Abstract
Neurofibromatosis type 1 (NF1) is caused by NF1 gene mutations. The phenotype is highly variable, with ‘modifiers' being discussed as potential determinants. Mismatch repair deficiency was shown to cause NF1 mutations, but constitutional mutation of mismatch repair genes was identified only once in a NF1 patient. We aimed to analyze whether DNA methylation of mismatch repair gene promoters, known to lead to transcriptional silencing, is associated with increased tumor load in NF1 defined by the number of cutaneous neurofibromas. Leukocyte DNA of 79 controls and 79 NF1 patients was investigated for methylation of mismatch repair genes MLH1, MSH2, MSH6, and PMS2 by methylation-specific PCR and pyrosequencing. MLH1, MSH6, and PMS2 promoters were not methylated. By contrast, we found promoter methylation of MSH2 with a higher rate of methylation in NF1 patients compared with controls. Furthermore, when comparing NF1 patients with a low vs those with a high number of cutaneous neurofibromas, MSH2 promoter methylation was significantly different. In patients with a high tumor burden, methylation of two (out of six) CpGs was enhanced. This finding was not confounded by age. In conclusion, enhanced methylation involving transcription start points of mismatch repair genes, such as MSH2 in NF1, has not been described so far. Methylation-induced variability of MSH2 gene expression may lead to variable mismatch repair capacity. Our results may point toward a role of MSH2 as a modifier for NF1, although the amount of DNA methylation and subsequent gene expression in other cell types of NF1 patients needs to be elucidated.
Keywords: neurofibromatosis type 1, NF1, mismatch repair, MSH2, neurofibroma, methylation, modifier
Introduction
Neurofibromatosis type 1 (NF1) is a common autosomal dominant disorder with an estimated incidence of 1:3000–1:2500. The major clinical manifestations are café-au-lait spots, axillary and inguinal freckling, multiple dermal and plexiform neurofibromas, and iris Lisch nodules. However, affected individuals may have any of a wide range of additional features including scoliosis, bone abnormalities, short stature, macrocephaly, and learning disabilities.1, 2 NF1 is caused by mutations of the NF1 gene (17q11.2). The gene product neurofibromin is known to act as a tumor suppressor, as loss-of-function mutations are associated with the development of benign and malignant tumors in neural crest-derived tissues3 and some myeloid malignancies.4
The great majority of known mutations are predicted to cause loss-of-function of neurofibromin. Affected individuals are heterozygous for a germline mutation of the NF1 gene. One additional loss-of-function mutation (second hit) in a cell of a susceptible tissue type is required to initiate tumor development (Knudson's two-hit hypothesis). For the typical cutaneous neurofibromas in NF1, it is assumed that every benign tumor shows its individual second NF1 hit, which affects the Schwann cell population.5, 6, 7, 8, 9
The NF1 phenotype is highly variable. Members of a family with NF1 often present with phenotypic variants, although they share an identical mutant allele. Hundreds of different mutations have been reported,10, 11 but there is no evidence for plausible genotype–phenotype correlations, with the exception of patients with microdeletions and c.2970–2972 delAAT mutations in exon 17.12, 13 Therefore, additional modifying factors may explain the clinical diversity and differences in tumor development. Easton et al14 were the first to provide evidence for modifying genes in NF1. Another study proposed modifying genes at unlinked loci or additional environmental factors.15 Thus, there is strong evidence for effects of unlinked modifying genes and linked modifying effects, as well as environmental influences.
No germline-modifying gene has yet been discovered in NF1, although some mechanisms have been proposed. An altered expression of non-linked mismatch repair genes may represent an essential modifying factor for NF1 phenotypic severity. The number of NF1-associated neurofibromas varies to a huge degree in NF1 patients and may be due to a variable amount of accumulating somatic NF1 gene mutations. Two research groups described the role of DNA mismatch repair for neurofibroma development in NF1.9, 16 Both provided evidence that a reduction of the mismatch repair capacity can result in point and small NF1 mutations in a high percentage of neurofibromas. It is interesting to note that patients with a constitutional biallelic mismatch repair gene alteration (called CMMR-D, CCS, MMR-D, or CoLoN) present some NF1 features as well as an early onset of cancer. This constellation points toward somatic NF1 mutations that may accumulate under mismatch repair deficiency.17, 18 From these findings, it can be speculated that early or constitutional alterations of mismatch repair genes in NF1 patients may lead to an altered mismatch repair gene expression, and thus to an accumulation of second hits of the NF1 gene being prone to mutation and showing one of the highest mutation rates known for human genes. Interestingly, beside one very recent report,19 constitutional mutations in human mismatch repair genes could not yet be detected in NF1 patients.8, 16, 18, 19
DNA methylation as an epigenetic modification of CpG dinucleotides is known to modulate gene transcription. CpG methylation can cause transcriptional silencing of many tumor suppressor genes.20 Mismatch repair genes can also be downregulated through methylation of their promoter regions.21, 22, 23 Mismatch repair deficiency is considered to be a modifier in NF1, but constitutional mutations of mismatch repair genes in NF1 patients seem to be very rare.19 Therefore, methylation may be speculated to be a mechanism explaining reduced mismatch repair activity in NF1 patients with a high tumor burden. This would represent an important modifying factor for NF1 phenotypic severity. The aim of this study was, therefore, to analyze if an increased tumor load in NF1, defined by the number of cutaneous neurofibromas, is associated with methylation of mismatch repair genes. First, we analyzed all DNA samples by a well-established and quick method, by methylation-specific PCR (MSP) for methylation of mismatch repair gene promoters most frequently involved in human cancers, namely MLH1, MSH6, PMS2, and MSH2. The findings were validated using a highly sensitive and specific technique, pyrosequencing, for MSH2 promoter methylation.
Materials and methods
Patients and materials
A total of 79 adult and unrelated NF1 patients (29 male and 50 female) were investigated. An intragenic, pathogenic germline NF1 mutation was detected in all but six of these patients. NF1 patients with microdeletions (large deletions of NF1) and patients with mosaic NF1 features were not included in this study. Diagnosis of NF1 was confirmed according to established criteria. Assessment of the number of neurofibromas was always carried out by two clinical geneticists with major NF1 expertise (S.T. and A.B.) during the examination of undressed patients. All cutaneous, subcutaneous, and plexiform neurofibromas of any size, which were visible or palpable by careful physical examination of the patients' skin, were counted and documented. All tumors and other features were marked in detailed drawings (data sheets) allowing the comparison of data from repeated examinations. Every tumor was counted individually up to the number of 100. Higher numbers of neurofibromas were calculated by counting the number within one defined area of the trunk and then projecting it to the area of the whole trunk (front and back trunk were evaluated separately). This method was applied to the trunk only. For all other parts of the body (such as, eg, arms, and legs), the number was assessed exactly. In addition, photographs were taken.
As controls, a total of 79 unrelated healthy non-NF1 probands (40 male and 39 female) were investigated. Owing to limited specimen volumes, we decided to divide the control group randomly into two subgroups. The first was used for MSP (n=39, 20 male and 19 female), whereas the second group (n=40, 20 male and 20 female) was used for pyrosequencing. DNA of peripheral blood samples was extracted by standard procedures and analyzed for methylation.
The study was carried out according to the Declaration of Helsinki and was approved by the local ethical committee. All patients provided written informed consent.
Bisulfite modification of DNA samples
Bisulfite modification of DNA was carried out using the EZ DNA methylation-gold kit (Zymo Research, Orange, CA, USA) following the manufacturer's instructions. Control PCRs using TP53 primers were carried out to check the quality of chemical conversion as described elsewhere.24
MSP
Methylation-specific PCR was carried out using published primer sequences and protocols with minor modifications. PCR reactions were carried out in triplicates (5 min, 95°C; 35 × (95°C, 30 s; annealing for 40 s; 72°C, 30 s); 10 min, 72°C, 4°C). PCR products were checked on a 2% agarose ethidium bromide-stained gel. For MLH1 (U 07418, GenBank) analysis, a 124 bp PCR product was amplified for unmethylated DNA (annealing at 60°C) as well as a 115 bp product for methylated DNA (annealing at 61°C).25 To investigate MSH6 (U 54777 GenBank), a 161 bp PCR product was amplified for unmethylated DNA (annealing at 58°C) and a 151 bp product for methylated DNA (annealing at 60°C).26 PMS2 (NM 000535 GenBank) was analyzed, amplifying a 148 bp PCR product for unmethylated DNA and a 121 bp product for methylated DNA using annealing at 60°C.27 For MSH2 (U 03911 GenBank) analysis, a 143 bp PCR product was amplified for unmethylated DNA (annealing at 62°C) and a 132 bp product for methylated DNA (annealing at 61°C).28
As controls, one sample of a completely methylated DNA (‘control 1', CpG methylated jurkat genomic DNA, New England Biolabs, Hitchin, UK) and one sample of an unmethylated DNA (‘control 2', CpGenome universal unmethylated DNA, Chemicon, Temecula, CA, USA) were amplified together with the samples in each MSP to ensure specificity of MSP, especially to exclude unspecific binding of the respective gene specific primers for methylated and unmethylated PCR products.
Pyrosequencing of MSH2
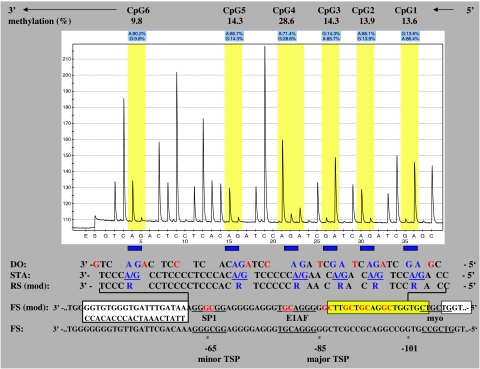
A fragment, spanning −68 nt to −101 nt of the MSH2 promoter and including six single CpG dinucleotides, was analyzed, namely: CpG1 (−98), CpG2 (−94), CpG3 (−91), CpG4 (−87), CpG5 (−80), and CpG6 (−66) (Figure 2). Both transcription start points were covered.29 Primers used to amplify a bisulfite-converted DNA fragment for pyrosequencing were: 5′-TTTGGAAGTTGATTGGGTGTGGT-biotin-3′ (forward) and a 1:1 mixture of four reverse oligonucleotides (5′-CCACTTCTCCCACATACCCTAAAAAAAAC-3′, 5′-CTACTTCTCCCACATACCCTAAAAAAAAC-3′, 5′-CCACTTCTCCTACATACCCTAAAAAAAAC-3′, 5′-CTACTTCTCCTACATACCCTAAAAAAAAC-3′) owing to the presence of two CpG sites within the primer sequence. PCR for bisulfite-treated DNA was carried using a HotStarTaq DNA polymerase kit (Qiagen, Hilden, Germany) following the manufacturer's instructions and 10 pmol of each primer. The cycling conditions were: 95°C 5 min, 45 × (94°C 30 s, 64.1°C 30 s; 72°C 30 s), 72°C 10 min. For pyrosequencing, biotin-labeled DNA strands of the 25 μl PCR product were first prepared using streptavidin-coated sepharose beads and binding buffer according to the manufacturer's instructions (PyroGoldReagens, Biotage, Uppsala, Sweden). The strands were separated using a Vacuum Prep Station (Biotage), and diluted in 40 μl annealing buffer containing the sequencing primer (5′-CCACACCCACTAAACTATT-3′ reverse) (PyroGoldReagens, Biotage). These were subsequently denatured at 85°C for 2 min. After filling the reagent PSQ cartridge with enzyme, substrate and nucleotides, the samples were placed in the pyrosequencer, and sequencing reaction was carried out (PSQ 96MA Biotage). The software PSQ 96 MA 2.1 (Biotage) was used for evaluation of peaks for methylation by SQA analysis. Experiments were replicated for each sample four to seven times and mean values were established.
Data management and statistics for MSH2 methylation
A grading system was introduced for results of MSH2 MSP analysis reflecting the band intensity of MSP products: 0=no band, 1=weak band, and 2=intense to thick band.
The following grading system was applied to the pyrosequencing values: 0–10%, >10–20%, >20–30%, and >30% of methylation.
The NF1 phenotype was defined by the number of cutaneous neurofibromas. NF1 patients were divided into patients with a ‘low number of neurofibromas' (⩽60 neurofibromas) and patients with a ‘high number of neurofibromas' (>100 neurofibromas). A total of 41 NF1 patients had a ‘low number of neurofibromas' ranging from 0 to 60 neurofibromas (15 male and 26 female). Thirty-eight NF1 patients showed a ‘high number of neurofibromas' ranging from 112 to 2200 neurofibromas (14 male and 24 female). The cutoff at >100 was done arbitrarily, but by chance there were no patients with tumor numbers between 61 and 100 in our cohort.
To investigate a possible relation between age and the number of neurofibromas, NF1 patients were divided into three groups (<40 years of age (23–39), n=41; 40–50 years of age, n=25; and >50 years of age (51–71), n=13).
Fisher's exact test and χ2 test were carried out to relate the percentage of patients of different phenotypic groups to the degree of methylation. Intraindividual comparisons of methylation patterns between different CpGs were carried out using κ statistics. A P-value of <0.05 was considered to be statistically significant. All statistical evaluations were carried out using SPSS for Windows 16.0 (SPSS, Munich, Germany).
Results
CpG methylation of MLH1, MSH6, MSH2, and PMS2
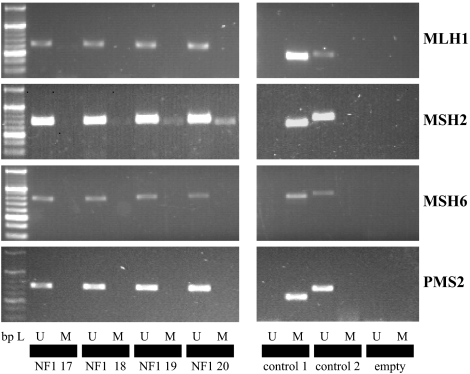
In the first approach, we used MSP to screen the promoter regions of MLH1, MSH6, MSH2, and PMS2 for methylation. We did not find any methylation in the MLH1, MSH6, and PMS2 promoters in blood DNA of both NF1 patients and normal controls (Figure 1). On the contrary, MSH2 was found to be methylated in NF1 patients (59/79) and controls (23/39) (Figure 1). Therefore, we decided to further investigate the MSH2 promoter by a sensitive, specific, and quantitative method that allows a spatial resolution of single CpGs methylated in the region analyzed. By pyrosequencing of bisulfite-modified DNA samples, the same site around the transcription start points of the MSH2 promoter was shown to be methylated (Figure 2). Percentages of methylation of six single CpGs ranged from 2.7 to 39.7% in NF1 patients and from 1.4 to 10.4% in controls.
Figure 1.
Methylation-specific PCR (MSP). Left side: A 20 bp ladder was used as the standard for gel analysis. U – unmethylated (primers to detect unmethylated DNA) and M – methylated (primers to detect methylated DNA). Samples 1–4 (left side) were for NF1 patients 17, 18, 19, and 20. Control reactions for primer specificity (right side): control 1 – a methylated control DNA was amplified with primers for methylated DNA and control 2 – an unmethylated DNA was amplified with primers for unmethylated DNA. Empty – PCR sample without DNA to exclude contamination.
Figure 2.
Example of a pyrogram (eg, sample 3). The percentage of CpG methylation is shown above and was directly taken from pyrogram evaluation. The sequencing direction is reverse (5′ → 3′) because of working with a reverse sequencing primer, whereas the forward strand was labeled with biotin. The sequence analyzed is presented below: dispensation order (DO) is the order of nucleotides pipetted by the pyrosequencer and red nucleotides served as control. CpGs analyzed are given in blue. sequence to analyze (STA) represents the analyzed DNA sequence from −101 nt to –68 nt. RS (mod) is the reverse sequence after bisulfite treatment. FS (mod) stands for forward sequence after bisulfite treatment. The yellow box marks the localization of the forward PCR primer used for methylation-specific PCR (MSP) analysis. The left white box marks the localization of the pyrosequencing primer (reverse). The right white box marks the site of the forward PCR primer for pyrosequencing. Major and minor transcription start points (TSP) as well as transcription factor-binding sites are given according to Iwahashi et al.29
Statistical analysis of MSP results of MSH2
On comparing the methylation levels between patients and control probands, 23/39 (59%) controls and 59/79 (75%) NF1 patients were found to be methylated indicating a non significantly higher percentage of methylation in the NF1 group (P=0.093; Fisher's exact test). Considering the degree of methylation by using the grading system for MSP band intensity, we detected a significant difference between NF1 patients and controls: a higher degree of methylation (grade 2 corresponding to intense up to thick bands) was more frequent in NF1 patients, whereas unmethylated bands were more frequent in controls (Table 1).
Table 1. Methylation of MSH2 promoter (methylation-specific PCR (MSP) analysis).
| Unmethylated grade 0 | Methylated grade 1 | Methylated grade 2 | P a | |
|---|---|---|---|---|
| Controls (n=39) | 16 (41%) | 22 (56.4%) | 1 (2.6%) | 0.001 |
| NF1 patients (n=79) | 20 (25.3%) | 33 (41.7%) | 26 (32.9) |
MSP grading system reflects band intensity of MSP products: 0=no band, 1=weak band, 2=intense to thick band.
χ2 test.
On comparing the methylation levels of NF1 patients with a ‘low' and a ‘high number of neurofibromas,' no significant difference was found (‘low number', methylated: 28/40 (70%); vs ‘high number,' methylated: 31/39 (80%); P=0.44, Fisher's exact test).
Statistical analysis of pyrosequencing values of MSH2
A total of 70 out of the 79 NF1 patients (27 male and 43 female) and 40 controls (20 male and 20 female) were analyzed by pyrosequencing. On comparing the methylation levels between the control group and NF1 patients, we found a significant difference indicating a higher methylation in NF1 patients in four out of six analyzed CpG dinucleotides: CpG1, CpG3, CpG4, and CpG5. Methylation of CpG dinucleotide 4 at −87 nt showed the most distinctive group differences (Table 2).
Table 2. Methylation of MSH2 promoter (pyrosequencing of CpG1–CpG6).
| Methylation <10% | Methylation 10<20% | Methylation 20<30% | Methylation >30% | P a | |
|---|---|---|---|---|---|
| CpG1 | |||||
| Controls | 40 (100%) | 0 | 0 | 0 | 0.001 |
| NF1 patients | 47 (67.1%) | 18 (25.7%) | 1 (1.4%) | 4 (5.7%) | |
| CpG2 | |||||
| Controls | 17 (42.5%) | 23 (57.5%) | 0 | 0 | 0.42 |
| NF1 patients | 39 (55.7%) | 16 (22.9%) | 12 (17.1%) | 3 (4.3%) | |
| CpG3 | |||||
| Controls | 40 (100%) | 0 | 0 | 0 | 0.005 |
| NF1 patients | 55 (78.6%) | 12 (17.1%) | 2 (2.9%) | 1 (1.4%) | |
| CpG4 | |||||
| Controls | 9 (22.5%) | 24 (60%) | 7 (17.5%) | 0 | <0.001 |
| NF1 patients | 2 (2.9%) | 32 (45.7%) | 25 (35.7%) | 11 (15.7%) | |
| CpG5 | |||||
| Controls | 39 (97.5%) | 1 (3.8%) | 0 | 0 | <0.001 |
| NF1 patients | 43 (61.4%) | 25 (35.7 %) | 1 (1.4%) | 1 (1.4%) | |
| CpG6 | |||||
| Controls | 39 (97.5%) | 1 (3.8%) | 0 | 0 | 0.20 |
| NF1 patients | 64 (91.4%) | 5 (7.1%) | 1 (1.4%) | 0 | |
Percentages of methylation were graded into 0–10%, >10–20%, >20–30%, and >30%.
χ2 test.
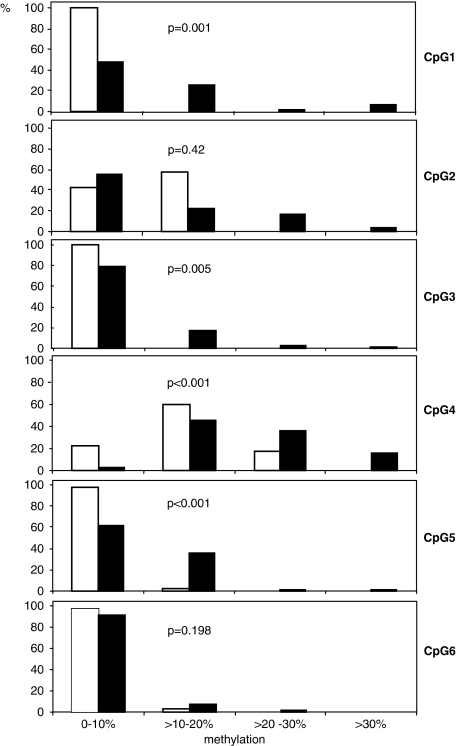
On analyzing the methylation levels among NF1 patients, we found a significant difference of methylation between NF1 patients with a ‘low number of neurofibromas' and a ‘high number of neurofibromas' for CpG3 and CpG4 (P<0.05, χ2 test for linear trend; Figure 3), indicating a higher methylation in the latter group. As neurofibroma manifestation can increase with age, we tested whether this result was confounded by age. On comparing the increasing number of neurofibromas with the increase in the age of patients, a significant correlation was seen (P=0.004, χ2 test for linear trend). But, on comparing the degree of methylation (0–10%, >10–20%, >20–30%, and >30%) of CpG3 as well as of CpG4 with increasing age, no significant correlation was seen (P=0.99 for CpG3, P=0.89 for CpG4; χ2 test for linear trend), indicating that age did not confound the relationship between the ‘high number of neurofibromas' and the higher methylation of CpG3 and CpG4. In addition, we investigated whether these relationships were also present in the younger NF1 patients (<40 years of age). To do so, we grouped them into three age groups (23–30 years of age, 31–35 years, and 36–40 years). Patient's age was significantly positively correlated to the number of neurofibromas (P=0.03, χ2 for linear trend). However, neither methylation of CpG 3 (P=0.57) nor methylation of CpG4 (P=0.79) showed a significant relationship to age of younger patients.
Figure 3.
Statistical analysis of MSH2 methylation (CpG1–CpG6) achieved by pyrosequencing comparing NF1 patients with a ‘low number of neurofibromas' (white bars) and a ‘high number of neurofibromas' (black bars). The proportions (%) of affected patients (n=70) are given. Methylation was graded into four ranks (0–10%, >10–20%, >20–30%, and >30%). A pathogenic germline NF1 mutation was detected in 31 out of 35 NF1 patients with a ‘low number of neurofibromas' and in 33 out of 35 NF1 patients with a ‘high number of neurofibromas' (4 out of 6 patients without detected NF1 mutations had parents with NF1). P-values are given for each CpG analysis.
As apparent from Figure 3, CpG4 (−87 nt) showed a methylation pattern that was different from those of the other CpGs. In most patients and all controls, this CpG showed higher methylation levels than the others. To obtain a quantitative measure of this difference, we calculated κ coefficients for concordance of methylation (⩽20 vs >20%). Comparisons between CpG1, CpG2, CpG3, and CpG5 in NF1 patients mostly revealed κ coefficients ranging from 0.10–0.79, (P<0.05). This indicates a significant concordance of the methylation pattern between these CpG sites. By contrast, a comparison of CpG4 with the other CpG sites revealed mostly κ coefficients ranging from 0.03–0.14 (P>0.05). This underlines that CpG4 shows no concordance of its methylation pattern with the other CpGs within the MSH2 promoter.
Discussion
The aim of our study was to analyze whether methylation of mismatch repair gene promoters is associated with an increased tumor load in NF1, as defined by the number of neurofibromas. Promoter methylation is known to function as an epimutation leading to gene silencing or decreased gene expression.20 For the first time, we could provide evidence that a promoter of a mismatch repair gene (MSH2) is significantly more methylated in NF1 patients compared with healthy persons by two different approaches. Furthermore, by applying a highly sensitive and quantitative method, we detected enhanced methylation of at least two CpG dinucleotides positively correlating with a ‘high number of neurofibromas' in NF1 patients. Provided that similar epigenetic modifications exist in NF1 Schwann cells, MSH2 could be discussed as a modifier for NF1 phenotypic severity, especially for excessive neurofibroma manifestation due to accumulating second hits. Such a role of MSH2 is supported by the finding of an MSH2 variant (p.G322D) that is present at a higher frequency among NF1 patients with multiple neurofibromas and an intragenic NF1 mutation compared with a control population.8 G322D is associated with a reduced efficiency of mismatch repair in the Saccharomyces cerevisiae model system, which can be interpreted as supporting the hypothesis that mismatch repair defects in MSH2 increase the likelihood of NF1 patients developing neurofibromas.8 As constitutional mutations of MSH2 have not yet been detected in NF1 patients in previous studies,8, 16 epigenetic alterations such as MSH2 promoter methylation could offer an alternative explanation for mismatch repair deficiency in NF1.
Our study is limited to DNA methylation in leukocytes. Therefore, our findings provide only suggestive evidence of a role for MSH2 in modifying the NF1 phenotype that will require further confirmation by investigations, such as MSH2 expression analysis in Schwann cells and other tissues of NF1 patients. Nevertheless, if these data will be reproducible for DNA from leukocytes in other patient cohorts, methylation of MSH2 in blood cells could serve as a marker in a presumable diagnostic blood test to evaluate the risk for a predicted higher tumor burden in NF1. This could have clinical implications for the management of NF1 patients.
To our knowledge, this is the first description of MSH2 methylation in DNA from leukocytes in normal persons. Therefore, it is essential to figure out in further studies at what values of MSH2 methylation mismatch repair capacity is affected in leukocytes and other tissues. Gutmann et al 30had already shown that MLH1 deficiency accelerated leukemogenesis in NF1 transgenic mice pointing toward a potential role of mismatch repair deficiency for NF1-associated malignancies such as leukemia.
Recently, an NF1 germline mutation (p.R1241X) was found in a child also carrying a homozygous MLH1 germline mutation, who developed atypical chronic myeloid leukemia at 12 months of age as well as café-au-lait spots and skin tumors.19 This seems to be the first case describing the co-occurrence of a constitutional NF1 germline mutation and a constitutional mismatch repair defect. Thus far, all other cases carrying mismatch repair gene mutations and presenting NF1 features were not found to carry NF1 germline mutations, although it is highly suggestive that they carry somatic NF1 gene mutations in certain cells giving rise to the early onset of cancers such as hematological malignancies and brain tumors.18 Wimmer and Etzler (2008) introduced the term CMMR-D syndrome for this phenotype and proposed that NF1 features in this syndrome are probably explained by mosaic NF1 mutations at an early developmental stage. The question arises whether other gene alterations than those seen in the biallelic inheritance of mismatch repair gene defects, such as in the CMMR-D syndrome, may constitute a modifier for the diverse phenotypes and tumor burdens in NF1 patients carrying a constitutional NF1 mutation.
One essential question is, if the MSH2 methylation we observed is a constitutional modification or is a secondary phenomenon developing during life. One recent report described the identification of a hereditary epimutation of MSH2.31 This might favor the inheritance of methylation at this site of MSH2 in NF1 patients.
Promoter methylation of MSH2 is a major contributor to mismatch repair deficiency in different types of cancer such as HNPCC and breast cancer. Naqvi et al32 studied the methylation of MSH2 genes in Indian female patients with breast cancer. Primers used for MSP in their study were identical to those used in our study. Those authors showed that the methylation of this MSH2 site is associated with undetectable protein and RNA expression in at least some specimens.
Only some of the CpGs investigated in our study showed methylation levels leading to significant differences between NF1 patients with ‘low' and ‘high numbers of neurofibromas' (CpG3 at −91 nt and CpG4 at −87 nt), as well as between NF1 patients compared with controls (by MSP and by pyrosequencing for CpG1 at −98 nt, CpG3 at −91 nt, CpG4 at −87 nt, and CpG5 at −80 nt). This may raise the question of a differential biological importance of individual CpG dinucleotides: for example, methylation-sensitive transcription factor binding at certain conserved sites or regulatory elements. Several binding sites such as for E1AF, an ETS oncogene family transcription factor, were found within the region we analyzed.29 The E1AF binding site was affected by significant differences in methylation of CpG5 at −80 nt between NF1 patients and controls. In fact, previous luciferase transporter assays pointed toward an 82% decrease of promoter activity when −80 nt was altered by mutation.33 This −80 Å insertion allele was described in HNPCC patients and showed novel transcription factor-binding abilities, which was interpreted either due to the presence of the transcription start point at −85 nt or due to a specific DNA-protein interaction between the −80 Å insertion oligomer and transcription factors.33 Whether E1AF binding is methylation sensitive because of variable methylation levels of its central CpG (CpG5 in our study) has not been investigated so far. Furthermore, another very close element was affected by variable methylation levels in our study: differential methylation appeared at CpG4 by comparing controls and NF1 patients, as well as by comparing NF1 patients with a ‘low number' and a ‘high number of neurofibromas'. CpG4 (−87 nt) is localized very close to the major transcription start point (−85 nt) of MSH2 and is located in a CGG. The probability that methylation of such an important site for gene transcription within a CpG island is relevant would be high, but needs further investigation.
The authors would like to admit that the definition of the NF1 phenotype by counting cutaneous neurofibromas may be limited. Volumetric measures and assessment of the internal tumor load by MRI would be more desirable. However, besides ethical problems, in most studies it is still not feasible to obtain data from a sufficient number of patients using such expensive and time-consuming methods.
To conclude, we found evidence of enhanced MSH2 promoter methylation close to the transcription start points in NF1 blood leukocytes. Differential methylation of MSH2 may lead to a variable MSH2 expression and therefore, to variable mismatch repair deficiency. Provided that similar epigenetic modifications exist in NF1 Schwann cells, MSH2 could be regarded as a modifier for NF1 phenotypic severity. In our opinion, it is also not yet ruled out that mutations of mismatch repair genes are relevant as modifiers for NF1. These genetic alterations may represent a spectrum other than the one seen in HNPCC and CMMR-D patients.
The identification of genetic modifiers would offer clues to the molecular pathogenesis of NF1 and indicates promising avenues of research for developing novel treatments.
Conflict of interest
The authors declare no conflict of interest.
Acknowledgments
The authors gratefully acknowledge the help of Monika Reißmann (Department of Animal Sciences, Institute of Agriculture and Horticulture, Humboldt University Berlin). AH holds a Rahel-Hirsch Fellowship of the Charité. This work was supported by a grant from the Berliner Krebsgesellschaft (HAFF200809) to AH.
References
- Gutmann DH, Aylsworth A, Carey JC, et al. The diagnostic evaluation and multidisciplinary management of neurofibromatosis 1 and neurofibromatosis 2. JAMA. 1997;278:51–57. [PubMed] [Google Scholar]
- Huson SM.Neurofibromatoss 1: a clinical and genetic overviewin Huson SM, Hughes RAC (eds): The Neurofibromatoses: A Pathogenetic and Clinical Overview London: Chapman and Hall; 1994160–203. [Google Scholar]
- Colman SD, Williams CA, Wallace MR. Benign neurofibromas in type 1 neurofibromatosis (NF1) show somatic deletions of the NF1 gene. Nat Genet. 1995;11:90–92. doi: 10.1038/ng0995-90. [DOI] [PubMed] [Google Scholar]
- Shannon KM, O'Connell P, Martin GA, et al. Loss of the normal NF1 allele from the bone marrow of children with type 1 neurofibromatosis and malignant myeloid disorders. N Engl J Med. 1994;330:597–601. doi: 10.1056/NEJM199403033300903. [DOI] [PubMed] [Google Scholar]
- Serra E, Rosenbaum T, Winner U, et al. Schwann cells harbor the somatic NF1 mutation in neurofibromas: evidence of two different Schwann cell subpopulations. Hum Mol Genet. 2000;9:3055–3064. doi: 10.1093/hmg/9.20.3055. [DOI] [PubMed] [Google Scholar]
- Rutkowski JL, Wu K, Gutmann DH, Boyer PJ, Legius E. Genetic and cellular defects contributing to benign tumor formation in neurofibromatosis type 1. Hum Mol Genet. 2000;9:1059–1066. doi: 10.1093/hmg/9.7.1059. [DOI] [PubMed] [Google Scholar]
- Kluwe L, Friedrich R, Mautner VF. Loss of NF1 allele in Schwann cells but not in fibroblasts derived from an NF1-associated neurofibroma. Genes Chromosomes Cancer. 1999;24:283–285. doi: 10.1002/(sici)1098-2264(199903)24:3<283::aid-gcc15>3.0.co;2-k. [DOI] [PubMed] [Google Scholar]
- Maertens O, Brems H, Vandesompele J, et al. Comprehensive NF1 screening on cultured Schwann cells from neurofibromas. Hum Mutat. 2006;27:1030–1040. doi: 10.1002/humu.20389. [DOI] [PubMed] [Google Scholar]
- Wiest V, Eisenbarth I, Schmegner C, Krone W, Assum G. Somatic NF1 mutation spectra in a family with neurofibromatosis type 1: toward a theory of genetic modifiers. Hum Mutat. 2003;22:423–427. doi: 10.1002/humu.10272. [DOI] [PubMed] [Google Scholar]
- Fahsold R, Hoffmeyer S, Mischung C, et al. Minor lesion mutational spectrum of the entire NF1 gene does not explain its high mutability but points to a functional domain upstream of the GAP-related domain. Am J Hum Genet. 2000;66:790–818. doi: 10.1086/302809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messiaen LM, Callens T, Mortier G, et al. Exhaustive mutation analysis of the NF1 gene allows identification of 95% of mutations and reveals a high frequency of unusual splicing defects. Hum Mutat. 2000;15:541–555. doi: 10.1002/1098-1004(200006)15:6<541::AID-HUMU6>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- Tinschert S.Clinical phenotypes in patients with microdeletionsin Kaufmann D (ed): Neurofibromatoses Basel, Switzerland: Karger; 2008. Vol. 16, 78–88. [Google Scholar]
- Upadhyaya M, Huson SM, Davies M, et al. An absence of cutaneous neurofibromas associated with a 3-bp inframe deletion in exon 17 of the NF1 gene (c.2970–2972 delAAT): evidence of a clinically significant NF1 genotype-phenotype correlation. Am J Hum Genet. 2007;80:140–151. doi: 10.1086/510781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Easton DF, Ponder MA, Huson SM, Ponder BA. An analysis of variation in expression of neurofibromatosis (NF) type 1 (NF1): evidence for modifying genes. Am J Hum Genet. 1993;53:305–313. [PMC free article] [PubMed] [Google Scholar]
- Szudek J, Joe H, Friedman JM. Analysis of intrafamilial phenotypic variation in neurofibromatosis 1 (NF1) Genet Epidemiol. 2002;23:150–164. doi: 10.1002/gepi.1129. [DOI] [PubMed] [Google Scholar]
- Wang Q, Montmain G, Ruano E, et al. Neurofibromatosis type 1 gene as a mutational target in a mismatch repair-deficient cell type. Hum Genet. 2003;112:117–123. doi: 10.1007/s00439-002-0858-4. [DOI] [PubMed] [Google Scholar]
- Kruger S, Kinzel M, Walldorf C, et al. Homozygous PMS2 germline mutations in two families with early-onset haematological malignancy, brain tumours, HNPCC-associated tumours, and signs of neurofibromatosis type 1. Eur J Hum Genet. 2008;16:62–72. doi: 10.1038/sj.ejhg.5201923. [DOI] [PubMed] [Google Scholar]
- Wimmer K, Etzler J. Constitutional mismatch repair-deficiency syndrome: have we so far seen only the tip of an iceberg. Hum Genet. 2008;124:105–122. doi: 10.1007/s00439-008-0542-4. [DOI] [PubMed] [Google Scholar]
- Alotaibi H, Ricciardone MD, Ozturk M. Homozygosity at variant MLH1 can lead to secondary mutation in NF1, neurofibromatosis type I and early onset leukemia. Mutat Res. 2008;637:209–214. doi: 10.1016/j.mrfmmm.2007.08.003. [DOI] [PubMed] [Google Scholar]
- Esteller M. Epigenetics in cancer. N Engl J Med. 2008;358:1148–1159. doi: 10.1056/NEJMra072067. [DOI] [PubMed] [Google Scholar]
- Noda H, Kato Y, Yoshikawa H, et al. Microsatellite instability caused by hMLH1 promoter methylation increases with tumor progression in right-sided sporadic colorectal cancer. Oncology. 2005;69:354–362. doi: 10.1159/000089768. [DOI] [PubMed] [Google Scholar]
- Tzao C, Hsu HS, Sun GH, et al. Promoter methylation of the hMLH1 gene and protein expression of human mutL homolog 1 and human mutS homolog 2 in resected esophageal squamous cell carcinoma. J Thorac Cardiovasc Surg. 2005;130:1371. doi: 10.1016/j.jtcvs.2005.06.004. [DOI] [PubMed] [Google Scholar]
- Hsu HS, Wen CK, Tang YA, et al. Promoter hypermethylation is the predominant mechanism in hMLH1 and hMSH2 deregulation and is a poor prognostic factor in nonsmoking lung cancer. Clin Cancer Res. 2005;11:5410–5416. doi: 10.1158/1078-0432.CCR-05-0601. [DOI] [PubMed] [Google Scholar]
- Harder A, Rosche M, Reuss DE, et al. Methylation analysis of the neurofibromatosis type 1 (NF1) promoter in peripheral nerve sheath tumours. Eur J Cancer. 2004;40:2820–2828. doi: 10.1016/j.ejca.2004.07.021. [DOI] [PubMed] [Google Scholar]
- Fleisher AS, Esteller M, Wang S, et al. Hypermethylation of the hMLH1 gene promoter in human gastric cancers with microsatellite instability. Cancer Res. 1999;59:1090–1095. [PubMed] [Google Scholar]
- Goldberg M, Rummelt C, Foja S, Holbach LM, Ballhausen WG. Different genetic pathways in the development of periocular sebaceous gland carcinomas in presumptive Muir-Torre syndrome patients. Hum Mutat. 2006;27:155–162. doi: 10.1002/humu.20281. [DOI] [PubMed] [Google Scholar]
- Truninger K, Menigatti M, Luz J, et al. Immunohistochemical analysis reveals high frequency of PMS2 defects in colorectal cancer. Gastroenterology. 2005;128:1160–1171. doi: 10.1053/j.gastro.2005.01.056. [DOI] [PubMed] [Google Scholar]
- Fang JY, Lu R, Mikovits JA, Cheng ZH, Zhu HY, Chen YX. Regulation of hMSH2 and hMLH1 expression in the human colon cancer cell line SW1116 by DNA methyltransferase 1. Cancer Lett. 2006;233:124–130. doi: 10.1016/j.canlet.2005.03.005. [DOI] [PubMed] [Google Scholar]
- Iwahashi Y, Ito E, Yanagisawa Y, et al. Promoter analysis of the human mismatch repair gene hMSH2. Gene. 1998;213:141–147. doi: 10.1016/s0378-1119(98)00187-5. [DOI] [PubMed] [Google Scholar]
- Gutmann DH, Winkeler E, Kabbarah O, et al. Mlh1 deficiency accelerates myeloid leukemogenesis in neurofibromatosis 1 (Nf1) heterozygous mice. Oncogene. 2003;22:4581–4585. doi: 10.1038/sj.onc.1206768. [DOI] [PubMed] [Google Scholar]
- Chan TL, Yuen ST, Kong CK, et al. Heritable germline epimutation of MSH2 in a family with hereditary nonpolyposis colorectal cancer. Nat Genet. 2006;38:1178–1183. doi: 10.1038/ng1866. [DOI] [PubMed] [Google Scholar]
- Naqvi RA, Hussain A, Deo SS, et al. Hypermethylation analysis of mismatch repair genes (hmlh1 and hmsh2) in locally advanced breast cancers in Indian women. Hum Pathol. 2008;39:672–680. doi: 10.1016/j.humpath.2007.09.011. [DOI] [PubMed] [Google Scholar]
- Shin KH, Shin JH, Kim JH, Park JG. Mutational analysis of promoters of mismatch repair genes hMSH2 and hMLH1 in hereditary nonpolyposis colorectal cancer and early onset colorectal cancer patients: identification of three novel germ-line mutations in promoter of the hMSH2 gene. Cancer Res. 2002;62:38–42. [PubMed] [Google Scholar]