Abstract
The adult mouse small intestinal epithelium undergoes perpetual regeneration, fueled by a population of multipotential stem cells and oligopotential daughters located at the base of crypts of Lieberkühn. Although the morphologic features of small intestinal epithelial progenitors (SiEPs) are known, their molecular features are poorly defined. Previous impediments to purification and molecular characterization of SiEPs include lack of ex vivo clonigenic assays and the difficulty of physically retrieving them from their niche where they are interspersed between their numerous differentiated Paneth cell daughters. To overcome these obstacles, we used germ-free transgenic mice lacking Paneth cells to obtain a consolidated population of SiEPs with normal proliferative activity. These cells were harvested by laser capture microdissection. Functional genomics analysis identified 163 transcripts enriched in SiEPs compared with Paneth cell-dominated normal crypt base epithelium. The dataset was validated by (i) correlation with the organellar composition of SiEPs versus Paneth cells, (ii) similarities to databases generated from recent mouse hematopoietic and neural stem cell genome anatomy projects, and (iii) laser capture microdissection/real-time quantitative RT-PCR studies of progenitor cell-containing populations retrieved from the small intestines, colons, and stomachs of conventionally raised mice. The SiEP profile has prominent representation of genes involved in c-myc signaling and in the processing, localization, and translation of mRNAs. This dataset, together with our recent analysis of gene expression in the gastric stem cell niche, discloses a set of molecular features shared by adult mouse gut epithelial progenitors.
Keywords: stem cells‖Paneth cells‖c-myc‖RNA binding proteins‖laser capture microdissection
The adult human gut contains one of the largest populations of epithelial stem cells in the body. Defining the properties of these cells offers an opportunity to develop new ways of classifying and treating common diseases that affect repair, regeneration, and transformation of this epithelium.
The mouse has served as the principal model organism for deciphering the features of gut stem cells. Thirty years have elapsed since 3H-thymidine labeling/electron microscopic (EM) autoradiographic studies of adult mice demonstrated that the small intestinal epithelium undergoes continuous renewal, fueled by multipotent stem cells located at the base of its numerous flask-shaped mucosal invaginations (crypts of Lieberkühn). These lineage tracing experiments provided a detailed morphologic description of small intestinal epithelial progenitors (SiEPs) (1).
Each active stem cell appears to gives rise to two populations of long-lived daughters, one committed to producing absorptive enterocytes, the other committed to producing secretory cell lineages (goblet, enteroendocrine, and Paneth) (2). Paneth cells complete their differentiation at the crypt base where they function as key components of innate immunity, producing a variety of antimicrobial proteins (e.g., cryptdins) (3). Each crypt contains 30–50 mature Paneth cells interposed between a much smaller population of SiEPs (4, 5). Members of the three other epithelial lineages complete their differentiation as they migrate out of crypts and move up adjacent finger-shaped villi to a cellular extrusion zone located near the tip of each villus. The cycle of renewal is completed every 3–5 days (6).
Recent work has provided insights about the factors that affect cell fate specification and proliferation of SiEPs in the developing mouse intestine. Hes1, the mammalian homologue of Drosophila Hairy and Enhancer of split genes, is an immediate downstream target of Notch and encodes a basic helix–loop–helix (bHLH) transcriptional repressor that opposes the activity of various bHLH transcriptional activators, including Math1 (7). Targeted disruption of Hes1 increases the fractional representation of secretory cell lineages and reduces the number of enterocytes, whereas Math1 disruption has the converse effect (8, 9). Tcf-4 encodes a high mobility group (HMG) box transcription factor that participates in Wnt/Apc/β-catenin signaling (10). Tcf4−/− mice die at birth without mitotically active intestinal stem cells (11). Genetic manipulation of Rac1 activity affects both proliferative status and epithelial differentiation (12, 13). Rac GTPases intersect with the Apc/β-catenin pathway through their interactions with the guanine nucleotide exchange factor, Asef (14).
In this study, we have used adult transgenic mice with a genetically engineered ablation of their Paneth cells to enhance the fractional representation of SiEPs at the crypt base so that they could be readily recovered by laser capture microdissection (LCM). Functional genomics studies of epithelial cells harvested from SiEP-enriched versus normal Paneth cell-dominated crypt bases yielded a dataset of 163 SiEP-associated mRNAs. This dataset provides a molecular profile of SiEPs within their niche. Components of c-myc signaling pathways, as well as RNA binding proteins involved in transcript processing, localization, and translation, are prominently represented. SiEPs share features with adult mouse colonic and gastric epithelial progenitors (GEPs), as well as with neural stem cells and hematopoietic stem cells (HSCs).
Materials and Methods
Mice.
A pedigree of FVB/N mice hemizygous for a transgene that directs expression of an attenuated diphtheria toxin A fragment to Paneth cells (CR2-tox176) was rederived as germ free (GF) and housed in sterile plastic gnotobiotic incubators (15, 16). Animals were kept under a strict 12-h light cycle and given an autoclaved chow diet (B & K Universal, East Yorkshire, U.K.). Conventionally raised normal FVB/N mice were maintained in microisolator cages in a specified pathogen-free state. Males were killed at 1200 hours on postnatal day 26 (GF) or postnatal days 56–70 (conventionally raised) by using protocols approved by our institution's Animal Studies Committee.
Transmission EM.
Tissue fragments (3 mm2), obtained from the junction of the middle and distal thirds of the small intestines of GF CR2-tox176 mice and their normal littermates, were prepared for EM (16). The identities of the five most basal epithelial cells of well-oriented crypts were scored by viewing 100-nm-thick sections with a JOEL 100C electron microscope (n = 30 crypts per specimen; three mice per group).
Quantification of Proliferation.
GF normal and CR2-tox176 littermates were given i.p. injections of a solution containing BrdUrd (120 mg/kg) and 5-fluro-2′-deoxyuridine (12 mg/kg). Mice were killed 1 h later. Eight-centimeter-long segments, spanning the junction of the middle and distal thirds of the small intestine, were removed, fixed in Bouin's solution, and embedded in paraffin, and 5-μm-thick sections were cut along the cephalocaudal axis. Deparaffinized sections were incubated with goat anti-BrdUrd (final dilution = 1:2,000 in PBS/1% BSA/0.3% Triton X-100), Alexa Fluor 594-conjugated donkey anti-goat Ig (1:1,000, Molecular Probes), FITC-conjugated Ulex europeaus agglutinin-1 (reacts with Fucα1,2Gal glycans expressed in Paneth cells; 1:100, Sigma), and bis-benzimide (nuclear stain; 50 ng/ml; Sigma). S-phase cells were scored in well-oriented crypts (defined as crypts that contain a single layer of epithelial cells surrounding their lumen and that are adjacent to the muscularis mucosa; n = 200 crypts scored per mouse; three mice per group).
GeneChip Analysis of LCM Crypt Base Epithelial Cells.
Eight-centimeter-long segments from the junction of the middle and distal thirds of the small intestine were prepared for LCM by using protocols described in ref. 17. The five most basal epithelial cells from well-oriented crypts were harvested by using a PixCell II LCM system (Arcturus, Mountain View, CA; 7.5-μm diameter laser spot) and CapSure HS LCM caps (Arcturus) (n = 700 crypt bases dissected per mouse; n = three mice per group). Total cellular RNA was extracted (PicoPure RNA isolation kit, Arcturus). Duplicate 15-ng aliquots of pooled RNA isolated from the crypt bases of CR2-tox176 or normal littermates were individually amplified ≈106-fold through two successive rounds of T7-mediated in vitro transcription (RiboAmp RNA Amplification kit, Arcturus). Amplification was performed according to the manufacturer's protocol with one exception: the T7 BioArray Transcript Kit (Enzo Biochem) was substituted for the last step so that the cRNAs could be biotinylated. Each cRNA was then hybridized to a set of Mu11K GeneChips (Affymetrix, Santa Clara, CA). To verify the linearity of amplification, four Bacillus subtilus transcripts (100,000 copies of Lys, 20,000 copies of Phe, 5,000 copies of Trp, and 1,000 copies of Thr) were added to each RNA sample before amplification (Mu11K GeneChips contain probesets for these bacterial mRNAs) (18).
Overall fluorescence for each GeneChip was scaled to a target intensity of 1,500 and pairwise comparisons were performed with Affymetrix microarray suite software (version 4.0). Genes were selected based on the following criteria: (i) for SiEP-enriched expression, chip-to-chip comparisons were performed with the normal adult GF mouse GeneChip designated as baseline; all mRNAs whose levels were called >1.5-fold increased over baseline by GeneChip software in duplicate chip-to-chip comparisons were culled; and (ii) for Paneth cell-enriched expression, comparisons were performed with the GF CR2-tox176 chip designated as baseline and transcripts >1.5-fold increased over baseline in duplicate comparisons were retrieved.
SYBR Green-Based Quantitative Real-Time RT-PCR.
RNA was purified from epithelial cells harvested by LCM from the crypt base, the upper half of their associated villi, the lower third of colonic crypts, or from the colonic villus homolog (the surface epithelial cuff) (n = 450 cells per compartment per mouse per mRNA assayed; three mice independently evaluated). cDNA corresponding to 300 pg of purified RNA (≈150 cells) was added to each 25-μl quantitative RT-PCR (qRT-PCR) mixture, which also contained 12.5 μl of 2× SYBR green master mix (Applied Biosystems), 900 nM gene-specific primers (see Table 1, which is published as supporting information on the PNAS web site, www.pnas.org), and 0.25 units of UDP-N-glycosidase (Invitrogen). A melting curve (15, 17) was used to identify a temperature where the amplicon and not primer-dimers were the source of SYBR green-bound fluorescence. Each assay per compartment per mouse was performed in triplicate and the data were normalized to GAPDH mRNA levels (ΔΔCT method).
Results and Discussion
A Transgenic Mouse Model with an Enriched Population of Small Intestinal Crypt Epithelial Progenitors.
Paneth cell lineage ablation was achieved by expressing an attenuated diphtheria toxin A fragment (tox176) under the control of transcriptional regulatory elements from the mouse cryptdin-2 gene (Defcr2; abbreviated CR2) (16). Paneth cell-deficient CR2-tox176 mice and their normal FVB/N littermates were rederived as GF to eliminate variations in their luminal microbial environment and to minimize the number of intraepithelial lymphocytes in their crypts (19).
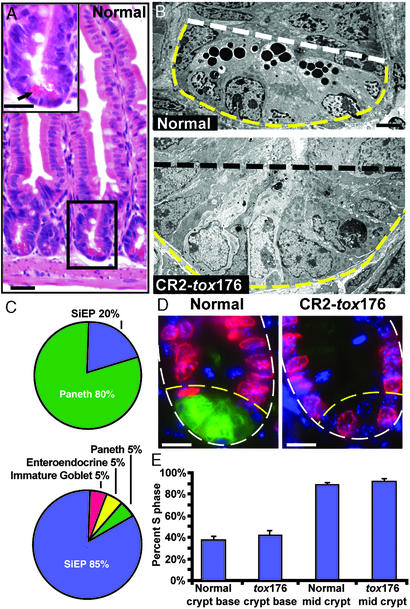
Removal of Paneth cells from the crypt base results in a marked increase in the fractional representation of cells with morphologic characteristics of undifferentiated SiEPs. Quantitative EM studies of crypts located at the junction between the middle and distal thirds of the small intestine (distal jejunum) disclosed that in normal GF mice SiEPs represent 15% of the crypt base population (defined as the five most basal epithelial cells), whereas 80% are mature Paneth cells. In age-matched GF CR2-tox176 animals, SiEPs comprise 85% of the crypt base epithelial population. The remaining 15% consist of differentiating goblet, enteroendocrine, and residual Paneth cells (5% per cell type; Fig. 1 A–C).
Figure 1.
The fractional representation of SiEPs is markedly increased in the crypt bases of Paneth cell-ablated CR2-tox176 mice. (A) Hematoxylin and eosin-stained section of the distal jejunum of a normal GF mouse. (Inset) An enlarged view of the boxed crypt. The crypt base is populated with Paneth cells that contain distinctive eosinophilic secretory granules (e.g., arrow). (B) EM images of distal jejunal crypt base epithelium from GF normal and transgenic CR2-tox176 mice. Paneth cells located at the normal crypt base (below white dashed line) contain characteristic electron-dense cytoplasmic granules. The CR2-tox176 crypt base is populated with poorly differentiated SiEPs. (C) Quantitative EM analysis of distal jejunal crypt base epithelial populations in normal (Upper) and CR2-tox176 (Lower) animals. (D) S-phase cells in GF normal and CR2-tox176 crypts. Mice were injected with BrdUrd 1 h before death. Sections of distal jejunum were stained with goat anti-BrdUrd, Alexa Fluor 594-labeled donkey anti-goat Ig (red), FITC-conjugated U. europeaus agglutinin-1 (to mark Paneth cells as green), and bis-benzimide (blue nuclear stain). The outer margin of each crypt is demarcated by a white dashed line and the base is outlined by yellow dashed lines. (E) Quantifying S-phase cells. Mean values ± SEM are plotted. See text for details. (Scale bars: A and D = 25 μm; B = 5 μm.)
Immunohistochemical studies using antibodies to the pan-lymphocytic marker CD45 revealed that GF normal and CR2-tox176 mice had <1 intraepithelial lymphocyte per 1,000 crypt base epithelial cells. In addition, histochemical and immunohistochemical assays (20) disclosed that GF CR2-tox176 mice had no obvious perturbations in crypt size or in the distribution and number of differentiated villus epithelial cell types, when compared with their normal littermates.
SiEPs are known to divide at a slower rate than their midcrypt transient amplifying descendants (1). To determine whether SiEPs in GF CR2-tox176 crypts retained this feature, normal and transgenic mice were given an i.p. injection of BrdUrd 1 h before death. S-phase cells were identified in sections of distal jejunum by using antibodies to BrdUrd (Fig. 1D), and the percentage of BrdUrd+ epithelial cells defined in two domains of well-oriented crypts: (i) cell layers 1–3 (the base) and (ii) layers 4–8. Paneth cells, marked by their reaction with FITC-conjugated Ulex europeaus agglutinin-1 (21), were excluded. Layers 4–8 in both GF CR2-tox176 and normal mice contained 85–90% BrdUrd+ cells (Fig. 1E). In contrast, the crypt base of CR2-tox176 animals contained only 38% BrdUrd+ cells. This latter value was not significantly different from the number of BrdUrd+ cells in the crypt bases of normal littermates (43%; P > 0.05; Student's t test) (Fig. 1E). We concluded that the morphologic and proliferative features of the expanded population of SiEPs in GF CR2-tox176 crypts were indistinguishable from those of normal littermate SiEPs.
Generating a Database of mRNAs Enriched in SiEPs.
The enrichment of SiEPs observed in an anatomically distinct region of CR2-tox176 crypts allowed us to define their molecular properties. To distinguish SiEPs from their differentiated progeny, we compared the population of mRNAs expressed in CR2-tox176 crypt base epithelium with mRNAs expressed in the Paneth cell-dominated crypt base epithelium of normal age-matched littermates. Because our goal was to perform this comparison without physical disruption of the stem niche, we retrieved each cell population by LCM of distal jejunal cryosections. Approximately 3,500 crypt base epithelial cells were harvested per mouse. Equivalent amounts of RNA were then pooled from each animal in each group (10 ng per mouse; n = three mice per group), and the pooled RNAs from each group were divided into two separate, but equal-sized, samples. Each sample was subsequently amplified ≈106-fold with two successive rounds of T7-mediated in vitro transcription (22, 23), and each cRNA product was used to interrogate Mu11K GeneChips containing probe sets representing 11,000 mouse genes and EST clusters.
We identified mRNAs from 163 known genes and 25 ESTs as enriched in SiEP-predominated CR2-tox176 crypt base epithelium (duplicate GeneChip comparisons; see Table 2, which is published as supporting information on the PNAS web site for a complete list). Transcripts from 55 genes (and eight ESTs) satisfied our selection criteria (see Materials and Methods) for enrichment in the Paneth cell-predominated crypt base epithelium of normal littermates (Table 3, which is published as supporting information on the PNAS web site).
Validation of the SiEP Dataset.
Morphology.
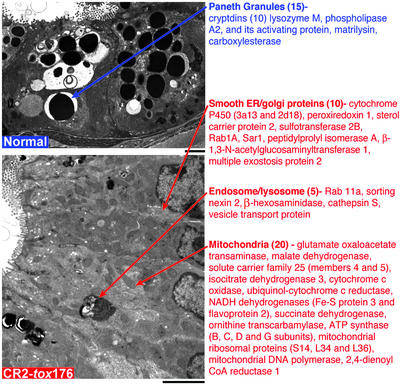
Differences in the cytoplasmic organelle composition of SiEPs and Paneth cells were reflected in the two datasets, providing a “morphologic validation” for the GeneChip results. Paneth cells have distinctive electron-dense secretory granules that are not represented in SiEPs. Fifteen of the 55 Paneth-enriched transcripts (27%) encode proteins associated with, or implicated in the function of these granules, including all 10 members of the cryptdin family of antimicrobial α-defensins that are represented in the Mu11K GeneChips (Fig. 2 Upper). SiEPs have relatively more mitochondria, smooth endoplasmic reticulum, and endosomal/lysosomal components than Paneth cells. Thirty-five of the 163 SiEP-enriched transcripts (21%) encode products that localize to these organelles (Fig. 2 Lower).
Figure 2.
Correlations between cytoplasmic organelle composition and mRNAs represented in SiEP and Paneth cell datasets. (Upper) Transmission EM of Paneth cells located at the base of normal GF crypts. Gene products that are enriched in Paneth cells and are affiliated with their distinctive electron dense secretory granules are listed. The number of genes per category is indicated in parentheses. (Lower) EM of SiEPs. mRNAs encoding proteins associated with organelles that are well represented in these progenitors are listed. (Scale bars: 5 μm.)
Comparisons with neural stem cell and HSC databases.
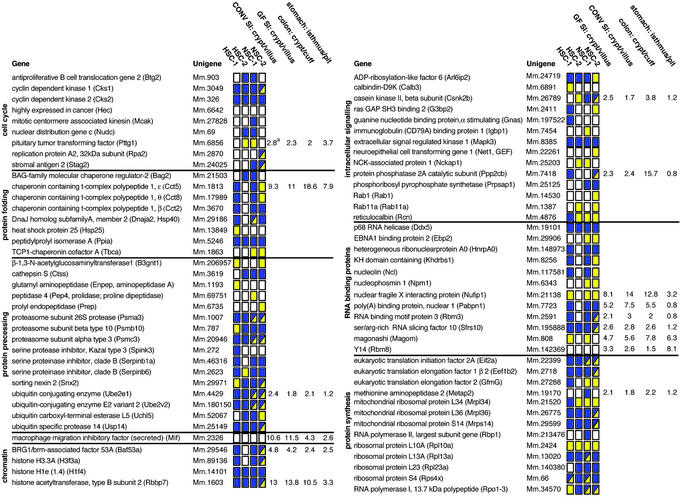
A second validation of the SiEP dataset came from comparisons with recently published databases of mRNAs enriched in neuronal stem cells (NSCs) and HSCs. Twenty-two percent and 27% of the entries in two independently generated HSC datasets were present in two independently generated NSC lists (refs. 24 and 25, respectively). Our SiEP dataset has 36% and 24% identity with these two NSC datasets and 10% and 7% identity with the two HSC profiles. If genes from the same family and with similar reported functions are included in the comparison, the SiEP dataset has 55% and 61% similarity with the two NSC datasets and 39% and 40% similarity with the two HSC datasets (Table 2 and Fig. 3).
Figure 3.
Representation of components of the SiEP database in mouse NSC and HSC databases. Gene abbreviations are in parentheses. HSC1/NSC1 datasets are from ref. 25. HSC2/NSC2 are from ref. 24. A yellow box indicates that the identical mRNA is present in the SiEP and HSC or NSC databases. A blue box indicates that one or more transcripts encoding functionally similar members of the gene family are expressed. A box with blue and yellow sectors means that the identical transcript plus one or more related family members are present. A white box indicates that the identical or similar mRNA is absent. See Table 2 for additional details. aAverage fold difference, defined by qRT-PCR, in progenitor versus differentiated epithelial populations harvested by LCM from the distal jejunum (SI), descending colon, and stomachs of GF or conventionally raised (CONV) normal adult male FVB/N mice.
We annotated all of the proteins in the SiEP, HSC, and NSC datasets by automated, algorithmic assignment of terms from the Gene Ontology (GO) Consortium (www.geneontology.org), and then compared the 10 GO terms with the highest fractional representation in each dataset. Fractional representation is defined as the number of proteins with a given GO term relative to the total number of proteins in that dataset with assigned GO terms. The results provided additional evidence for the functional similarities between the databases and independently demonstrated that the highest concordance was between SiEPs and NSCs (Table 4, which is published as supporting information on the PNAS web site).
LCM/real-time qRT-PCR studies involving GF and conventionally raised normal mice.
The third form of validation of the SiEP database entailed an analysis of whether selected mRNAs were enriched in the crypt base versus differentiated villus epithelium of GF postnatal day 26 normal mice. Epithelial cells located in the lower third of distal jejunal crypts (progenitor compartment) and in the upper half of neighboring villi (differentiated compartment) were harvested by LCM, and the relative levels of 15 mRNAs, representing each of the eight functional categories listed in Fig. 3, were quantified by qRT-PCR. The results confirmed that the levels of all 15 mRNAs were higher in the progenitor compartment (range = 2- to 14-fold; Fig. 3).
Our previous functional genomics studies of GF mice colonized with single or multiple components of the normal small intestinal microbiota revealed that these indigenous microorganisms influence gene expression in both the crypt and villus epithelium (26, 27). Therefore, a second round of LCM/qRT-PCR analysis, analogous to that performed with GF mice, compared the relative levels of these 15 mRNAs in distal jejunal crypt base and upper villus epithelium harvested from normal, adult (8–10 weeks old), conventionally raised FVB/N mice (i.e., animals with a normal gut microbiota). Remarkably, the presence or absence of a microbiota was not associated with statistically significant differences in the crypt/villus distribution of these 15 transcripts (Fig. 3). We concluded that the enhanced expression of these mRNAs in SiEPs occurs independently of the microbiota and that the SiEP profile obtained from GF transgenic mice lacking Paneth cells is representative of the intact, unperturbed crypt base progenitor niche of normal conventionally raised animals.
Functional Interpretation of the SiEP Dataset.
The prominence of c-myc pathways.
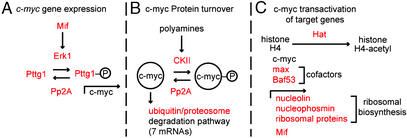
As noted in the Introduction, Tcf4−/− mice die at birth without mitotically active intestinal stem cells (11). C-myc is a downstream target of the Wnt/β-catenin/Tcf-4 signaling pathway (28). The contribution of c-myc to SiEP biology has yet to be deciphered in vivo: e.g., c-myc−/− mice die at embryonic day 10, 5 days before the first appearance of the small intestinal epithelium. However, recent studies have shown that Tcf-4 signaling through c-myc plays a key role in imposing an undifferentiated state in cultured intestinal epithelial cells and that loss of β-catenin/Tcf-4 activity results in decreased expression of c-myc, induction of p21CIP1/WAF1, cell cycle arrest in G1, and differentiation (29). Our dataset provides evidence of the importance of c-myc signaling in SiEPs, i.e., the list includes transcripts encoding proteins involved in regulating c-myc transcription, the stability of c-myc protein, and c-myc transactivation of its target genes.
SiEPs express the mitogen-activated protein kinase family member Erk1 (extracellular-regulated kinase 1). Erk1 stimulates cell proliferation in response to a variety of factors, including Mif (macrophage migration inhibitory factor; also in the SiEP list) (30). Activated Erk1 phosphorylates another member of the SiEP dataset, pituitary tumor transforming factor-1 (Pttg1), allowing phospho-Pttg1 to translocate to the nucleus (31) where it transactivates c-myc (32) (Fig. 4A).
Figure 4.
Overview of SiEP-associated factors that affect c-myc signaling. Members of the SiEP dataset are indicated in red. See text for additional details.
The SiEP dataset also includes casein kinase II (CkII) mRNA. C-myc is phosphorylated by CkII in response to polyamines, which are known SiEP mitogens (33). CkII-mediated phosphorylation of c-myc prevents its degradation via the ubiquitin/proteasomal pathway (34), which is well represented in the SiEP dataset (ubiquitin-conjugating enzymes 2e1 and 2v2; ubiquitin carboxyl-terminal esterase; ubiquitin-specific protease 14; proteasome subunits β type 10, α type 3, and 26S protease regulatory subunit 6A) (Fig. 4B).
Loss-of-function mutations of protein phosphatase 2A components are found in human intestinal adenocarcinomas (35). SiEP express Ppp2cb mRNA (encodes the β isoform of the catalytic subunit of the protein phosphatase 2A complex; ref. 36) CkII substrates are dephosphorylated by Pp2A (37). In the case of c-myc, this dephosphorylation promotes degradation by the ubiquitin/proteasomal pathway (37). Erk substrates are also dephosphorylated by Pp2A (37). Pp2A-mediated dephosphorylation of the c-myc transactivator phospho-Pttg1 represents a potential second route by which c-myc activity could be modulated in SiEPs by this phosphatase (Fig. 4 A and B).
A final level of c-myc signaling represented in the SiEPs involves regulation of expression of its downstream target genes. C-myc binding to chromatin induces histone acetylation by histone acetyltransferase (Hat; ref. 38). Hat type B (Rbbp7), which targets histone 4, is present in the dataset. The c-myc transactivation complex contains two SiEP-expressed components, max (basic helix–loop–helix coactivator of c-myc; ref. 39) and Baf53a (related to the yeast SWI/SNF complex that functions in nucleosome remodeling during transcription; refs. 40 and 41). C-myc transactivates a number of genes represented in the SiEP dataset, nucleolin and nucleophosmin (rRNA binding proteins), several ribosomal proteins (42), as well as Mif (Figs. 3 and 4C).
Control of the cell cycle.
As anticipated from the proliferative activity of SiEPs, mRNAs encoding components of the cell cycle, such as cyclin-dependent kinase regulatory subunits 1 and 2, are represented in the dataset (Fig. 3 and Table 2). The dataset also includes several members of the chaperonin-containing t-complex polypeptide 1 (Cct β, θ, and ɛ) that binds newly synthesized cyclin E to cyclin-dependent kinase 2 (43) plus transcripts encoding proteins involved in sister chromatid separation during anaphase (e.g., centromere-associated kinesin and Pttg1; refs. 44 and 45).
Meticulous control of the rate of SiEP division is critical if crypt-villus units are to maintain a constant epithelial cell census. This control is not only reflected by the SiEP-associated factors that modulate c-myc expression, c-myc stability, and transactivation of c-myc targets, but also by the presence of Btg2 (B cell translocation gene 2) that inhibits cycD1 (cyclin D1) transcription (46), and the ubiquitin/proteasome complex that mediates destruction of cyclins (47), Ccts (48), and Btg2 (49), and regulates sister chromatid separation (47).
Heparin sulfate proteoglycans also serve as regulatory cofactors (stimulators and inhibitors) for a number of growth factors (50). The SiEP dataset contains three transcripts encoding proteins that affect proteoglycan bioavailability/activity: syndecan binding protein, glypican 1, and multiple exostosis protein 2 (a glycosytransferase required for heparan sulfate addition that is also present in the NSC and HSC databases). Interestingly, knockout of the winged helix transcription factor gene Foxl1 (normally expressed in the pericryptal mesenchyme) results in increased SiEP proliferation and increased production of syndecan (epithelial heparin sulfate proteoglycan that acts as a Wnt receptor; ref. 51).
Control of mRNA processing, trafficking, and translation.
Dividing cells increase their apparatus for transcription and translation, in part in response to c-myc signaling (see above and refs. 52 and 53). Our previous analysis of mRNAs enriched in adult mouse GEPs revealed a prominent representation of transcripts encoding proteins required for nuclear RNA processing, cytoplasmic localization, and translation (53). This feature is recapitulated in the SiEP dataset that contains mRNAs encoding Ser/Arg-rich RNA splicing factor 10, nuclear poly(A) binding protein 1, two mouse homologs of Drosophila proteins involved in mRNA processing, trafficking, and establishment of polarity in developing oocytes (mago-m and Y14), four ribosomal proteins (L10A, L13A, L23, S4), plus initiation and elongation factors that are downstream targets of CkII signaling cascades (eIF2A, eEF1β2, eEF2) (Fig. 3). Control of mRNA processing and localization in gut epithelial progenitors may play an important role in regulating their communications with other cellular components of their niches, as well as influencing their capacity for asymmetric division, or instigating polarized migration of their offspring. Notably, HSCs and NCSs also express identical and/or functionally equivalent genes involved in mRNA processing, localization, and translation (Fig. 3).
Comparisons of SiEPs with Epithelial Progenitors in the Stomach and Distal Colon of Adult Conventionally Raised Normal Mice.
The 15 mRNAs used for LCM/qRT-PCR validation of the SiEP dataset (see above) were used to compare the features of small intestinal and colonic progenitors. LCM was utilized to recover epithelial cells from the lower third of crypts located in the descending colon and from the surface epithelial cuffs that ring the orifice of these crypts (cuffs are homologues of small intestinal villi). qRT-PCR analysis revealed a strong similarity between colonic epithelial progenitors and SiEPs: levels of all 15 SiEP-enriched transcripts were higher in colonic crypt base epithelium compared with the surface epithelial cuff (range 1.5- to 19-fold; Fig. 3).
Epithelial cells were then harvested from two regions of gastric units: the midportion or isthmus, which contains multipotent stem cells and their immediate committed daughters (54), and the upper third, which contains differentiated mucus-producing pit cells and acid-producing parietal cells. Eight of the 15 mRNAs that were enriched in SiEPs were enriched in the isthmal GEP niche, including three of the six transcripts encoding mRNA binding proteins (Fig. 3). Moreover, none of the 15 mRNAs showed statistically significant decreases in GEPs compared with their differentiated progeny.
The concordance between SiEP and GEP gene expression prompted a more detailed analysis of the degree of overlap between our recently reported GEP gene expression profile (54) and that of SiEPs. Eleven of the 147 genes in the GEP dataset were present in the SiEP dataset. An additional 22 genes encoded related family members or additional subunits of multisubunit complexes. The observed degree of overlap between lists is striking. Both the SiEP and GEP lists were identified by using GeneChips containing probe sets representing ≈11,000 genes. Thus, based on chance alone, we would expect no more than two of the 163 SiEP transcripts to be present in the GEP list. Furthermore, there was no overlap between the GEP and the Paneth cell datasets. Together, these findings provide the beginnings of a molecular definition of features shared by epithelial progenitors distributed along the length of the adult mouse gastrointestinal tract.
Prospectus.
The approach used in this study to enrich for normally rare epithelial progenitors in the adult mouse small intestine involved an engineered ablation of the principal differentiated cell type that normally coexists with SiEPs at the crypt base. Remarkably, ablation did not affect the proliferative status of SiEPs, indicating that Paneth cells and their associated factors are dispensable when it comes to maintaining proper regulation of cell division rates in the crypt stem cell niche. This finding contrasts with the situation in the adult mouse stomach, where tox176-mediated ablation of parietal cells, the principal epithelial lineage that differentiates in the gastric stem niche, results in marked enhancement of GEP proliferation (54).
Enrichment of SiEPs in the crypt base epithelium through erasure of differentiated Paneth cells permitted their direct retrieval by LCM, thereby avoiding prolonged periods of separation from their niche before mRNA extraction. Evidence for the validity of the SiEP profile includes correlation with their organellar content, compositional/functional similarities to databases obtained from recent NSC and HSC genome anatomy projects, plus direct LCM/qRT-PCR comparisons of gene expression in crypt base and villus epithelium of normal, conventionally raised mice. This validation sets the stage for the next phase of characterization where normalized micro-cDNA libraries are prepared from LCM SiEPs harvested from CR2-tox176 crypt bases and then sequenced. Such an approach is likely to yield an expression profile that includes very low abundance transcripts. In addition, cDNA library-based genome anatomy projects do not have to limit themselves to mRNAs that exceed a set threshold difference in expression levels between progenitor and reference control populations. Thus, they should provide a more comprehensive view of the biological properties of these formerly elusive cells.
Supplementary Material
Acknowledgments
We thank David O'Donnell, Maria Karlsson, Sabrina Wagoner, Chris Chen, and Diane Redmond for superb technical assistance. This work was funded by National Institutes of Health Grants DK30292, DK58529, and DK63483. T.S.S. is the recipient of a National Institutes of Health K08 Career Development Award.
Abbreviations
- EM
electron microscopy
- SiEP
small intestinal epithelial progenitor
- LCM
laser capture microdissection
- GF
germ free
- qRT-PCR
quantitative RT-PCR
- NSC
neuronal stem cell
- HSC
hematopoietic stem cell
- GEP
gastric epithelial progenitor
References
- 1.Cheng H, Leblond C P. Am J Anat. 1974;141:461–479. doi: 10.1002/aja.1001410403. [DOI] [PubMed] [Google Scholar]
- 2.Bjerknes M, Cheng H. Gastroenterology. 1999;116:7–14. doi: 10.1016/s0016-5085(99)70222-2. [DOI] [PubMed] [Google Scholar]
- 3.Ouellette A J. Am J Physiol. 1999;277:G257–G261. doi: 10.1152/ajpgi.1999.277.2.G257. [DOI] [PubMed] [Google Scholar]
- 4.Cheng H. Am J Anat. 1974;141:521–535. doi: 10.1002/aja.1001410406. [DOI] [PubMed] [Google Scholar]
- 5.Bjerknes M, Cheng H. Am J Anat. 1981;160:51–63. doi: 10.1002/aja.1001600105. [DOI] [PubMed] [Google Scholar]
- 6.Wright N A, Irwin M. Cell Tissue Kinet. 1982;15:595–609. doi: 10.1111/j.1365-2184.1982.tb01066.x. [DOI] [PubMed] [Google Scholar]
- 7.Zheng J L, Shou J, Guillemot F, Kageyama R, Gao W Q. Development (Cambridge, UK) 2000;127:4551–4560. doi: 10.1242/dev.127.21.4551. [DOI] [PubMed] [Google Scholar]
- 8.Jensen J, Pedersen E E, Galante P, Hald J, Heller R S, Ishibashi M, Kageyama R, Guillemot F, Serup P, Madsen O. Nat Genet. 2000;24:36–44. doi: 10.1038/71657. [DOI] [PubMed] [Google Scholar]
- 9.Yang Q, Bermingham N A, Finegold M J, Zoghbi H Y. Science. 2001;294:2155–2158. doi: 10.1126/science.1065718. [DOI] [PubMed] [Google Scholar]
- 10.Korinek V, Barker N, Willert K, Molenaar M, Roose J, Wagenaar G, Markman M, Lamers W, Destree O, Clevers H. Mol Cell Biol. 1998;18:1248–1256. doi: 10.1128/mcb.18.3.1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Korinek V, Barker N, Moerer P, van Donselaar E, Huls G, Peters P J, Clevers H. Nat Genet. 1998;19:379–383. doi: 10.1038/1270. [DOI] [PubMed] [Google Scholar]
- 12.Stappenbeck T S, Gordon J I. Development (Cambridge, UK) 2001;128:2603–2614. doi: 10.1242/dev.128.13.2603. [DOI] [PubMed] [Google Scholar]
- 13.Stappenbeck T S, Gordon J I. Development (Cambridge, UK) 2000;127:2629–2642. doi: 10.1242/dev.127.12.2629. [DOI] [PubMed] [Google Scholar]
- 14.Kawasaki Y, Senda T, Ishidate T, Koyama R, Morishita T, Iwayama Y, Higuchi O, Akiyama T. Science. 2000;289:1194–1197. doi: 10.1126/science.289.5482.1194. [DOI] [PubMed] [Google Scholar]
- 15.Hooper L V, Mills J C, Roth K A, Stappenbeck T S, Wong M H, Gordon J I. Mol Cell Microbiol. 2002;31:559–589. [Google Scholar]
- 16.Garabedian E M, Roberts L J J, McNevin M S, Gordon J I. J Biol Chem. 1997;272:23729–23740. doi: 10.1074/jbc.272.38.23729. [DOI] [PubMed] [Google Scholar]
- 17.Stappenbeck T S, Hooper L V, Manchester J K, Wong M H, Gordon J I. Methods Enzymol. 2002;356:168–196. doi: 10.1016/s0076-6879(02)56932-9. [DOI] [PubMed] [Google Scholar]
- 18.Luzzi V, Holtschlag V, Watson M A. Am J Pathol. 2001;158:2005–2010. doi: 10.1016/S0002-9440(10)64672-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mysorekar I U, Lorenz R G, Gordon J I. J Biol Chem. 2002;277:37811–37819. doi: 10.1074/jbc.M205300200. [DOI] [PubMed] [Google Scholar]
- 20.Hermiston M L, Gordon J I. J Cell Biol. 1995;129:489–506. doi: 10.1083/jcb.129.2.489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Falk P G, Roth K A, Gordon J I. Am J Physiol. 1994;266:G987–G1003. doi: 10.1152/ajpgi.1994.266.6.G987. [DOI] [PubMed] [Google Scholar]
- 22.Luo L, Salunga R C, Guo H, Bittner A, Joy K C, Galindo J E, Xiao H, Rogers K E, Wan J S, Jackson M R, Erlander M G. Nat Med. 1999;5:117–122. doi: 10.1038/4806. [DOI] [PubMed] [Google Scholar]
- 23.Baugh L R, Hill A A, Brown E L, Hunter C P. Nucleic Acids Res. 2001;29:E29. doi: 10.1093/nar/29.5.e29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ramalho-Santos M, Yoon S, Matsuzaki Y, Mulligan R C, Melton D A. Science. 2002;298:597–600. doi: 10.1126/science.1072530. [DOI] [PubMed] [Google Scholar]
- 25.Ivanova N B, Dimos J T, Schaniel C, Hackney J A, Moore K A, Lemischka I R. Science. 2002;298:601–604. doi: 10.1126/science.1073823. [DOI] [PubMed] [Google Scholar]
- 26.Hooper L V, Wong M H, Thelin A, Hansson L, Falk P G, Gordon J I. Science. 2001;291:881–884. doi: 10.1126/science.291.5505.881. [DOI] [PubMed] [Google Scholar]
- 27.Stappenbeck T S, Hooper L V, Gordon J I. Proc Natl Acad Sci USA. 2002;99:15451–15455. doi: 10.1073/pnas.202604299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.He T C, Sparks A B, Rago C, Hermeking H, Zawel L, da Costa L T, Morin P J, Vogelstein B, Kinzler K W. Science. 1998;281:1509–1512. doi: 10.1126/science.281.5382.1509. [DOI] [PubMed] [Google Scholar]
- 29.van de Wetering M, Sancho E, Verweij C, de Lau W, Oving I, Hurlstone A, van der Horn K, Battle E, Coudreuse D, Haranis A P, et al. Cell. 2002;111:241–250. doi: 10.1016/s0092-8674(02)01014-0. [DOI] [PubMed] [Google Scholar]
- 30.Mitchell R A, Metz C N, Peng T, Bucala R. J Biol Chem. 1999;274:18100–18106. doi: 10.1074/jbc.274.25.18100. [DOI] [PubMed] [Google Scholar]
- 31.Pei L. J Biol Chem. 2000;275:31191–31198. doi: 10.1074/jbc.M002451200. [DOI] [PubMed] [Google Scholar]
- 32.Pei L. J Biol Chem. 2001;276:8484–8491. doi: 10.1074/jbc.M009654200. [DOI] [PubMed] [Google Scholar]
- 33.Leroy D, Heriche J K, Filhol O, Chambaz E M, Cochet C. J Biol Chem. 1997;272:20820–20827. doi: 10.1074/jbc.272.33.20820. [DOI] [PubMed] [Google Scholar]
- 34.Channavajhala P, Seldin D C. Oncogene. 2002;21:5280–5288. doi: 10.1038/sj.onc.1205640. [DOI] [PubMed] [Google Scholar]
- 35.Wang S S, Esplin E D, Li J L, Huang L, Gazdar A, Minna J, Evans G A. Science. 1998;282:284–287. doi: 10.1126/science.282.5387.284. [DOI] [PubMed] [Google Scholar]
- 36.Sontag E. Cell Signalling. 2001;13:7–16. doi: 10.1016/s0898-6568(00)00123-6. [DOI] [PubMed] [Google Scholar]
- 37.Han M H, Han D K, Aebersold R H, Glomset J A. J Biol Chem. 2001;276:27698–27708. doi: 10.1074/jbc.M101983200. [DOI] [PubMed] [Google Scholar]
- 38.Frank S R, Schroeder M, Fernandez P, Taubert S, Amati B. Genes Dev. 2001;15:2069–2082. doi: 10.1101/gad.906601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Eisenman R N. Genes Dev. 2001;15:2023–2030. doi: 10.1101/gad928101. [DOI] [PubMed] [Google Scholar]
- 40.Park J, Wood M A, Cole M D. Mol Cell Biol. 2002;22:1307–1316. doi: 10.1128/mcb.22.5.1307-1316.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhao K, Wang W, Rando O J, Xue Y, Swiderek K, Kuo A, Crabtree G R. Cell. 1998;95:625–636. doi: 10.1016/s0092-8674(00)81633-5. [DOI] [PubMed] [Google Scholar]
- 42.Watson J D, Oster S K, Shago M, Khosravi F, Penn L Z. J Biol Chem. 2002;277:36921–36930. doi: 10.1074/jbc.M201493200. [DOI] [PubMed] [Google Scholar]
- 43.Won K, Schumacher R J, Farr G W, Horwich A L, Reed S I. Mol Cell Biol. 1998;18:7584–7589. doi: 10.1128/mcb.18.12.7584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Maney T, Hunter A W, Wagenbach M, Wordeman L. J Cell Biol. 1998;142:787–801. doi: 10.1083/jcb.142.3.787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang Z, Yu R, Melmed S. Mol Endocrinol. 2001;15:1870–1879. doi: 10.1210/mend.15.11.0729. [DOI] [PubMed] [Google Scholar]
- 46.Tirone F. J Cell Physiol. 2001;187:155–165. doi: 10.1002/jcp.1062. [DOI] [PubMed] [Google Scholar]
- 47.Peters J-M. Mol Cell. 2002;9:931–943. doi: 10.1016/s1097-2765(02)00540-3. [DOI] [PubMed] [Google Scholar]
- 48.Yokota S, Kayano T, Ohta T, Kurimoto M, Yanagi H, Yura T, Kubota H. Biochem Biophys Res Commun. 2000;279:712–717. doi: 10.1006/bbrc.2000.4011. [DOI] [PubMed] [Google Scholar]
- 49.Sasajima H, Nakagawa K, Yokosawa H. Eur J Biochem. 2002;269:3596–3604. doi: 10.1046/j.1432-1033.2002.03052.x. [DOI] [PubMed] [Google Scholar]
- 50.Tumova S, Woods A, Couchman J R. Int J Biochem Cell Biol. 2000;32:269–288. doi: 10.1016/s1357-2725(99)00116-8. [DOI] [PubMed] [Google Scholar]
- 51.Perreault N, Katz J P, Sackett S D, Kaestner K H. J Biol Chem. 2001;276:43328–43333. doi: 10.1074/jbc.M104366200. [DOI] [PubMed] [Google Scholar]
- 52.Levens D. Proc Natl Acad Sci USA. 2002;99:5757–5799. doi: 10.1073/pnas.102173199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shiio Y, Donohoe S, Yi E C, Goodlet D R, Aebersold R, Eisenman R N. EMBO J. 2002;19:5088–5096. doi: 10.1093/emboj/cdf525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mills J C, Andersson N, Hong C V, Stappenbeck T S, Gordon J I. Proc Natl Acad Sci USA. 2002;99:14819–14824. doi: 10.1073/pnas.192574799. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.