. As for the IL31RA gene, we sequenced 24 individuals from PCA families, 91 cases of sporadic PCA, and 142 control subjects by examining all exons and flanking introns of the IL31RA gene using 21 pairs of primers for nine splice isoforms of IL31RA. We also sequenced the IL6ST gene in 22 individuals from PCA families and two cases of sporadic PCA by using 18 pairs of primers for two splice isoforms of IL6ST (NM_002184 and NM_175767). Detailed information of primer sequences is available on request. The sequencing reactions were performed using the ABI BigDye Terminator reagents and were electrophorezed on the ABI PRISM 3730 DNA sequencer (Applied Biosystems, Foster City, CA, USA). The sequence data were analyzed by using the PolyPhred software (v5.04).25
Genotyping with the Illumina Human CNV370-Duo DNA Analysis BeadChip
Genome-wide genotyping on 75 Taiwanese subjects, including 23 PCA subjects, was performed with the Illumina Human CNV370-Duo DNA Analysis BeadChip (Illumina, San Diego, CA, USA). We also applied the same procedure to determine the genotypes for selected targets on chromosome 5 for seven individuals of one PCA family from Chile (see mutation data results below). The chip contains 318 237 haplotype tagging SNPs, with an additional 52 167 markers designed to specifically target nearly 14 000 copy number variant (CNV) regions of the genome, for a total of 370 404 markers. Genotyping was performed according to the manufacturer's protocols.
Haplotype and statistical analysis
We investigated and compared the haplotype structure for the chromosome 5 region containing the genes for four cytokine receptors (LIFR, OSMR, IL31RA, IL6ST), using genotype data of the International HapMap Project (http://www.hapmap.org).26
Two-point and multipoint linkage analysis was conducted using the MLINK and LINKMAP options of the LINKAGE packages v5.1, respectively (ftp://linkage.rockefeller.edu/software/linkage).27 Multipoint lod score, non-parametric linkage, and haplotype construction were performed by the GENEHUNTER program v2.5 (http://www.broad.mit.edu/ftp/distribution/software/genehunter/).28
The Haploview software v4. 0 (www.broad.mit.edu/personal/jcbarret/haploview/) was used for halpotype block identification, and haplotype association test.29 The GENECOUNTING program v2.1 (http://www.mrc-epid.cam.ac.uk/Personal/jinghua.zhao/software/) was used for estimating the frequencies of haplotypes in different haplotype blocks.30, 31 P-values less than 0.01 were considered to be statistically significant.
Results
Linkage replication study
In the initial genome-wide scan, we conducted genotype analysis on 15 families and identified 10 pedigrees (A, B, E, G, I, J, K, L, M, N) that showed positive linkage to the chromosome 5 markers.21 On this basis, we recruited seven additional pedigrees for linkage replication (see Figure 1 for details of pedigrees/individuals studied). Three of them (PCA006, PCA011, PCA031) gave positive lod scores, with a maximum multipoint lod score of 1.07 for the marker D5S623. The remaining four pedigrees (R, PCA016, PCA025, PCA033), however, showed no evidence of linkage because of small size of the pedigrees. Nevertheless, when we combined these seven pedigrees with the 15 pedigrees from our initial study for linkage analysis, a maximum heterogeneity lod score of 5.10 (α=0.60) was found for the marker D5S623 (Supplementary Table 1).
The Affymetrix 10K Array was also used for genome-wide linkage scan on nine families. This alternative mapping tool helped identify one family (pedigree J) that had a positive lod score of 1.81. Furthermore, additional regions giving lod>1 were detected for chromosomes 2, 11, 15, 16, and 21 with the SNP markers (Supplementary Table 2).
OSMR mutations in PCA
We applied PCR sequencing to screen exonic mutation in the OSMR gene in PCA patients. As shown in Table 1 and Figure 1, we identified three new heterozygous mutations in the OSMR gene in the Taiwanese patients. One mutation, c.2081C>T (p.P694L), was identified in six FPCA pedigrees, whereas the mutation c.2090A>C (p.K697T) was found in three families and a third mutation (c.1940A>T; p.D647V) was detected in one pedigree. Interestingly, the p.P694L mutation was also detected in two Taiwanese sporadic cases, who had reported earlier no other affected individual in their families. We also identified this mutation in a Chilean family with FPCA (unreported data). None of the 142 control subjects from Taiwan (or over 250 control chromosomes from other populations) showed presence of p.P694L or the other missense mutations. All these amino-acid substitutions occur within the fibronectin III-like domains of OSMRβ, in a similar location to the mutations reported by Arita et al.22 No mutations in OSMR were identified in the other 19/29 pedigrees, including eight pedigrees that showed linkage to the chromosome 5 interval.
Table 1. OSMR mutations in Taiwan.
| Familial PCA | Sporadic PCA | Control | |||
|---|---|---|---|---|---|
| Location | Nucleotide | Codon | n=73 | n=91 | n=142 |
| Exon 14 | c.1940A>T | p.D647V | 3 (4.1%) | 0 (0.0%) | 0 (0.0%) |
| Exon 15 | c.2081C>T | p.P694 L | 20 (27.4%) | 2 (2.2%) | 0 (0.0%) |
| Exon 15 | c.2090A>C | p.K697T | 4 (5.5%) | 0 (0.0%) | 0 (0.0%) |
Haplotype block structure of the OSMR region
To define the genetic backgrounds that are associated with the recurrent OSMR mutations in Taiwan, we conducted haplotype analysis on affected individuals from the multiplex families using the Illumina Human CNV370-Duo DNA Analysis BeadChip array. This array contains 370 404 probes and it can detect 21 068 SNP variations located on human chromosome 5. One representative case from each of the six families with the p.P694L mutation (B, K, N, PCA008, PCA023, PCA027), the three families with the p.K697T mutation (A, F, PCA025), the single family with the p.D647V mutation (G), as well as from the eight families with no detectable OSMR gene mutation (C, E, I, J, L, M, Q, PCA006) (Figure 1) was selected for genotype analysis. On the basis of haplotype block deduced from the genotype data of Taiwanese individuals, we investigated, in different populations, the haplotype block patterns of the region on chromosome 5 containing the genes for four cytokine receptors (LIFR, OSMR, IL31RA, IL6ST) (Supplementary Figure 1a and 1b). For reference, SNP genotype data from 52 other Taiwainese (non-PCA family subjects) were also collected for haplotype analysis. Using the combined genotype data of the controls and the PCA patients, 37 and 32 blocks could be identified for the Taiwanese population in the regions covering LIFR-OSMR genes (38–39.5 b) and IL31RA-IL6ST genes (54.5–56 b), respectively. Haplotype analysis showed that blocks 4 and 25 in the region of the LIFR–OSMR genes produced significant results with P-values of 0.0007, and 0.0080, respectively. Another two blocks (21 and 30) in the region of the IL31RA–IL6ST genes were also found to differ significantly between PCA cases and controls (P-values=0.0099 and 0.0022, respectively).
Haplotypes associated with OSMR mutations
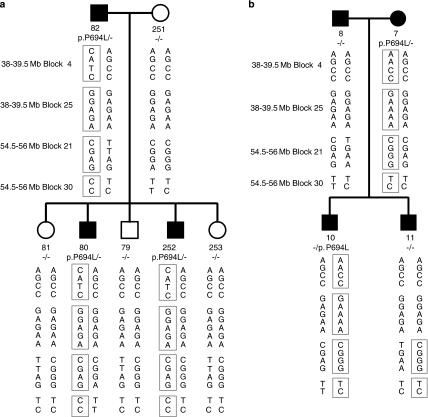
We then investigated whether the recurrent OSMR mutations in Taiwanese subjects are associated with specific chromosome 5 haplotypes. If so, this could indicate a common ancestor for all the respective families. Using the haplotype block information deduced from the genotype data, we investigated the chromosome 5 region containing the genes for four cytokine receptors (LIFR, OSMR, IL31RA, IL6ST) and looked for haplotypes that were associated with the OSMR mutations. As shown in the Table 2, we focused on four haplotype blocks, 4, 25, 21, 30, and found that the 4-AACC and 21-CGGG haplotypes were overrepresented in the group with the mutation p.K697T, whereas the 25-GAAAA and 30-CC haplotypes were overrepresented in the group with the mutation p.P694L. The association between the 25-GAAAA haplotype and the mutation p.P694L was striking; affected individuals from five of six families with the mutation p.P694L carried this haplotype. Moreover, we could confirm this association in two sporadic PCA cases (PCA007-1 and PCA020-1) that were found to have the mutation p.P694L (Figure 1): both of them had the 25-GAAAA haplotype. Taken together, our data indicate that a major proportion of PCA from Taiwan associated with the mutation p.P694L originates from the same specific chromosome background represented by the 25-GAAAA haplotype. We then went on to determine the haplotypes for members of an FPCA pedigree from Chile some of which also harbored the mutation p.P694L. As shown in Figure 2a, the affected individuals (80, 82 and 252) in this family were found to have the clinical manifestation of PCA. We found that the three Chilean individuals carrying the OSMR mutation have the same haplotypes of 4-CATC, 25-GGAGA, 21-CGAG, and 30-CC. Comparison with the Taiwanese data indicates that the Chilean FPCA has occurred on different genetic backgrounds.
Table 2. Haplotypes associated with OSMR mutations.
| Genomic region | Block number | Associated haplotype | With mutation p.P694L (n=12) | With mutation p.K697T (n=6) | With mutation p.D647V (n=2) | No detectable mutation (n=16) | Control (n=104) | P-value |
|---|---|---|---|---|---|---|---|---|
| 38–39.5 b | 4a | AACC | 3 (25.0%) | 3 (50.0%) | — | 2 (12.5%) | 4 (3.8%) | 0.001 |
| 25b | GAAAA | 5 (41.7%) | — | — | — | 2 (1.9%) | 0.00041 | |
| 54.5–56 b | 21c | CGGG | 1 (8.3%) | 2 (33.3%) | — | 3 (18.8%) | 4 (3.8%) | 0.023 |
| 30d | CC | 4 (33.3%) | — | — | 2 (12.5%) | 3 (2.9%) | 0.006 |
Defined by rs10512674, rs270592, rs10055239, and rs1494645.
Defined by rs386994, rs451298, rs1501742, rs3805558, and rs420444.
Defined by rs286007, rs11741905, rs286002, and rs158214.
Defined by rs4129542 and rs4700382.
Figure 2.
Haplotype analysis of a Chilean family and a Taiwanese family with the OSMR mutation p.P694L. (a) Three individuals of the Chilean family, including individuals 82, 80, and individual 252, have the clinical manifestation of PCA and were found to share the same haplotypes. (b) All four individuals of the Taiwanese family had clinical manifestation of PCA. However, only two (individual 7 and individual 10) were found to share the same haplotypes. In individual 11, the paternal allele was marked with 4-AGCC, 25-GGAGA, 21-TGAA, and 30-TC, and the maternal allele was determined to be a recombinant of the two alleles from the mother.
We also encountered a unique pedigree originated from Taiwan, in which both parents and their two sons were all diagnosed with PCA (Figure 2b). To trace the genetic origin(s) of PCA in the family, we conducted genotyping and sequencing to identify the mutant allele(s) associated with the disease for each patient. SNP genotyping showed that the mother (individual 7) and one affected son (individual 10) carried the prototypic haplotype of the p.P694L mutation and DNA sequence confirmed the presence of the mutation. On the other hand, the father (individual 8) and the other affected son (individual 11) did not have this OSMR mutation or the associated haplotype (or indeed any other OSMR mutation). Thus, the data indicate that two different genes were involved in the PCA etiology of this family. Interestingly, we noticed that those who have p.P694L mutation showed greater severity of PCA. Although all four patients presented typical clinical pictures, individuals 7 and 10 had larger areas of skin lesion (upper back, forearms, thighs, and lower legs) and a higher density of amyloid papules, as compared with those without the mutation (individuals 8 and 11).
IL31RA mutation in FPCA
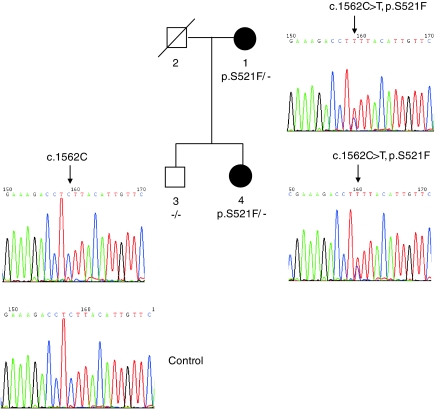
So far we have found OSMR coding mutation in 5 out of the 13 pedigrees that showed linkage to the chromosome 5 region. We also investigated the other two cytokine genes (IL31RA, IL6ST) for sequence change in the other eight families that mapped to the chromosome 5 region. No alteration was found in the coding sequence of IL6ST for these eight families. However, as indicated in Figure 3, a p.S521F mutation (c.1562C>T, codon 521 TCT>TTT) was found in the IL31RA gene in a FPCA family. The p.S521F variation was detected in the affected individual but not found in unaffected individual in Pedigree L. This variant neither exists in other PCA patients nor in 142 controls we examined, and the codon change occurs in a codon position that is well conserved in mammals (Supplementary Figure 2). No other alteration was identified in the coding sequence and exon/intron boundary except a cSNP at codon 529 (p.S529N, c.1586G>A, in dbSNP: rs161704) for this family.
Figure 3.
Genetic analysis of the IL31RA mutation p.S521F in a FPCA pedigree. Sequencing of the exon 12 of the IL31RA gene in Pedigree L revealed c.1562C>T (NM_139017) heterozygous transition in two affected individuals (#1 and #4) whereas unaffected individual (#3) did not harbor this variation.
Discussion
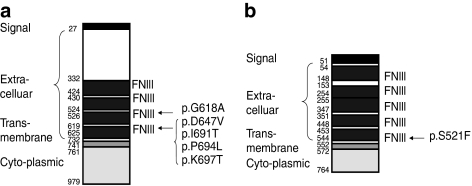
Clinically, it is evident that PCA is a common disease in Taiwan and South America. Although environmental factors may have contributed to the high PCA prevalence in these regions, we have taken a genetic approach and identified chromosomal backgrounds that are uniquely associated with the disease in Taiwan. In this study, we have identified three new mutations in the OSMR gene that underlie PCA. Moreover, we have found out that IL31RA, another gene mapped to the same chromosome and functionally related to the IL-6 cytokines, is involved in FPCA. Two recurrent mutations (p.P694L and p.K697T) are located in exon 15, whereas the other mutation, p.D647V, occurs in exon 14 of the OSMR gene. Including the published p.G618A and p.I691T mutations reported by Arita et al,22 substitutions of five different highly conserved amino-acid residues in the fibronectin III-like domains of OSMRβ have now been determined as the molecular basis of FPCA. As shown in Figure 4a and 4b, the mutations reported in this paper are all located in the fibronectin III-like domains of OSMRβ and IL-31RA. Earlier studies have indicated that these particular domains are critical for receptor dimerization for both gp130 homodimers and gp130-LIFR heterodimers.32, 33 Therefore, Arita et al22 have proposed that the amino-acid substitutions in the fibronectin type III-like domain may interfere with receptor coupling of OSMRβ with gp130 or OSMRβ with IL31RA. Another effect of the pathogenic OSMR mutations could be premature degradation of the mutant proteins leading to reduced availability of OSMRβ to interact with either gp130 or IL31RA. Although the precise cellular mechanism of PCA pathogenesis remains to be established, Arita et al22 have shown, in cultured keratinocytes from FPCA patients, decreased phosphorylation of STATs, Erk1/2, and Akt after OSM stimulation, and no phosphorylation after IL-31 stimulation. Thus, mutations involving members of the IL-6 receptor gene family can cause dysfunction of the downstream signals and lead to the development of cutaneous diseases, here manifesting as FPCA.
Figure 4.
Diagrammatic representation of OSMRβ (a) and IL-31RA (b), showing the functional domains of the encoded proteins and the positions of mutated codons. Amino-acid numbers are shown to the left of the diagram and the sites of the missense mutations to the right. The p.G618A and p.I691T mutations were reported earlier by Arita et al. The figure of IL-31RA is based on the amino-acid sequence of isoform 2. FNIII, fibronectin type III-like domain. Adapted from http://www.uniprot.org/uniprot/Q99650 (OSMRβ) and http://www.expasy.org/uniprot/q8ni17 (IL-31RA).
Data from the linkage analyses have provided new evidence that FPCA is genetically heterogeneous. As well as other putative genes within the region of linkage on chromosome 5 being possible candidate genes in some pedigrees (including other IL-6 family cytokine receptors), other undisclosed loci may be relevant to disease pathogenesis. Of note, two families (D and P) gave negative lod scores, and as such the FPCA in these cases is likely to be caused by genes outside the chromosome 5 region. Consistent with our knowledge of the complex signaling pathways elicited by the IL-6-related cytokines (IL-6, IL-31, and OSM) and the now-proven genetic heterogeneity, the pedigree analyzed in Figure 2b illustrates that FPCA can be traced to different genetic causes even within the same family.
In this study, we have shown that the recurrent OSMR mutation p.P694L is present on similar chromosomal backgrounds (haplotypes) in several Taiwanese families with FPCA. We have also analyzed the OSMR sequence in 91 sporadic PCA and identified 2 cases (PCA007 and PCA020) with the same mutation and the same haplotype. It is likely, therefore, that these two subjects and those in five of the six families with this mutation share a common ancestor. The observation that six Taiwanese families and a family from Chile both had this same mutation, but occurring on three different genetic backgrounds, suggests that the mutation p.P694L is both an ancestral and a recurrent mutation. The mutation p.P694L is the most frequent mutation underlying FPCA and is the only one that has been observed in two different ethnic groups. This may be partly explained by the sequence change (CCG>CTG) occurring at a CpG dinucleotide, a hotspot for mutagenesis.
By taking a positional candidate approach, two genes (OSMR and IL31RA) of the IL-6 cytokine pathway are now recognized to be associated with FPCA. It remains to be determined how the signaling changes induced by OSMR and IL31RA mutations led to the PCA phenotypes. Severe itching is a hallmark of PCA19 and prolonged scratching might induce apoptosis and lead to PCA.5 It has been shown that normal human epidermal keratinocyte cell lines expressed gp130, IL-31RA, and OSMRβ, and responded to OSM and IL-31 activation.34, 35 In skin, IL-31 induces severe pruritus and dermatitis in transgenic mice.34 High IL-31 levels were detected in prurigo nodularis, one of the most pruritic forms of chronic skin inflammation.36 Moreover, dorsal root ganglion expressed very high level of IL-31RA mRNA36 and OSMRβ has been detected on a specific subset of nociceptive neurons in dorsal root ganglion,37, 38 suggesting that IL-31 might induce pruritus by directly modulating the function of sensory neurons.
The genetic heterogeneity for PCA presented by mutations in two physically adjacent and functionally related genes is intriguing. OSMRβ is a common component of both the OSM type II receptor and the IL-31 receptor. It will be of interest to dissect the signaling pathways mediated by the OSM and IL-31 receptors under different cultured conditions to investigate the molecular mechanism(s) leading to PCA pathogenesis. It is known that the two cytokine receptors mediate distinct signaling reactions and response patterns in lung epithelial cells.39 IL-31 was significantly over-expressed in skin samples from patients with atopic dermatitis but not with psoriasis.36 On the contrary, expression of both OSM type II receptor and OSM were enhanced in both psoriatic and atopic dermatitic lesions, two common inflammatory skin disorders.35 Mainly secreted by activated T cells, monocytes, and dendritic cells, OSM is also a potent inducer of keratinocyte migration and triggers hyperplasia of the reconstituted human epidermis.35 Although PCA itself is not considered as an inflammatory skin disease, further investigation of the OSMRβ-related signaling pathways might lead to better understanding of the pathogenesis and to the identification of new therapeutic targets for PCA.
In summary, the genetic data collected from this investigation clearly support multi-origins of OSMR mutations in different populations, and we conclude that haplotype comparison between PCA families in Taiwan, a region of high prevalence, offers a case for studying the evolution and dissemination of a mutant allele in this population. In view of cytokine-receptors-mediated signaling in the skin, our finding on the frequent OSMR mutations and the discovery of IL31RA mutation in PCA patients might shed light on future study of disease pathophysiology. New knowledge about the abnormal cellular signaling in PCA is a key to developing novel therapeutics for this relatively common skin disease for our patients.
Acknowledgments
We thank Dr Tsung-Sheng Su for critical reading of the manuscript, the patients, and members of the PCA families who participated in this study, Ms Li-Ru Chen, Ms Ying-Chen Chang, and Ms Ying-Yen Weng for excellent technical support, and the staff of the Department of Dermatology, Taipei Veterans General Hospital, for referral of PCA subjects. This work was supported in part by grants VGH 93-354, VGHUST94-P1-14, NSC94-3112-B-010-019, NSC95-3112-B-010-001, NSC96-3112-B-010-001, NSC97-2314-B-010-016-MY2, and a grant from The Ministry of Education, Taiwan, Aim for the Top University Plan. The linkage analysis server was supported by grants VGH 94-368-2, V95S2-003, and V96S2-005. The sequencing services and Affymetrix 10K SNP assays were provided by the Sequencing Core Facility and Gene Expression Core Facility of the National Research Program for Genomic Medicine supported by grants from the National Science Council, Taiwan, respectively. Studies on PCA in the UK were kindly supported by a grant from Action Medical Research.
Footnotes
Supplementary Information accompanies the paper on European Journal of Human Genetics website (http://www.nature.com/ejhg)
Supplementary Material
References
- Wong CK. Lichen amyloidosus: a relatively common skin disorder in Taiwan. Arch Dermatol. 1974;110:438–440. doi: 10.1001/archderm.110.3.438. [DOI] [PubMed] [Google Scholar]
- Ollague W, Ollague J, Ferretti H. Epidemiology of primary cutaneous amyloidoses in South America. Clin Dermatol. 1990;8:25–29. doi: 10.1016/0738-081x(90)90084-e. [DOI] [PubMed] [Google Scholar]
- Tan T. Epidemiology of primary cutaneous amyloidoses in Southeast Asia. Clin Dermatol. 1990;8:20–24. doi: 10.1016/0738-081x(90)90083-d. [DOI] [PubMed] [Google Scholar]
- Wong CK, Lin CS. Friction amyloidosis. Int J Dermatol. 1988;27:302–307. doi: 10.1111/j.1365-4362.1988.tb02357.x. [DOI] [PubMed] [Google Scholar]
- Chang YT, Wong CK, Chow KC, Tsai CH. Apoptosis in primary cutaneous amyloidosis. Br J Dermatol. 1999;40:210–215. doi: 10.1111/j.1365-2133.1999.02651.x. [DOI] [PubMed] [Google Scholar]
- Chang YT, Liu HN, Wong CK, Chow KC, Chen KY. Detection of Epstein-Barr virus in primary cutaneous amyloidosis. Br J Dermatol. 1997;136:823–826. [PubMed] [Google Scholar]
- Porto JA, Posse FA. Amiloidose cutanea genuina familial. Bol Da Soc Brasil Dermatol E Sif. 1960;35:102–103. [Google Scholar]
- de Souza AR. Amiloidose cutanea bohlosa familial. Observacao de 4 casos. Rev Hosp Clin Fac Med S Paolo. 1963;18:413–417. [PubMed] [Google Scholar]
- Sagher F, Shanon J. Amyloidosis cutis. Arch Dermatol. 1963;87:171–175. doi: 10.1001/archderm.1963.01590140033005. [DOI] [PubMed] [Google Scholar]
- Tay CH. Genodermatoses in Singapore – a genetic study of certain skin diseases. Asian J Med. 1971;7:413–422. [Google Scholar]
- Rajagopalan K, Tay CH. Familial lichen amyloidosis. Report of 19 cases in 4 generations of a Chinese family in Malaysia. Br J Dermatol. 1972;87:123–129. doi: 10.1111/j.1365-2133.1972.tb16186.x. [DOI] [PubMed] [Google Scholar]
- Vasily DB, Bhatia SG, Uhlin SR. Familial primary cutaneous amyloidosis. Arch Dermatol. 1978;114:1173–1176. [PubMed] [Google Scholar]
- de Pietro WP. Primary familial cutaneous amyloidosis. Arch Dermatol. 1981;117:639–642. doi: 10.1001/archderm.117.10.639. [DOI] [PubMed] [Google Scholar]
- Ozaki M. Familial lichen amyloidosis. Int J Dermatol. 1984;23:190–193. doi: 10.1111/j.1365-4362.1984.tb04509.x. [DOI] [PubMed] [Google Scholar]
- Newton JA, Jagjivan A, Bhogal B, McKee PH, McGibbon DH. Familial primary cutaneous amyloidosis. Br J Dermatol. 1985;112:201–208. doi: 10.1111/j.1365-2133.1985.tb00084.x. [DOI] [PubMed] [Google Scholar]
- Lee DD, Huang JY, Wong CK, Gagel RF, Tsai SF. Genetic heterogeneity of familial primary cutaneous amyloidosis: lack of evidence for linkage with the chromosome 10 pericentromeric region in Chinese families. J Invest Dermatol. 1996;107:30–33. doi: 10.1111/1523-1747.ep12297840. [DOI] [PubMed] [Google Scholar]
- Bergamo F, Annessi G, Ribuffo M. Familial lichen amyloidosis. Chron Dermatol. 1997;6:959–961. [Google Scholar]
- Hartshorne ST. Familial primary cutaneous amyloidosis in a South African family. Clin Exp Dermatol. 1999;24:438–442. doi: 10.1046/j.1365-2230.1999.00526.x. [DOI] [PubMed] [Google Scholar]
- Wang WJ, Chang YT, Huang CY, Lee DD. Clinical and histopathological characteristics of primary cutaneous amyloidosis in 794 Chinese patients. Zhonghua Yi Xue Za Zhi (Taipei) 2001;64:101–107. [PubMed] [Google Scholar]
- Lin MW, Lee DD, Lin CH, et al. Suggestive linkage of familial primary cutaneous amyloidosis to a locus on chromosome 1q23. Br J Dermatol. 2005;152:29–36. doi: 10.1111/j.1365-2133.2004.06254.x. [DOI] [PubMed] [Google Scholar]
- Lee DD, Lin MW, Chen IC, et al. Genome-wide scan identifies a susceptibility locus for familial primary cutaneous amyloidosis on chromosome 5p13.1-q11.2. Br J Dermatol. 2006;155:1201–1208. doi: 10.1111/j.1365-2133.2006.07524.x. [DOI] [PubMed] [Google Scholar]
- Arita K, South AP, Hans-Filho G, et al. Oncostatin M receptor-β mutations underlie familial primary localized cutaneous amyloidosis. Am J Hum Genet. 2008;82:73–80. doi: 10.1016/j.ajhg.2007.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheller J, Ohnesorge N, Rose-John S. Interleukin-6 trans-signalling in chronic inflammation and cancer. Scand J Immunol. 2006;63:321–329. doi: 10.1111/j.1365-3083.2006.01750.x. [DOI] [PubMed] [Google Scholar]
- Heinrich PC, Behrmann I, Haan S, Hermanns HM, Muller-Newen G, Schaper F. Principles of interleukin (IL)-6-type cytokine signalling and its regulation. Biochem J. 2003;374:1–20. doi: 10.1042/BJ20030407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens M, Sloan JS, Robertson PD, Scheet P, Nickerson DA. Automating sequence-based detection and genotyping of SNPs from diploid samples. Nat Genet. 2006;38:375–381. doi: 10.1038/ng1746. [DOI] [PubMed] [Google Scholar]
- The International HapMap Consortium The International HapMap Project. Nature. 2003;426:789–796. doi: 10.1038/nature02168. [DOI] [PubMed] [Google Scholar]
- Lathrop GM, Lalouel JM, Julier C, Ott J. Multilocus linkage analysis in humans: detection of linkage and estimation of recombination. Am J Hum Genet. 1985;37:482–498. [PMC free article] [PubMed] [Google Scholar]
- Kruglyak L, Daly MJ, Reeve-Daly MP, Lander ES. Parametric and nonparametric linkage analysis: unified multipoint approach. Am J Hum Genet. 1996;58:1347–1363. [PMC free article] [PubMed] [Google Scholar]
- Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- Zhao JH, Lissarrague S, Essioux L, Sham PC. GENECOUNTING: haplotype analysis with missing genotypes. Bioinformatics. 2002;18:1694–1695. doi: 10.1093/bioinformatics/18.12.1694. [DOI] [PubMed] [Google Scholar]
- Zhao JH. 2LD, GENECOUNTING and HAP: computer programs for linkage disequilibrium analysis. Bioinformatics. 2004;20:1325–1326. doi: 10.1093/bioinformatics/bth071. [DOI] [PubMed] [Google Scholar]
- Kurth I, Horsten U, Pflanz S, et al. Importance of the membrane-proximal extracellular domains for activation of the signal transducer glycoprotein 130. J Immunol. 2000;164:273–282. doi: 10.4049/jimmunol.164.1.273. [DOI] [PubMed] [Google Scholar]
- Timmermann A, Küster A, Kurth I, Heinrich PC, Müller-Newen G. A functional role of the membrane-proximal extracellular domains of the signal transducer gp130 in heterodimerization with the leukemia inhibitory factor receptor. Eur J Biochem. 2002;269:2716–2726. doi: 10.1046/j.1432-1033.2002.02941.x. [DOI] [PubMed] [Google Scholar]
- Dillon SR, Sprecher C, Hammond A, et al. Interleukin 31, a cytokine produced by activated T cells, induces dermatitis in mice. Nat Immunol. 2004;5:752–760. doi: 10.1038/ni1084. [DOI] [PubMed] [Google Scholar]
- Boniface K, Diveu C, Morel F, et al. Oncostatin M secreted by skin infiltrating T lymphocytes is a potent keratinocyte activator involved in skin inflammation. J Immunol. 2007;178:4615–4622. doi: 10.4049/jimmunol.178.7.4615. [DOI] [PubMed] [Google Scholar]
- Sonkoly E, Muller A, Lauerma AI, et al. IL-31: a new link between T cells and pruritus in atopic skin inflammation. J Allergy Clin Immunol. 2006;117:411–417. doi: 10.1016/j.jaci.2005.10.033. [DOI] [PubMed] [Google Scholar]
- Morikawa Y, Tamura S, Minehata K-i, Donovan PJ, Miyajima A, Senba E. Essential function of oncostatin M in nociceptive neurons of dorsal root ganglia. J Neurosci. 2004;24:1941–1947. doi: 10.1523/JNEUROSCI.4975-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura S, Morikawa Y, Miyajima A, Senba E. Expression of oncostatin M receptor in a specific subset of nociceptive sensory neurons. Eur J Neurosci. 2003;617:2287–2298. doi: 10.1046/j.1460-9568.2003.02681.x. [DOI] [PubMed] [Google Scholar]
- Chattopadhyay S, Tracy E, Liang P, Robledo O, Rose-John S, Baumann H. Interleukin-31 and oncostatin-M mediate distinct signaling reactions and response patterns in lung epithelial cells. J Biol Chem. 2007;282:3014–3026. doi: 10.1074/jbc.M609655200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.