Abstract
The association between common variants in the FTO gene with weight, adiposity and body mass index (BMI) has now been widely replicated. Although the causal variant has yet to be identified, it most likely maps within a 47 kb region of intron 1 of FTO. We performed a genome-wide association study in the Sorbian population and evaluated the relationships between FTO variants and BMI and fat mass in this isolate of Slavonic origin resident in Germany. In a sample of 948 Sorbs, we could replicate the earlier reported associations of intron 1 SNPs with BMI (eg, P-value=0.003, β=0.02 for rs8050136). However, using genome-wide association data, we also detected a second independent signal mapping to a region in intron 2/3 about 40–60 kb away from the originally reported SNPs (eg, for rs17818902 association with BMI P-value=0.0006, β=−0.03 and with fat mass P-value=0.0018, β=−0.079). Both signals remain independently associated in the conditioned analyses. In conclusion, we extend the evidence that FTO variants are associated with BMI by putatively identifying a second susceptibility allele independent of that described earlier. Although further statistical analysis of these findings is hampered by the finite size of the Sorbian isolate, these findings should encourage other groups to seek alternative susceptibility variants within FTO (and other established susceptibility loci) using the opportunities afforded by analyses in populations with divergent mutational and/or demographic histories.
Keywords: FTO, BMI, Sorbs
Introduction
Common variants in the FTO gene have repeatedly been shown to be associated with body mass index (BMI) and fat mass in populations of European descent1, 2, 3 and recently also in Asian populations.4, 5, 6, 7 FTO is therefore considered to be the first replicated signal at which common variants contribute to common obesity,8 although details of the molecular and physiological processes involved remain unclear.
On the basis of the available genome-wide and fine-mapping association data, the associated SNPs (and therefore, by implication, the still unidentified causal variant) map to a 47-kb region, bounded by flanking recombination hotspots, mainly in intron 1. It has been shown that the FTO protein shares sequence motifs with Fe(II)- and 2-oxoglutarate-dependent oxygenases.9 FTO mRNA is widely expressed in all tissues but particularly highly in hypothalamic nuclei generally implicated in energy balance and feeding behaviour.10 In mice, levels of FTO in the arcuate nucleus are regulated by feeding and fasting.9, 11 Despite this accumulating evidence, it remains to be established that FTO is the causal gene. For example, the similarity of expression profiles between FTO and the adjacent gene RPGRIP1L (KIAA1005, FTM) may indicate joint transcriptional regulation.12 The product of RPGRIP1L is involved in ciliary function.13, 14 Although this raises parallels with the Bardet–Biedl obesity syndrome12, 15, 16 (which is known to result from mutations in genes with similar roles), severe loss-of-function mutations in FTM result in Joubert syndrome type B and Meckel syndrome, but do not cause serious disturbances of body weight.13 FTO variants seem to influence body weight by modulation of food intake rather than energy expenditure.17, 18, 19 Effects on glucose metabolism independent of obesity have yet to be shown.20, 21, 22
Identification of a wider spectrum of susceptibility alleles can be a valuable tool in efforts to relate DNA sequence variation to clinical phenotypes,23, 24 and to this end, studies of populations with a divergent mutational and/or demographic history (including those from a different ethnic background or defined population isolates) offer particular advantages. Analysis of such samples provides an opportunity to detect novel allelic or haplotypic associations and may offer valuable clues to additional, independent causal variants as well as helping to map earlier described variants.
We performed a genome-wide scan for traits related to the metabolic syndrome in the Sorbs, a self-contained population of Slavonic origin resident in Germany. We specifically focus the analyses to effects of FTO variants on BMI in this population and report findings that suggest possible allelic heterogeneity across populations.
Materials and methods
Subjects and phenotyping
All subjects are part of a sample from an extensively phenotyped self-contained population from Eastern Germany, the Sorbs. The Sorbs are of Slavonic origin, and lived in ethnic isolation among the Germanic majority during the past 1100 years. Today, the Sorbian-speaking, Catholic minority comprises ∼15 000 full-blooded Sorbs resident in about 10 villages in rural Upper Lusatia (Oberlausitz), Eastern Saxony. This historical evidence establishing the Sorbs as a genetic isolate is substantiated by extended homozygosity and differences in allele frequency distribution compared with an outbred population.
At present, about 1000 Sorbian individuals are enrolled in the study. Sampling comprised unrelated subjects as well as families. Mean IBD sharing in the pairwise comparison was 0.008, median <10−6 (25th percentile <10−6, 75th percentile: 0.012). Extensive phenotyping included standardised questionnaires for past medical history and family history, collection of anthropometric data (weight, height, waist-to-hip-ratio (WHR), body impedance analysis (BIA)) and a 75-g glucose-tolerance-test (OGTT). BIA was performed with BIA-2000-S (Data Input GmbH, Darmstadt, Germany) and evaluated with the software Nutri3 (Data Input GmbH). A total of 948 subjects were available for this study, which was approved by the ethics committee of the University of Leipzig and all subjects gave the written informed consent before taking part in the study.
For comparison of allele and haplotype frequencies of the FTO region, we used the WTCCC T2D case set.25, 26 For the evaluation of linkage disequilibrium (LD) patterns, we selected 427 subjects with IBD sharing <0.15 from the Sorbs sample and compared them with 60 CEU HapMap samples.27
DNA extraction, genotyping and SNP selection
Genomic DNA was extracted using QIAmp DNA Blood Midi Kit (Qiagen Inc., Valencia, CA, USA) according to the manufacturer's protocol. Genotyping was performed using the 500 K Affymetrix GeneChip and the Affymetrix Genome-Wide Human SNP Array 6.0 (Affymetrix Inc., Santa Clara, CA, USA) by the Microarray Core Facility of the Interdisciplinary Centre for Clinical Research, University of Leipzig, Germany, and by ATLAS Biolabs GmbH, Berlin, Germany. Genotypes were determined with GeneChip Genotyping Analysis Software (GTYPE, Affymetrix Inc.) using the BRLMM algorithm for the 500 K arrays and the Birdseed Algorithm for Genome-Wide Human SNP Array 6.0 (Affymetrix Inc.). Only arrays with call rates >93% were selected for further analyses and only SNPs fulfilling the following criteria were included: missing rate per SNP <5%, Hardy–Weinberg equilibrium (HWE) P>0.0001 and minor allele frequency (MAF) >0.01. The average genotyping rate was 99%. In all, 390619 (380408 autosomal, 10211 X-chromosomal) markers overlapping between the 500 K Affymetrix GeneChip and the Affymetrix Genome-Wide Human SNP Array 6.0 were included in the analyses. All alleles were standardised to the forward strand.
Statistical methods and software
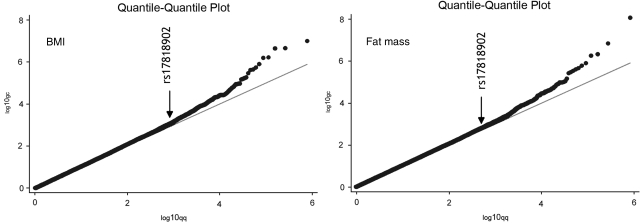
Call rates per sample and cluster-plots were checked using the GeneChip Genotyping Analysis Software (GTYPE). The calculation of minor allele frequencies, HWE, and missing rates per SNP was performed with PLINK.28 Pairwise IBD estimation was performed with the --genome command in PLINK. To perform multidimensional scaling analysis on the N × N matrix of genome-wide IBS pairwise distances, we used the --mds-plot option in conjunction with --cluster.28 BMI and fat mass were non-normally distributed and therefore ln-transformed before analysis and genome-wide association was assessed by linear regression in PLINK. We corrected for age and gender and furthermore for cryptic relatedness and possible population substructure by using genomic control (λ=1.16 for BMI and 1.18 for fat mass) and by using the first four vectors of multiple dimensional scaling as a covariate. We examined the distribution of test statistics and the deviation from the expected distribution under the null hypothesis of no association in a quantile–quantile plot (Figure 1). Out of the 390619 SNPs that passed, QC 22 showed evidence of association with BMI and fat mass on a significance level of P<0.0001 for both traits simultaneously in the Sorbian population (Table 4). Furthermore, we assessed the associations of FTO-SNPs with BMI in a subset of subjects with IBD sharing <0.15 and in a subset of non-diabetic individuals (according to the current ADA criteria29).
Figure 1.
QQ-plots for association with BMI and fat mass in the Sorbian population. rs17818902 represents the position of the SNP with the strongest association with each trait.
To analyse haplotypic association with BMI, we applied the GENEBPM algorithm30 in a sliding window of five overlapping SNPs across the region flanking FTO. The method was originally developed to test for association between a binary disease phenotype and SNP haplotypes, but has been adapted here to model association with a quantitative trait in a linear regression framework. The strength of evidence in favour of the association of haplotypes within each window with BMI was assessed by means of Bayes factor. For a direct comparison of the Bayes factors of the single point and haplotype-based analyses, we also applied the GENEBPM algorithm to each SNP alone in a single-locus analysis of the region.
Conditional analyses were conducted in a similar manner with inclusion of the conditioned SNP as an additional covariate. Linkage disequilibrium metrics were calculated in Haploview 4.1.31 Power calculations were performed in Quanto 1.1.32, 33 Given the effect size of rs8050136 in the WTCCC T2D case set (β=0.012), we would have a power of 30% to detect an effect on BMI (additive model) at an α of 0.05 in the whole Sorbian sample set. Allowing for the more stringent genome-wide α of 5 × 10−8, we would need a sample size of about 17 800 subjects to achieve a power of 80% to detect an effect of rs8050136 on BMI under the additive model.
Results
Genome-wide association with BMI and fat mass in 948 subjects
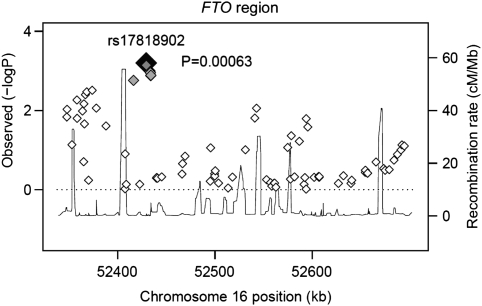
There were 27 SNPs available in introns 1, 2 and 3 of the FTO region in our data set. Seven SNPs out of these gave modest evidence (P<0.01) of association with BMI within the earlier reported 47-kb region of introns 1 and 2 (Table 1, eg, rs1861868: P-value=0.009; rs8050136: P-value=0.003; rs17817288: P-value=0.007). Furthermore, in contrast to the earlier reported data, another eight SNPs that map downstream of this region in introns 2 and 3 showed moderate effects on BMI (Figure 2, Table 1, eg, rs10852522: P-value=0.002; rs17818902: P-value=0.0006; rs8053367: P-value=0.001). In addition, the SNPs in the region between 52.41 and 52.43 Mb had consistent effects on fat mass (eg, rs10852522: P-value=0.01; rs17818902: P-value=0.002; rs8053367: P-value=0.007), whereas the presented SNPs in intron 1 showed a trend but no effects on the significance level of 0.05 (Table 1). The independent effect of both signals was confirmed by the conditional analyses (Table 2), in which adjustment for SNPs in the other cluster resulted in modest attenuation but not obliteration of the signal. The effects on BMI also remained after exclusion of closely related subjects. In a subset of 427 individuals with IBD sharing <0.15 rs8050136 (P-value=0.0029) and rs17818902 (P-value=0.0089) likewise showed modest evidence of association with BMI. Furthermore, we evaluated the effect of the FTO-SNPs in a subset of 802 non-diabetic subjects (λ=1.14). Again, the results were consistent with the effects in the whole Sorbian sample (rs8050136: P-value=0.014 and rs17818902: P-value=0.0029).
Table 1. Association of SNPs in FTO-introns 1–3 with BMI and fat mass in the Sorbs.
| BMI | Fat mass | |||||||
|---|---|---|---|---|---|---|---|---|
| CHR | SNP | BP | MAF | A1 | P-value | β | P-value | β |
| Intron 1 | ||||||||
| 16 | rs1861869 | 52 347 682 | 0.497 | G | 0.0147 | 0.020 | 0.1742 | 0.027 |
| 16 | rs1861868 | 52 347 903 | 0.475 | T | 0.0094 | 0.020 | 0.1391 | 0.029 |
| 16 | rs9940700 | 52 352 910 | 0.191 | C | 0.0735 | −0.018 | 0.7145 | −0.009 |
| 16 | rs9939973 | 52 358 069 | 0.454 | A | 0.0054 | 0.022 | 0.1243 | 0.030 |
| 16 | rs9940128 | 52 358 255 | 0.461 | A | 0.0158 | 0.019 | 0.2208 | 0.024 |
| 16 | rs9922047 | 52 363 781 | 0.429 | C | 0.0104 | −0.021 | 0.1029 | −0.033 |
| 16 | rs16952522 | 52 364 999 | 0.039 | G | 0.1976 | 0.026 | 0.6636 | 0.022 |
| 16 | rs17817288 | 52 365 265 | 0.441 | A | 0.0068 | −0.022 | 0.0657 | −0.037 |
| 16 | rs1477196 | 52 365 759 | 0.341 | A | 0.0218 | −0.019 | 0.0182 | −0.049 |
| 16 | rs1121980 | 52 366 748 | 0.452 | A | 0.0040 | 0.023 | 0.0888 | 0.033 |
| 16 | rs7193144 | 52 368 187 | 0.422 | C | 0.0034 | 0.024 | 0.0562 | 0.038 |
| 16 | rs16945088 | 52 370 025 | 0.091 | G | 0.5796 | −0.007 | 0.6606 | −0.015 |
| 16 | rs8050136 | 52 373 776 | 0.428 | A | 0.0031 | 0.023 | 0.0574 | 0.038 |
| 16 | rs9939609 | 52 378 028 | 0.418 | A | 0.0087 | 0.021 | 0.0748 | 0.036 |
| 16 | rs9930506 | 52 387 966 | 0.462 | G | 0.0247 | 0.017 | 0.2296 | 0.023 |
| Intron 2 | ||||||||
| 16 | rs11075994 | 52 407 580 | 0.351 | A | 0.9537 | 0.001 | 0.9977 | 6e−05 |
| 16 | rs1421090 | 52 407 671 | 0.190 | G | 0.1254 | −0.015 | 0.0544 | −0.048 |
| 16 | rs9972717 | 52 408 805 | 0.156 | A | 0.7354 | −0.004 | 0.9985 | 5e−05 |
| 16 | rs10852522 | 52 416 278 | 0.359 | A | 0.0017 | −0.025 | 0.0092 | −0.053 |
| Intron 3 | ||||||||
| 16 | rs10521308 | 52 422 627 | 0.067 | A | 0.7355 | 0.005 | 0.4853 | 0.027 |
| 16 | rs17818902 | 52 429 307 | 0.182 | G | 0.0006 | −0.035 | 0.0018 | −0.079 |
| 16 | rs17818920 | 52 429 404 | 0.182 | C | 0.0007 | −0.034 | 0.0017 | −0.079 |
| 16 | rs8053367 | 52 432 985 | 0.392 | T | 0.0010 | −0.027 | 0.0070 | −0.056 |
| 16 | rs8053740 | 52 433 213 | 0.400 | C | 0.0010 | −0.026 | 0.0109 | −0.051 |
| 16 | rs7203051 | 52 433 650 | 0.401 | C | 0.0011 | −0.026 | 0.0120 | −0.050 |
| 16 | rs7205009 | 52 433 945 | 0.402 | T | 0.0013 | −0.026 | 0.0132 | −0.050 |
| 16 | rs7205213 | 52 434 067 | 0.403 | T | 0.0013 | −0.026 | 0.0099 | −0.051 |
27 Affymetrix-GeneArray-SNPs within introns 1, 2 and 3 of the FTO-gene, which passed QC in the Sorbs. All positions (BP) are based on NCBI build 35. P-values were calculated under the additive model of inheritance and adjusted for genomic control factor λ (=1.16 for BMI and =1.18 for fat mass), age, gender and first four vectors of MDS.
Reference allele is the minor allele, which is indicated as A1. The minor allele frequency is given as MAF.
Figure 2.
Associations of SNPs in FTO with BMI in the Sorbs. Positions are based on NCBI Build 35. Estimated recombination rates (HapMap27) reflect the local LD structure around the associated SNPs and their correlated proxies. The SNP with the lowest P-value for association with BMI (rs17818902) is given as a black diamond, correlated SNPs according to their LDs in a scale from grey (r2=1) to white (r2=0) based on the pairwise r2 values estimated in the Sorbian population.
Table 2. Conditioned analyses for rs8050136 and rs17818902.
| SNP | N | β | P | P adj BMI |
|---|---|---|---|---|
| BMI (kg/m2) | ||||
| rs8050136 | 937 | 0.021 | 0.0036 | |
| rs17818902 | 937 | −0.033 | 0.0006 | |
| Fat mass (%) | ||||
| rs8050136 | 937 | 0.033 | 0.0723 | 0.1359 |
| rs17818902 | 937 | −0.076 | 0.0011 | 0.3998 |
| Waist (cm) | ||||
| rs8050136 | 941 | 1.132 | 0.0287 | 0.2039 |
| rs17818902 | 941 | −2.323 | 0.0005 | 0.1931 |
P-values represent results of the conditioned analysis, which adjusted each SNP for the other and vice versa. (a) rs8050136 conditioned for rs17818902, (b) rs17818902 conditioned for rs8050136. P-values were calculated in a linear regression analysis with age, gender and first four MDS vectors as covariates and for fat mass and waist additionally with BMI as covariate. The minor allele is the reference allele.
Effect sizes and directions of effects of the published FTO-SNPs in the Sorbian sample were consistent with the reported data in the other European populations. The observed allele frequencies and haplotype frequencies in the Sorbs, HapMap CEU sample and WTCCC T2D cases were similar (Table 3, Supplementary Tables 1 and 2).
Table 3. Haplotype frequencies in the Sorbs and the WTCCC T2D case sample.
| Rank | Haplotype | Frequency (%) Sorbs | Posterior mean lnBMI | Posterior SD lnBMI | Frequency (%) WTCCC T2D cases |
|---|---|---|---|---|---|
| 1 | GTGTA | 57.1 | 1.65 | 0.005 | 53.5 |
| 2 | GAGGC | 18.2 | 1.61 | 0.010 | 21.7 |
| 3 | AAGTA | 15.8 | 1.64 | 0.007 | 16.9 |
| 4 | GTATA | 6.4 | 1.64 | 0.005 | 4.1 |
| 5 | GAGTA | 2.3 | 1.65 | 0.007 | 2.7 |
| 6 | GTGGC | 0.2 | 1.63 | 0.069 | 0.4 |
| 7 | AAGTC | 0.1 | 1.62 | 0.023 | <0.1 |
| 8 | AAGGC | — | 0.6 | ||
| 9 | ATGTA | — | 0.1 | ||
| 10 | GAGTC | — | <0.1 | ||
| 11 | GTAGC | — | <0.1 |
Haplotype frequencies, posterior mean and standard deviation lnBMI for SNPs rs9972717, rs10852522, rs10521308, rs17818902, rs17818920 in the Sorbian population and haplotype frequencies in the WTCCC T2D cases. The haplotypes are presented in order of frequency in the Sorbs, where rank 1 is the most common, and are used as labels in Supplementary Figure 1.
The strongest haplotypic association with BMI in the FTO/RPGRIP1L region, after adjustment for the effects of age, gender and four vectors of multidimensional scaling as covariates, was obtained for a window containing SNPs rs9972717, rs10852522, rs10521308, rs17818902, rs17818920. These SNPs cover a region of about 21 kb across parts of intron 2, exon 3 and intron 3. Visual inspection of the clustering of haplotypes within this window suggests that carriers of the combination of alleles G and C at SNPs rs10521308 and rs17818920, respectively, have the lowest mean BMI, and occur with relative frequency of ∼18% in the population (Table 3 and Supplementary Figure 1).
None of the SNPs in the region between 52.41 and 52.43 Mb was associated with BMI in 1913 subjects from the WTCCC T2D case sample set (Supplementary Table 1) and nor was any association evident using haplotype-based analyses. In contrast, rs8050136 is not significantly associated with BMI in 1479 subjects of the British 58 Birth Cohort (P=0.21), but SNPs in intron 2/3 show a moderate trend with the same effect direction as in the Sorbs (rs17818902: P-value=0.046, β=−0.4246; rs17818920: P-value=0.035181, β=−0.4488), (http://www.b58cgene.sgul.ac.uk/index.php). In the Diabetes Genetics Initiative (DGI) sample, the cluster of SNPs in the 47-kb region in intron 1/2 does not show any significant associations with BMI either in the T2D cases or in the control set (Supplementary Table 1). The SNPs in intron 2/3 show modest effects on BMI in the DGI control set but in the opposite direction than in the Sorbian sample (Supplementary Table 1).
Based on a subset of 427 Sorbian subjects with IBD sharing <0.15, LD between rs8050136 and the three representative tagging SNPs, which show moderate evidence of association with BMI in the intron 2/3 region in the Sorbian dataset was modest (to rs10852522: D′=0.046, r2=0.001, to rs17818902: D′=0.192, r2=0.006 and to rs8053367: D′=0.16, r2=0.012) suggesting that the intron 1 and intron 3 effects are independent.
Discussion
Recently, a genome-wide association study for type 2 diabetes showed an association of common FTO variants with weight, BMI, fat mass and risk of obesity.2, 26 This finding has been replicated in several independent samples of European origin.1, 3 In line with these findings, this study also identifies evidence of association with the same cluster of SNPs in intron 1.
Beyond this replication, we have identified a novel association mapping to the region between 52.41 and 52.43 Mb, which spans parts of intron 2 and intron 3. These events seem to be unrelated and statistically independent. One possible explanation of this is that the intron 3 signal represents a second mutational event segregating in the Sorbian population. Our hypothesis is that we may have detected a causal novel variant on an existing haplotype in the Sorbian population. An alternative explanation is that the BMI effect attributable to the intron 3 SNPs is the result of interaction with environmental factors and/or genetic variants outside the FTO region that are specific to the Sorbian isolate.
Our findings to some extent echo those of a recent study in the Amish, similar to the Sorbs, an isolate of European descent.34 A low-density genome-wide association scan in this population also showed evidence for FTO association with BMI. The strongest signals, however, were seen at SNPs in intron 1 (rs1861869, rs1861868 and rs147796), which were not in high LD with the recently published SNPs, and the effect was modified by physical exercise.
Table 4. SNPs associated with BMI and fat mass in the Sorbian population at P<0.0001.
| Sorbs | British 1958 Birth Cohort | DGI | Cases | Controls | All samples | ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| BMI | Fat mass | BMI | BMI | Fat mass | |||||||||||||||||
| CHR | SNP | BP | A1 | MAF | P-value | β | P-value | β | GENE | A1 | MAF | P-value | β | A1 | MAF | P-value | β | P-value | β | P-value | β |
| 7 | rs7800783 | 21 544 705 | A | 0.156 | 9.80E−08 | −0.072 | 1.45E−07 | −0.176 | DNAH11 | A | 0.030 | 0.7336 | 0.190 | A | 0.035 | 0.3527 | −0.106 | 0.4343 | 0.078 | 0.5181 | −0.069 |
| 4 | SNP_A-2058578 | 172 165 262 | T | 0.178 | 2.15E−07 | −0.066 | 2.52E−06 | −0.149 | GALNT17 | — | — | — | — | T | 0.040 | 0.1274 | 0.163 | 0.4368 | −0.069 | 0.5626 | −0.057 |
| 18 | rs4799668 | 28 670 898 | A | 0.150 | 2.25E−07 | −0.073 | 4.70E−07 | −0.176 | KLHL14 | A | 0.010 | 0.5615 | 0.461 | A | 0.023 | 0.4797 | 0.100 | 0.1386 | 0.188 | 0.7529 | 0.043 |
| 6 | rs2253802 | 30 792 665 | T | 0.153 | 5.96E−07 | −0.071 | 8.43E−09 | −0.202 | MDC1 | T | 0.040 | 0.7124 | 0.164 | — | — | — | — | — | — | — | — |
| 15 | rs7173766 | 52 790 731 | A | 0.010 | 6.29E−07 | 0.188 | 8.88E−05 | 0.370 | C15orf15 | — | — | — | — | — | — | — | — | — | — | — | — |
| 11 | rs17106280 | 106 364 880 | T | 0.146 | 1.23E−06 | −0.072 | 3.38E−06 | −0.170 | GUCY1A2 | — | — | — | — | — | — | — | — | — | — | — | — |
| 10 | rs7092248 | 104 252 332 | G | 0.142 | 1.82E−06 | −0.070 | 1.27E−06 | −0.175 | ACTR1A | — | — | — | — | — | — | — | — | — | — | — | — |
| 3 | rs1533747 | 133 328 591 | G | 0.240 | 2.26E−06 | 0.044 | 4.24E−05 | 0.095 | CPNE4 | 0.8288 | −0.042 | G | 0.299 | 0.6078 | 0.023 | 0.7721 | 0.011 | 0.8008 | 0.011 | ||
| 12 | rs2238082 | 2 356 972 | A | 0.142 | 2.37E−06 | −0.071 | 2.16E−06 | −0.178 | CACNA1C | — | — | — | — | — | — | — | — | — | — | — | — |
| 3 | rs9288907 | 96 629 152 | C | 0.018 | 5.36E−06 | 0.131 | 6.88E−05 | 0.287 | ENST00000332119 | C | 0.020 | 0.3445 | 0.588 | C | 0.012 | 0.7915 | 0.040 | 0.4179 | −0.128 | 0.3050 | 0.147 |
| 2 | rs16831005 | 135 371 064 | C | 0.170 | 6.38E−06 | −0.056 | 1.64E−06 | −0.147 | ACMSD | — | — | — | — | — | — | — | — | — | — | — | — |
| 2 | rs10183748 | 48 109 619 | T | 0.414 | 1.11E−05 | 0.035 | 1.03E−05 | 0.086 | MSH6, FBXO11 | T | 0.370 | 0.4231 | 0.146 | T | 0.382 | 0.3655 | −0.036 | 0.9802 | 0.001 | 0.9466 | −0.003 |
| 2 | rs10207319 | 48 109 700 | T | 0.414 | 1.11E−05 | 0.035 | 1.03E−05 | 0.086 | MSH6, FBXO11 | T | 0.370 | 0.4231 | 0.146 | — | — | — | — | — | — | — | — |
| 2 | rs6717617 | 48 104 613 | G | 0.407 | 1.47E−05 | 0.035 | 9.80E−06 | 0.088 | MSH6, FBXO11 | G | 0.370 | 0.4013 | 0.153 | G | 0.383 | 0.3639 | −0.037 | 0.9851 | −0.001 | 0.9539 | −0.002 |
| 2 | rs6729916 | 48 085 850 | T | 0.412 | 1.71E−05 | 0.034 | 2.06E−05 | 0.083 | MSH6, FBXO11 | T | 0.370 | 0.4613 | 0.134 | T | 0.380 | 0.3321 | −0.039 | 0.7107 | 0.013 | 0.9473 | 0.002 |
| 8 | rs2083437 | 62 492 578 | C | 0.102 | 2.31E−05 | 0.058 | 3.72E−06 | 0.157 | MGC34646 | — | — | — | — | — | — | — | — | — | — | — | — |
| 8 | rs12164179 | 26 347 353 | A | 0.068 | 4.36E−05 | −0.065 | 8.45E−05 | −0.156 | BNIP3L | A | 0.050 | 0.6763 | −0.170 | — | — | — | — | — | — | — | — |
| 13 | rs4773396 | 111 039 718 | G | 0.495 | 4.45E−05 | −0.032 | 7.88E−05 | −0.077 | ENST00000375713 | G | 0.480 | 0.0071 | −0.478 | G | 0.445 | 0.0342 | −0.083 | 0.4680 | −0.026 | 0.3484 | −0.035 |
| 10 | rs12245255 | 49 985 086 | G | 0.107 | 5.02E−05 | 0.053 | 6.17E−05 | 0.130 | C10orf72 | G | 0.120 | 0.9636 | −0.013 | — | — | — | — | — | — | — | — |
| 22 | rs5759236 | 41 983 689 | A | 0.232 | 6.59E−05 | −0.042 | 2.85E−05 | −0.108 | SCUBE1 | A | 0.180 | 0.8256 | 0.051 | A | 0.184 | 0.8193 | 0.011 | 0.9697 | 0.002 | 0.6543 | 0.021 |
| 11 | rs11231111 | 62 016 581 | T | 0.161 | 7.39E−05 | −0.053 | 5.48E−07 | −0.165 | AHNAK | T | 0.030 | 0.0692 | 0.971 | — | — | — | — | — | — | — | — |
| 1 | rs16843298 | 195 250 203 | A | 0.036 | 9.90E−05 | −0.084 | 2.90E−06 | −0.250 | ATP6V1G3, PTPRC | A | 0.080 | 0.0019 | −1.026 | — | — | — | — | — | — | — | — |
P-values were calculated under the additive model of inheritance and adjusted for genomic control factor λ (=1.16 for BMI and =1.18 for fat mass), age, gender and first four vectors of MDS for the Sorbian cohort. P-values for the DGI sample are adjusted for genomic control.
Reference allele is the minor allele (A1) for all cohorts. For SNPs not within a gene, the closest gene is given. Only SNPs with both a P-value<0.0001 for BMI and fat mass in the Sorbs are presented (sorted by P-value for association with BMI). Positions (BP) are based on NCBI Build 35 annotation. MAF (minor allele frequency) is given for each cohort.
As the molecular mechanisms relating the earlier reported association signal in FTO with the functional disruption of nearby genes remain uncertain and the causal gene has not yet been unequivocally identified, the detection of additional association signals and independent causal variants may prove extremely useful. The study of population isolates provides a convenient tool for seeking out independent events that may reflect the specific mutational and/or demographic history of that population. At the same time, the limited size of many isolates imposes a burden in terms of the capacity to generate findings that reach stringent levels of genome-wide significance, and in terms of the intrinsic limitations with respect to seeking replication in samples from other populations.
Certainly, the additional associations we have detected in FTO do not, despite the analysis of all Sorbian samples for which we currently have DNA and phenotypic data, attain levels of significance that make them entirely convincing on the genome-wide scale.
Furthermore, there remain some limitations of the study despite all quality control and consistency checks. Despite being a robust and widely applicable tool to adjust for population structure in association studies, there remain restrictions of the correction by genomic control under certain settings.35, 36, 37 However, in this situation, the existing evidence implicating FTO variation in the regulation of weight, as well as the substantial increase in the earlier expectation that additional variants in the gene that affect function and/or expression will have similar phenotypic consequences, should be taken into account as the genome-wide significance threshold may be too conservative.
So far, it remains unclear which factors contribute to the association observed in the Sorbs. The signal in intron 2/3 cannot be replicated in the WTCCC T2D cases or the DGI T2D cases. rs17818902 shows a moderate trend of association with BMI in the DGI control set but with an opposite effect direction. However, it should be stated that the earlier published SNP rs8050136 is not significantly associated with BMI in about 1500 subjects of the British 58 Birth Cohort (P-value=0.21), whereas the FTO-SNP with strongest evidence for an effect on BMI in the Sorbs shows a moderate trend with a consistent effect direction in the British 58 Birth Cohort (P-value=0.046 for rs17818902, http://www.b58cgene.sgul.ac.uk/index.php).
Viewed in this light, our findings (allied to those in the Amish) provide qualified support for the notion that a detailed examination of this gene region in a range of populations will show additional independent alleles implicated in body weight regulation. As the causal variants underlying these are identified, these should provide valuable clues to the molecular mechanisms by which sequence variation in this region affects clinical phenotypes. Our study should act as a stimulus to similar efforts in other ethnic groups. Such efforts should be directed towards a search for the full allelic spectrum of FTO regional variation using complementary approaches (indirect LD-mapping, deep resequencing, structural variant detection) designed to capture variants of all frequencies and types.
Acknowledgments
We thank all those who participated in the studies. This work was supported by grants from the Interdisciplinary Centre for Clinical Research at the University of Leipzig (B27 to MS, PK and AT; N06 to PK, BE, JB and DS, Z03 to KK), from the German Diabetes Association (to YB, AT and PK) and a Travel Grant from BIF (to AT). MS is supported by a grant from the DFG (KFO 152). EZ is a Wellcome Trust Research Career Development Fellow. APM is a Wellcome Trust Senior Research Fellow. Furthermore, we would like to thank John Broxholm from the Bioinformatics Core Unit of the Wellcome Trust Centre for Human Genetics.
Footnotes
Supplementary information accompanies the paper on European Journal of Human Genetics website (http://www.nature.com/ejhg)
Supplementary Material
References
- Dina C, Meyre D, Gallina S, et al. Variation in FTO contributes to childhood obesity and severe adult obesity. Nat Genet. 2007;39:724–726. doi: 10.1038/ng2048. [DOI] [PubMed] [Google Scholar]
- Frayling TM, Timpson NJ, Weedon MN, et al. A common variant in the FTO gene is associated with body mass index and predisposes to childhood and adult obesity. Science. 2007;316:889–894. doi: 10.1126/science.1141634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scuteri A, Sanna S, Chen WM, et al. Genome-wide association scan shows genetic variants in the FTO gene are associated with obesity-related traits. PLoS Genet. 2007;3:e115. doi: 10.1371/journal.pgen.0030115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan JT, Dorajoo R, Seielstad M, et al. FTO variants are associated with obesity in the Chinese and Malay populations in Singapore. Diabetes. 2008;57:2851–2857. doi: 10.2337/db08-0214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cha SW, Choi SM, Kim KS, et al. Replication of genetic effects of FTO polymorphisms on BMI in a Korean population. Obesity (Silver Spring) 2008;16:2187–2189. doi: 10.1038/oby.2008.314. [DOI] [PubMed] [Google Scholar]
- Chang YC, Liu PH, Lee WJ, et al. Common variation in the fat mass and obesity-associated (FTO) gene confers risk of obesity and modulates BMI in the Chinese population. Diabetes. 2008;57:2245–2252. doi: 10.2337/db08-0377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hotta K, Nakata Y, Matsuo T, et al. Variations in the FTO gene are associated with severe obesity in the Japanese. J Hum Genet. 2008;53:546–553. doi: 10.1007/s10038-008-0283-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loos RJ, Bouchard C. FTO: the first gene contributing to common forms of human obesity. Obes Rev. 2008;9:246–250. doi: 10.1111/j.1467-789X.2008.00481.x. [DOI] [PubMed] [Google Scholar]
- Gerken T, Girard CA, Tung YC, et al. The obesity-associated FTO gene encodes a 2-oxoglutarate dependent nucleic acid demethylase. Science. 2007;318:1469–1472. doi: 10.1126/science.1151710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters T, Ausmeier K, Ruther U. Cloning of Fatso (fto), a novel gene deleted by the Fused toes (Ft) mouse mutation. Mamm Genome. 1999;10:983–986. doi: 10.1007/s003359901144. [DOI] [PubMed] [Google Scholar]
- Fredriksson R, Hagglund M, Olszewski PK, et al. The obesity gene, FTO, is of ancient origin, up-regulated during food deprivation and expressed in neurons of feeding-related nuclei of the brain. Endocrinology. 2008;149:2062–2071. doi: 10.1210/en.2007-1457. [DOI] [PubMed] [Google Scholar]
- Stratigopoulos G, Padilla SL, LeDuc CA, et al. Regulation of Fto/Ftm gene expression in mice and humans. Am J Physiol Regul Integr Comp Physiol. 2008;294:R1185–R1196. doi: 10.1152/ajpregu.00839.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delous M, Baala L, Salomon R, et al. The ciliary gene RPGRIP1L is mutated in cerebello-oculo-renal syndrome (Joubert syndrome type B) and Meckel syndrome. Nat Genet. 2007;39:875–881. doi: 10.1038/ng2039. [DOI] [PubMed] [Google Scholar]
- Vierkotten J, Dildrop R, Peters T, Wang B, Ruther U. Ftm is a novel basal body protein of cilia involved in Shh signalling. Development. 2007;134:2569–2577. doi: 10.1242/dev.003715. [DOI] [PubMed] [Google Scholar]
- Tayeh MK, Yen HJ, Beck JS, et al. Genetic interaction between Bardet–Biedl syndrome genes and implications for limb patterning. Hum Mol Genet. 2008;17:1956–1967. doi: 10.1093/hmg/ddn093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis RE, Swiderski RE, Rahmouni K, et al. A knockin mouse model of the Bardet–Biedl syndrome 1 M390R mutation has cilia defects, ventriculomegaly, retinopathy, and obesity. Proc Natl Acad Sci USA. 2007;104:19422–19427. doi: 10.1073/pnas.0708571104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speakman JR, Rance KA, Johnstone AM. Polymorphisms of the FTO gene are associated with variation in energy intake, but not energy expenditure. Obesity (Silver Spring) 2008;16:1961–1965. doi: 10.1038/oby.2008.318. [DOI] [PubMed] [Google Scholar]
- Berentzen T, Kring SI, Holst C, et al. Lack of association of fatness-related FTO gene variants with energy expenditure or physical activity. J Clin Endocrinol Metab. 2008;93:2904–2908. doi: 10.1210/jc.2008-0007. [DOI] [PubMed] [Google Scholar]
- Wardle J, Carnell S, Haworth CM, Farooqi IS, O'Rahilly S, Plomin R. Obesity associated genetic variation in FTO is associated with diminished satiety. J Clin Endocrinol Metab. 2008;93:3640–3643. doi: 10.1210/jc.2008-0472. [DOI] [PubMed] [Google Scholar]
- Jacobsson JA, Klovins J, Kapa I, et al. Novel genetic variant in FTO influences insulin levels and insulin resistance in severely obese children and adolescents. Int J Obes (Lond) 2008;32:1730–1735. doi: 10.1038/ijo.2008.168. [DOI] [PubMed] [Google Scholar]
- Freathy RM, Timpson NJ, Lawlor DA, et al. Common variation in the FTO gene alters diabetes-related metabolic traits to the extent expected given its effect on BMI. Diabetes. 2008;57:1419–1426. doi: 10.2337/db07-1466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Do R, Bailey SD, Desbiens K, et al. Genetic variants of FTO influence adiposity, insulin sensitivity, leptin levels, and resting metabolic rate in the Quebec Family Study. Diabetes. 2008;57:1147–1150. doi: 10.2337/db07-1267. [DOI] [PubMed] [Google Scholar]
- Worman HJ, Bonne G. Laminopathies': a wide spectrum of human diseases. Exp Cell Res. 2007;313:2121–2133. doi: 10.1016/j.yexcr.2007.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capell BC, Collins FS. Human laminopathies: nuclei gone genetically awry. Nat Rev Genet. 2006;7:940–952. doi: 10.1038/nrg1906. [DOI] [PubMed] [Google Scholar]
- Wellcome Trust Case Control Consortium Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447:661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeggini E, Weedon MN, Lindgren CM, et al. Replication of genome-wide association signals in UK samples reveals risk loci for type 2 diabetes. Science. 2007;316:1336–1341. doi: 10.1126/science.1142364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The International HapMap Consortium The International HapMap Project. Nature. 2003;426:789–796. doi: 10.1038/nature02168. [DOI] [PubMed] [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- American Diabetes Association Diagnosis and classification of diabetes mellitus. Diabetes Care. 2006;29 Suppl 1:S43–S48. [PubMed] [Google Scholar]
- Morris AP. A flexible Bayesian framework for modeling haplotype association with disease, allowing for dominance effects of the underlying causative variants. Am J Hum Genet. 2006;79:679–694. doi: 10.1086/508264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- Gauderman WJ. Sample size requirements for association studies of gene–gene interaction. Am J Epidemiol. 2002;155:478–484. doi: 10.1093/aje/155.5.478. [DOI] [PubMed] [Google Scholar]
- Gauderman WJ. Sample size requirements for matched case–control studies of gene–environment interaction. Stat Med. 2002;21:35–50. doi: 10.1002/sim.973. [DOI] [PubMed] [Google Scholar]
- Rampersaud E, Mitchell BD, Pollin TI, et al. Physical activity and the association of common FTO gene variants with body mass index and obesity. Arch Intern Med. 2008;168:1791–1797. doi: 10.1001/archinte.168.16.1791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bacanu SA, Devlin B, Roeder K. The power of genomic control. Am J Hum Genet. 2000;66:1933–1944. doi: 10.1086/302929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devlin B, Roeder K, Bacanu SA. Unbiased methods for population-based association studies. Genet Epidemiol. 2001;21:273–284. doi: 10.1002/gepi.1034. [DOI] [PubMed] [Google Scholar]
- Bacanu SA, Devlin B, Roeder K. Association studies for quantitative traits in structured populations. Genet Epidemiol. 2002;22:78–93. doi: 10.1002/gepi.1045. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.