Abstract
Abdominal aortic aneurysm (AAA) is a multifactorial condition. The transforming growth factor β (TGF-β) pathway regulates vascular remodeling and mutations in its receptor genes, TGFBR1 and TGFBR2, cause syndromes with thoracic aortic aneurysm (TAA). The TGF-β pathway may be involved in aneurysm development in general. We performed an association study by analyzing all the common genetic variants in TGFBR1 and TGFBR2 using tag single nucleotide polymorphisms (SNPs) in a Dutch AAA case–control population in a two-stage genotyping approach. In stage 1, analyzing 376 cases and 648 controls, three of the four TGFBR1 SNPs and nine of the 28 TGFBR2 SNPs had a P<0.07. Genotyping of these SNPs in an independent cohort of 360 cases and 376 controls in stage 2 confirmed association (P<0.05) for the same allele of one SNP in TGFBR1 and two SNPs in TGFBR2. Joint analysis of the 736 cases and 1024 controls showed statistically significant associations of these SNPs, which sustained after proper correction for multiple testing (TGFBR1 rs1626340 OR 1.32 95% CI 1.11–1.56 P=0.001 and TGFBR2 rs1036095 OR 1.32 95% CI 1.12–1.54 P=0.001 and rs4522809 OR 1.28 95% CI 1.12–1.46 P=0.0004). We conclude that genetic variations in TGFBR1 and TGFBR2 associate with AAA in the Dutch population. This suggests that AAA may develop partly by similar defects as TAA, which in the future may provide novel therapeutic options.
Keywords: abdominal aortic aneurysm, association study, TGFBR1, TGFBR2, transforming growth factor-β pathway, vascular remodeling
Introduction
Aneurysms of the abdominal aorta (AAA) affect 5–8% of men above 60 years of age.1 In addition to male gender and age, other well-known risk factors of AAA are Caucasian race, smoking, the presence of other atherosclerotic conditions (myocardial infarction, cerebrovascular and peripheral arterial disease) and a positive family history.2 AAA is therefore considered a multifactorial disorder in which both environmental and genetic factors contribute to its development.3
AAA develops through a not well-defined mechanism. Ultimately, vascular extracellular matrix (ECM) changes, accompanied by transmural inflammation, destructive remodeling of the elastic media, and depletion of medial smooth muscle cells, are observed.4 This weakening of the vessel wall increases the risk of rupture with often a fatal outcome. Rupture rates increase with the size of the AAA, and therefore surgical intervention is considered when the AAA exceeds the 55 mm diameter threshold.5
Variations in genes that regulate ECM stability have been described to increase the susceptibility to AAA. Association has been reported with matrix-metalloproteinases6, 7 and their inhibitors (tissue inhibitors of metalloproteinases).7 However, attempts to replicate these associations in different populations have led to conflicting results.8, 9 In contrast, association of AAA with a sequence variant on 9p21 has consistently been replicated.10, 11 This variant was shown to associate with AAA, coronary artery disease (CAD), and intracranial aneurysms (IA) in different populations.10 This variant did not predispose to the atherosclerotic conditions peripheral artery disease and large atherosclerotic or cardiogenic shock. Although atherosclerosis is a major determinant for development of CAD and AAA, it has no or a modest role in IA formation. Therefore, it was suggested that the variant on 9p21 influences another, shared mechanism in the development of CAD, AAA and IA, like vascular remodeling.
The transforming growth factor (TGF) β-pathway is an important regulator of vascular remodeling, and has effects on both ECM synthesis and ECM degradation.12 Dysregulated TGF-βsignaling by mutations in the receptor genes, TGFBR1 and TGFBR2, causes thoracic aortic aneurysm (TAA) syndromes, including Marfan syndrome type II (OMIM 154705),13, 14 Loeys–Dietz syndrome (OMIM 609192),14, 15 and thoracic aortic aneurysms leading to type A dissections.16, 17 Although these syndromes are clinically distinct, their phenotypes overlap, with TAA and aortic dissections as the common denominator. In families with TAA caused by mutations in the TGFBR2 gene, family members had aneurysms at different locations of the vascular system. Most affected members had TAA of the ascending aorta but some also had aneurysms of the descending aorta, the carotid, brachiocephalic, subclavian or popliteal arteries whereas others had ruptured or unruptured IA.17, 18 Furthermore, detailed characterization revealed that half of the probands with Loeys-Dietz aortic aneurysm syndrome not only had TAA but also aneurysms at different locations, mainly consisting of AAA and aneurysms of head and neck.15 It is therefore conceivable that dysregulated TGF-β signaling has a role in aneurysm formation in general. Genetics variants in TGFBR1 and TGFBR2 may influence their signaling capacity and thereby the quality of vascular remodeling induced by the TGF-β.
In contrast to TAA syndromes, AAA is usually not caused by a single gene defect, but multiple genetic and environmental factors are thought to participate in its development. We hypothesized that variants in TGFBR1 and TGFBR2 may contribute to AAA formation. To investigate this, we performed a genetic association study to analyze all the common TGFBR1 and TGFBR2 variants using tag single nucleotide polymorphisms (SNPs) in a Dutch case–control population.
Materials and methods
Patient collection and controls
We included Dutch Caucasian cases with a proven AAA (>30 mm) who visited their vascular surgeon from May 2007 until December 2007 in eight large centers in the Netherlands. Controls comprised healthy blood bank volunteers (828 samples) in whom an AAA was not excluded, and men between 60–80 years of age in whom an AAA was excluded by ultrasonography (196 control samples from age-matched controls out of an AAA-screening study).19 The study was approved by the Medical Ethics Committee of the University Medical Center Utrecht, and by each local review board.
Study design, SNP selection, and genotyping
Tag SNPs from the TGFBR1 and TFGBR2 genes spanning the entire genes and 5 kb upstream and downstream of the genes were selected from the International HapMap Project using the aggressive tagger option of the program Tagger (Paul de Bakker, http://www.broadinstitute.org/mpg/tagger/, NCBI Build 35/UCSC hg17/May 2004) so that all SNPs with a minor allele frequency of 10% were captured with r2>0.8. One tag SNP in TGFBR2 with low Illumina quality design scores was excluded. In total, five tag SNPs were derived for genotype analysis of TGFBR1 and 30 tag SNPs for genotype analysis of TGFBR2. SNP genotyping was performed using a two-stage genotyping approach. For stage 1, the tagging SNPs were genotyped in 376 Dutch Caucasian cases and 652 Dutch Caucasian controls (bloodbank volunteers) using the GoldenGate assay on an Illumina BeadStation 500 GX (Illumina Inc., San Diego, CA, USA). All tag SNPs were examined for their quality and one tag SNP in TGFBR1 and two tag SNPs in TGFBR2 that had low signals were excluded. DNA samples with low signals for most SNPs were also excluded (n=4, all controls). For stage 2, all SNPs with arbitrary low P-values (P<0.07) in stage 1 were genotyped in a second independent cohort of 360 cases and 376 controls (180 bloodbank volunteers and 196 men aged 60–80 without an AAA). For these selected SNPs we obtained Taqman Assays on Demand (Applied Biosystems, Foster City, CA, USA), which were genotyped on an ABI7900HT instrument (Applied Biosystems). DNA samples with low signals were excluded (varying between 0.3 and 4% per assay).
Statistical analysis
Association χ2 with two-tailed P-values and Hardy–Weinberg equilibriums were calculated using the HAPLOVIEW program (available at http://www.hapmap.org). Differences in allele frequencies were assessed as an odds ratio (OR) with corresponding 95% confidence intervals (CI) and P-values, using the allele with the lower frequency in the controls as opposed to the allele frequency in cases as the reference allele. Significant associations in the joined analysis were corrected for multiple testing. We genotyped 32 SNPs in total, therefore associations with a P-value smaller than 0.0015 (0.05/32) were considered statistically significant after correction for multiple testing. With the AAA sample size from both stages, the study had 73% power to detect a susceptibility locus with a relative risk of 1.3, and 88% power to detect a susceptibility locus with a relative risk of 1.4, at a significance level of 0.05 assuming an additive model (genetic power calculator, SGDP statistical genetics group, http://statgen.iop.kcl.ac.uk). The population attributable risk (PAR) of the SNPs that remained significantly associated after correction for multiple testing was calculated using the following formula: PAR=1–1/(f_wt+(f_het*OR_het)+(f_hom*OR_hom)), where f_wt, f_het and f_hom are the population frequencies of the wild-type, the heterozygous and the homozygous carriers, respectively. Similarly, OR_het and OR_hom are the odds ratios for heterozygous and homozygous carriers, respectively.
Results
The clinical data of the AAA cases analyzed in stages 1 and 2 are shown in Table 1.
Table 1. Clinical data of the analyzed cases with abdominal aortic aneurysms.
| Stage 1 (N=376) | Stage 2 (N=360) | |
|---|---|---|
| Male sex – no. (%) | 337 (89.6) | 326 (90.6) |
| Mean age – year (±SD) | 72.1 (7.5) | 71.3 (7.3) |
| Diameter AAA – mm (±SD) | 57.1 (15.3) | 57.6 (16.0) |
| Ruptured AAA – no. (%) | 13 (3.5) | 22 (6.1) |
| Surgical intervention – no. (%) | 240 (63.8) | 216 (60.0) |
| Smoking status – no. (%) | ||
| Currently | 107 (28.5) | 108 (30.0) |
| Previously | 242 (64.4) | 229 (63.6) |
| Never | 27 (7.1) | 23 (6.4) |
| Hypertension – no. (%) | 227 (60.4) | 216 (60.0) |
| Other cardiovascular disease – no. (%) | 233 (62.0) | 214 (59.4) |
| Family history of AAA – no. (%) | 87 (23.1) | 79 (21.9) |
In stage 1 no tag SNPs showed deviation from Hardy–Weinberg equilibrium. In this stage four tag SNPs of TGFBR1 and 28 tag SNPs of TGFBR2 were successfully genotyped in 376 cases and 648 controls. The association data of these tag SNPs and corresponding 95% confidence intervals and P-values, are shown in the Supplementary Table online. Three of the four analyzed SNPs in TGFBR1 and nine of the 28 analyzed SNPs in TGFBR2 showed arbitrary low P-values (P<0.07) (strongest association for TGFBR1 SNP rs1571590, P=0.032 and strongest association for TGFBR2 SNP rs3087465, P=0.002) (Supplementary Table online and Table 2) and were selected for additional genotyping in an independent cohort of 360 cases and 376 controls.
Table 2. The analysis of the TGFBR1 and TGFBR2 genes in abdominal aortic aneurysm cases and controls.
| Gene | SNP | Associated allele | Stage 1: frequency allele P<0.07 (%) | Stage 2: frequency allele P<0.07 in stage 1 (%) | Stage 1+ 2: allele frequencies combined (%) | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Cases (n=376) | Controls (n=648) | OR (95% CI) | P-value | Cases (n=360) | Controls (n=376) | OR (95% CI) | P-value | Cases (n=736) | Controls (n=1024) | OR (95% CI) | P-value | |||
| TGFBR1 | rs10819634 | A | 18.5 | 15.3 | 1.25 (0.98–1.67) | 0.066 | 20.2 | 16.4 | 1.28 (0.98–1.69) | 0.064 | 19.3 | 15.7 | 1.28 (1.08–1.54) | 0.006 |
| rs1571590 | C | 20.6 | 16.8 | 1.28 (1.02–1.61) | 0.032 | 21.9 | 18.6 | 1.23 (0.94–1.59) | 0.123 | 21.2 | 17.4 | 1.28 (1.08–1.52) | 0.005 | |
| rs1626340* | A | 21.7 | 18.4 | 1.23 (0.98–1.54) | 0.069 | 24.0 | 18.2 | 1.43 (1.10–1.82) | 0.007 | 22.8 | 18.3 | 1.32 (1.11–1.56) | 0.001 | |
| TGFBR2 | rs764522 | A | 21.7 | 17.3 | 1.32 (1.05–1.67) | 0.014 | 20.6 | 18.3 | 1.16 (0.89–1.49) | 0.271 | 21.1 | 17.6 | 1.25 (1.05–1.49) | 0.01 |
| rs3087465 | A | 25.1 | 19.4 | 1.39 (1.12–1.72) | 0.002 | 22.5 | 19.9 | 1.16 (0.90–1.49) | 0.239 | 23.9 | 19.6 | 1.28 (1.10–1.52) | 0.002 | |
| rs1036095* | C | 26.6 | 21.1 | 1.35 (1.10–1.67) | 0.005 | 24.6 | 19.9 | 1.32 (1.02–1.69) | 0.035 | 25.7 | 20.6 | 1.32 (1.12–1.54) | 0.001 | |
| rs4522809* | A | 58.6 | 51.8 | 1.32 (1.10–1.58) | 0.003 | 56.4 | 51.1 | 1.24 (1.01–1.53) | 0.042 | 57.6 | 51.5 | 1.28 (1.12–1.46) | 0.0004 | |
| rs13075948 | C | 75.1 | 70.0 | 1.29 (1.05–1.58) | 0.015 | 69.8 | 69.6 | 1.01 (0.81–1.26) | 0.924 | 72.5 | 70.3 | 1.12 (0.96–1.29) | 0.152 | |
| rs9831477 | A | 46.5 | 41.0 | 1.25 1.05–1.50) | 0.014 | 43.1 | 40.9 | 1.09 (0.89–1.35) | 0.42 | 44.9 | 41.0 | 1.17 (1.03–1.35) | 0.021 | |
| rs1346907 | A | 47.3 | 42.0 | 1.23 (1.03–1.49) | 0.020 | 47.3 | 45.1 | 1.10 (0.88–1.35) | 0.402 | 47.3 | 43.1 | 1.18 (1.03–1.35) | 0.014 | |
| rs9843143 | A | 53.3 | 48.5 | 1.22 (1.02–1.46) | 0.034 | 53.0 | 52.4 | 1.02 (0.83–1.26) | 0.8287 | 53.2 | 49.9 | 1.14 (0.99–1.46) | 0.059 | |
| rs304839 | C | 20.7 | 17.9 | 1.25 (1.00–1.57) | 0.052 | 19.7 | 19.2 | 1.03 (0.79–1.34) | 0.824 | 20.2 | 18.0 | 1.16 (0.98–1.37) | 0.096 | |
*Significant association after correction for multiple testing.
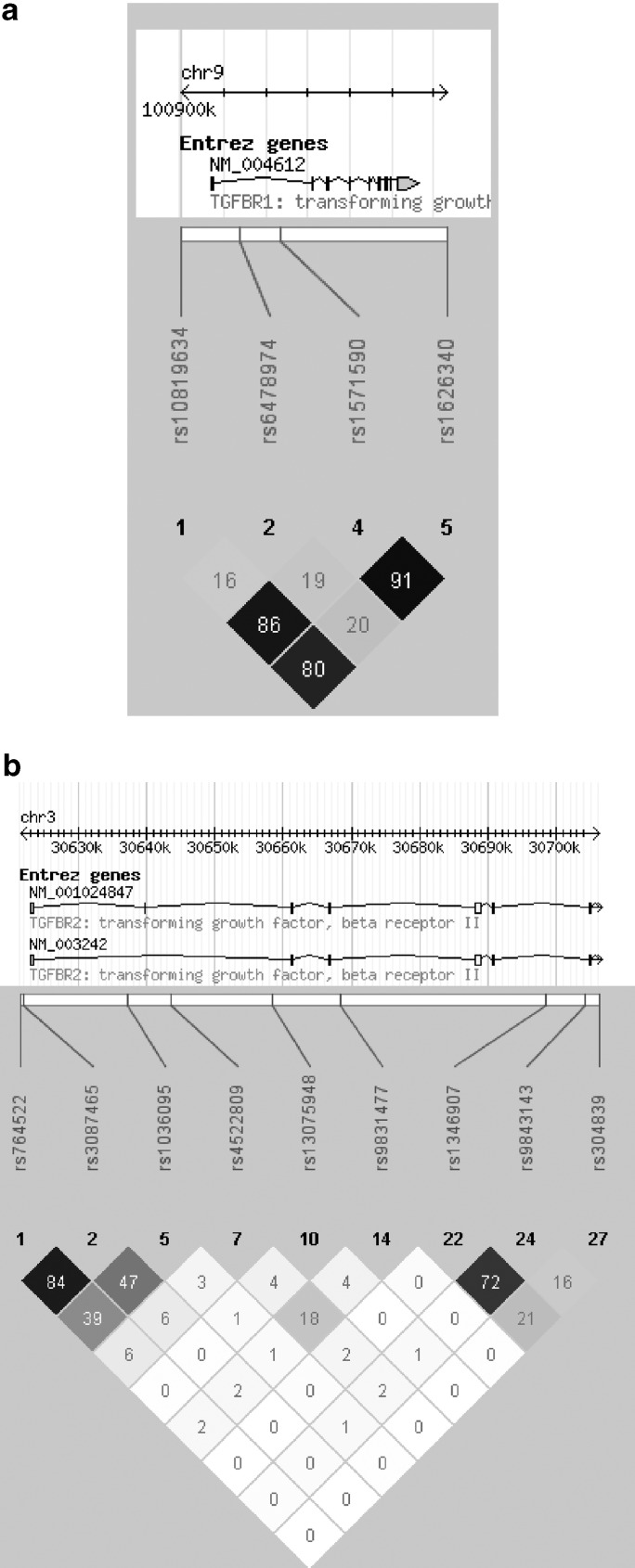
In stage 2 there was also no evidence of a deviation from Hardy–Weinberg equilibrium. Association (P<0.05) for the same allele of one SNP in TGFBR1 (rs1626340, P=0.007) with AAA (Table 2) was shown. Combining both cohorts (stage 1 and stage 2) strengthened the association of this SNP as seen in stage 2 (OR 1.32, 95% CI 1.11–1.56, P=0.001). This association remained statistically significant after correcting for multiple testing (ie, P<0.0015, 0.05/32). The PAR of rs1626340 was estimated to be 10%. For the two remaining TGFBR1 SNPs rs1571590 and rs10819634 association was shown on combined analyses of both cohorts (rs1571590 OR 1.28, 95% CI 1.08–1.52, P=0.005 and rs10819634 OR 1.28, 95% CI 1.08–1.54, P=0.006) (Table 2). The three SNPs that were associated with AAA in the joined analysis were in strong LD (Figure 1a). None of the constructed haplotypes had a stronger association with AAA than the independent SNPs (data not shown).
Figure 1.
Block-based haplotype analysis using HAPLOVIEW (R2 displayed) of all tag SNPs tested in stage 1 of TGFBR1 (a) and the tag SNPs with a P<0.07 in stage 1 of TGFBR2 (b).
In stage 2, association was observed for the same allele of two of the nine TGFBR2 SNPs (rs4522809, P=0.042; rs1036095, P=0.035) (Table 2). Combining both cohorts strengthened the association of these two SNPs as seen in stage 2 (rs4522809 OR 1.28, 95% CI 1.12–1.46, P=0.0004 and rs1036095 OR 1.32, 95% CI 1.12–1.54, P=0.001). These associations remained statistically significant after correcting for multiple testing (ie, P<0.0015, 0.05/32). The PARs of rs4522809 and rs1036095 were estimated to be 3 and 6%, respectively. No association was shown for SNPs rs1346907, rs764522, rs3087465, and rs9834177 in stage 2, but on combined analyses of both cohorts association of these SNPs was statistically significant (rs1346907 OR 1.18, 95% CI 1.03–1.35, P=0.014; rs764522 OR 1.25, 95% CI 1.05–1.49, P=0.01, rs3087465 OR 1.28, 95% CI 1.10–1.52, P=0.002, rs9831477 OR 1.17, 95% CI 1.03–1.35, P=0.021) (Table 2). For the remaining SNPs rs13075948, rs9843143, and rs304839 no association in stage 2 nor in the combined cohorts was shown (Table 2). The nine TGFBR2 SNPs that were tested in stage 2 were scattered throughout the gene and there was no obvious LD between the SNPs (Figure 1b). Therefore, no haplotypes were constructed.
Discussion
By analyzing genetic variants in the TGFBR1 and TGFBR2 genes, involved in the development of TAA and possibly involved in the development of aneurysms in general, we identified SNPs in TGFBR1 and TGFBR2 that associate with AAA in the Dutch population.
A common feature of AAA is fragmentation of the elastic laminae and smooth muscle cell loss.4 This may be the result of defective vascular remodeling, which under physiological circumstances induces adaptive changes within the vessel wall upon hemodynamic stress or as a response to vascular injury. Vascular remodeling includes a tightly regulated balance between degradation and rebuilding of the ECM. A pathological shift towards excessive ECM degradation results in loss of vascular wall integrity, which may precede aneurysm formation.12 One of the upstream regulators of vascular remodeling is the TGF-β signaling pathway, which has been extensively studied in light of TAA syndromes.12 TGF-β transduces its signals by two transmembrane receptors, which are encoded by the TGFBR1 and TGFBR2 genes.20 Mutations in these genes are responsible for Marfan syndrome type II (OMIM 154705),13 Loeys-Dietz syndrome (OMIM 609192)14, 15 and thoracic aortic aneurysms leading to type A dissections (TAA; OMIM 608967).16, 17 Marfan syndrome type I is clinically similar to Marfan type II syndrome, but it is caused by mutations in the fibrillin-1 gene.21 Fibrillin-1 encodes the structural ECM protein fibrillin-1, which is thought to regulate the availability of active TGF-β by binding to TGF-β in its latent form.22 In mouse models of Marfan syndrome type I increased TGF-β activity was observed in different organs, including aortic tissue.23 Similarly, aortic tissues of Loeys–Dietz syndrome patients showed increased TGF-β signaling activity.24 Paradoxically, mutations in the receptor genes are primarily located within its intracellular kinase domain, resulting in kinase inactivity.12 This is believed to impair intracellular signaling. Apparently, fine-tuning of TGF-β signaling is required for optimal vascular wall structure maintenance. In light of this, it is interesting to note that in AAA expression of TGF-β1 was found to be upregulated,25 whereas with gene therapy in rats the already formed AAA was shown to stabilize after overexpression of TGF-β1.26
Our findings that variations within the TGF-β receptor genes, which may result in different functional activity or expression, associate with AAA suggest that the TGF-β pathway has a role in AAA pathogenesis, similar to that seen in TAA. This may offer novel therapeutic options, because aortic root dilatation in Marfan mouse models could be prevented by administration of TGF-β antagonists.27 It is unlikely that aneurysm formation is preventable in AAA by inhibiting a single pathway, because in contrast to the TAA syndromes, AAA is considered as a complex disease. Not a single gene defect, but multiple genetic and environmental factors contribute to its pathogenesis. However, targeting different molecular players as well as aiming for efficient risk factor reduction (ie, smoking cessation, tension control) may prove to be worth while in future therapies.
The three SNPs that were found to associate significantly with AAA after correction for multiple testing together had a PAR of 19%. The PAR of rs10757278, previously found to be associated with AAA in a part of the current cohort, was estimated to be 26%.10 Therefore, these SNPS combined can explain almost half of the increased genetic risk.
A previous study analyzing one polymorphism of the TGFBR1 gene (9A6A) in 201 Italian AAA patients and 252 controls failed to show an association with AAA.28 In the presence of the angiotensin-converting enzyme DD genotype they did show that the TGFBR1 6A allele affected susceptibility to AAA. The Italian study only analyzed one sequence variant in TGFBR1, whereas we selected tag SNPs on the basis of known patterns of LD to capture the maximum coverage of the gene. This difference in approach may explain the discrepant results of the two studies. Furthermore, the power of the previous study may have been too small to detect the association.
A recent study reported no association of tag SNPs of TGFBR1 and TGFBR2 with AAA in 640 cases and 1071 controls.29 This is an interesting finding, because those AAA cases were identified from population-based screening, and had much smaller AAAs (36.1 versus 57.3 mm in our cohort). This suggests that the TGFBR1 and TGFBR2 SNPs we found to be associated with AAA predispose to a kind of AAA that proceeds to a severe form, often requiring surgery. In our group over 60% of the patients had a surgical intervention. In addition, genetic heterogeneity may explain part of the discrepant results.
In complex conditions as AAA, the contribution of a single gene variation is expected to be modest, and low OR are expected to be found. Therefore, substantial sample sizes are needed to reach statistical significant values. As we used a two-stage approach, our sample size in stage 1 was not appropriate to select only those SNPs with a P<0.05. We therefore selected the SNPs in stage 1 with arbitrary low P-values; <0.07. Association of some these selected SNPs in the second stage, and associations of other SNPs on combined analyses of both stages, suggests that they are true associations. Moreover, three SNPs remained significantly associated after proper correction for multiple testing of the joint analysis, which strongly indicates that they are true associations. The ultimate proof of these associations will be consistent replication in other patient populations and the identification of the functional variants, which can be achieved by sequencing risk haplotypes in a succeeding study. Recently, TGF-β receptor genes were directly sequenced in patients with familiar forms of intracranial aneurysms.30 No mutations in TGFBR1 and TGFBR2 were identified, but novel variants were discovered in the co-receptor genes β-glycan (TGFBR3) and endoglin (ENG).
A shortcoming of our study is that we were not able to control for confounding, because no information of the common AAA risk factors, like hypertension and smoking, was available in our control group. Demographically, the cases and controls are similar because they are both from Dutch origin, and genome-wide association data of the control group did not reveal population stratification.
In conclusion, we identified several SNPs in the TGFBR1 and TGFBR2 genes that associate with AAA. These are intriguing findings that give new insights in AAA pathogenesis and in the future may provide novel therapeutic options.
Conflict of interest
The authors declare no conflict of interest.
Acknowledgments
AF Baas was supported by grants from the Novartis Foundation for Cardiovascular Excellence and the foundation ‘De Drie Lichten'. YM Ruigrok was supported by a grant from the Dr E. Dekker program of the Netherlands Heart Foundation (2005T014).
Footnotes
Supplementary Information accompanies the paper on European Journal of Human Genetics website (http://www.nature.com/ejhg)
Supplementary Material
References
- Sakalihasan N, Limet R, Defawe OD. Abdominal aortic aneurysm. Lancet. 2005;365:1577–1589. doi: 10.1016/S0140-6736(05)66459-8. [DOI] [PubMed] [Google Scholar]
- Cornuz J, Sidoti PC, Tevaearai H, Egger M. Risk factors for asymptomatic abdominal aortic aneurysm: systematic review and meta-analysis of population-based screening studies. Eur J Public Health. 2004;14:343–349. doi: 10.1093/eurpub/14.4.343. [DOI] [PubMed] [Google Scholar]
- Sandford RM, Bown MJ, London NJ, Sayers RD. The genetic basis of abdominal aortic aneurysms: a review. Eur J Vasc Endovasc Surg. 2007;33:381–390. doi: 10.1016/j.ejvs.2006.10.025. [DOI] [PubMed] [Google Scholar]
- Annambhotla S, Bourgeois S, Wang X, Lin PH, Yao Q, Chen C. Recent advances in molecular mechanisms of abdominal aortic aneurysm formation. World J Surg. 2008;32:976–986. doi: 10.1007/s00268-007-9456-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The UK Small Aneurysm Trial Participants Mortality results for randomised controlled trial of early elective surgery or ultrasonographic surveillance for small abdominal aortic aneurysms. Lancet. 1998;352:1649–1655. [PubMed] [Google Scholar]
- Jones GT, Phillips VL, Harris EL, Rossaak JI, van Rij AM. Functional matrix metalloproteinase-9 polymorphism (C-1562T) associated with abdominal aortic aneurysm. J Vasc Surg. 2003;38:1363–1367. doi: 10.1016/s0741-5214(03)01027-9. [DOI] [PubMed] [Google Scholar]
- Ogata T, Shibamura H, Tromp G, et al. Genetic analysis of polymorphisms in biologically relevant candidate genes in patients with abdominal aortic aneurysms. J Vasc Surg. 2005;41:1036–1042. doi: 10.1016/j.jvs.2005.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smallwood L, Allcock R, van Bockxmeer F, et al. Polymorphisms of the matrix metalloproteinase 9 gene and abdominal aortic aneurysm. Br J Surg. 2008;95:1239–1244. doi: 10.1002/bjs.6345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armani C, Curcio M, Barsotti MC, et al. Polymorphic analysis of the matrix metalloproteinase-9 gene and susceptibility to sporadic abdominal aortic aneurysm. Biomed Pharmacother. 2007;61:268–271. doi: 10.1016/j.biopha.2006.12.002. [DOI] [PubMed] [Google Scholar]
- Helgadottir A, Thorleifsson G, Magnusson KP, et al. The same sequence variant on 9p21 associates with myocardial infarction, abdominal aortic aneurysm and intracranial aneurysm. Nat Genet. 2008;40:217–224. doi: 10.1038/ng.72. [DOI] [PubMed] [Google Scholar]
- Thompson AR, Golledge J, Cooper JA, Hafez H, Norman PE, Humphries SE.Sequence variant on 9p21 is associated with the presence of abdominal aortic aneurysm disease but does not have an impact on aneurysmal expansion Eur J Hum Genet 2008. Oct 15. [DOI] [PMC free article] [PubMed]
- Jones JA, Spinale FG, Ikonomidis JS. Transforming growth factor-beta signaling in thoracic aortic aneurysm development: a paradox in pathogenesis. J Vasc Res. 2008;46:119–137. doi: 10.1159/000151766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuguchi T, Collod-Beroud G, Akiyama T, et al. Heterozygous TGFBR2 mutations in Marfan syndrome. Nat Genet. 2004;36:855–860. doi: 10.1038/ng1392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh KK, Rommel K, Mishra A, et al. TGFBR1 and TGFBR2 mutations in patients with features of Marfan syndrome and Loeys-Dietz syndrome. Hum Mutat. 2006;27:770–777. doi: 10.1002/humu.20354. [DOI] [PubMed] [Google Scholar]
- Loeys BL, Schwarze U, Holm T, et al. Aneurysm syndromes caused by mutations in the TGF-beta receptor. N Engl J Med. 2006;355:788–798. doi: 10.1056/NEJMoa055695. [DOI] [PubMed] [Google Scholar]
- Pannu H, Fadulu VT, Chang J, et al. Mutations in transforming growth factor-beta receptor type II cause familial thoracic aortic aneurysms and dissections. Circulation. 2005;112:513–520. doi: 10.1161/CIRCULATIONAHA.105.537340. [DOI] [PubMed] [Google Scholar]
- Law C, Bunyan D, Castle B, et al. Clinical features in a family with an R460H mutation in transforming growth factor beta receptor 2 gene. J Med Genet. 2006;43:908–916. doi: 10.1136/jmg.2006.042176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasham SN, Willing MC, Guo DC, et al. Mapping a locus for familial thoracic aortic aneurysms and dissections (TAAD2) to 3p24–25. Circulation. 2003;107:3184–3190. doi: 10.1161/01.CIR.0000078634.33124.95. [DOI] [PubMed] [Google Scholar]
- Boll AP, Verbeek AL, van de Lisdonk EH, van der Vliet JA. High prevalence of abdominal aortic aneurysm in a primary care screening programme. Br J Surg. 1998;85:1090–1094. doi: 10.1046/j.1365-2168.1998.00814.x. [DOI] [PubMed] [Google Scholar]
- ten Dijke P, Miyazono K, Heldin CH. Signaling via hetero-oligomeric complexes of type I and type II serine/threonine kinase receptors. Curr Opin Cell Biol. 1996;8:139–145. doi: 10.1016/s0955-0674(96)80058-5. [DOI] [PubMed] [Google Scholar]
- Lee B, Godfrey M, Vitale E, et al. Linkage of Marfan syndrome and a phenotypically related disorder to two different fibrillin genes. Nature. 1991;352:330–334. doi: 10.1038/352330a0. [DOI] [PubMed] [Google Scholar]
- Dietz HC, Loeys B, Carta L, Ramirez F. Recent progress towards a molecular understanding of Marfan syndrome. Am J Med Genet C Semin Med Genet. 2005;139C:4–9. doi: 10.1002/ajmg.c.30068. [DOI] [PubMed] [Google Scholar]
- Mizuguchi T, Matsumoto N. Recent progress in genetics of Marfan syndrome and Marfan-associated disorders. J Hum Genet. 2007;52:1–12. doi: 10.1007/s10038-006-0078-1. [DOI] [PubMed] [Google Scholar]
- Boileau C, Jondeau G, Mizuguchi T, Matsumoto N. Molecular genetics of Marfan syndrome. Curr Opin Cardiol. 2005;20:194–200. doi: 10.1097/01.hco.0000162398.21972.cd. [DOI] [PubMed] [Google Scholar]
- Fukui D, Miyagawa S, Soeda J, Tanaka K, Urayama H, Kawasaki S. Overexpression of transforming growth factor beta1 in smooth muscle cells of human abdominal aortic aneurysm. Eur J Vasc Endovasc Surg. 2003;25:540–545. doi: 10.1053/ejvs.2002.1857. [DOI] [PubMed] [Google Scholar]
- Dai J, Losy F, Guinault AM, et al. Overexpression of transforming growth factor-beta1 stabilizes already-formed aortic aneurysms: a first approach to induction of functional healing by endovascular gene therapy. Circulation. 2005;112:1008–1015. doi: 10.1161/CIRCULATIONAHA.104.523357. [DOI] [PubMed] [Google Scholar]
- Habashi JP, Judge DP, Holm TM, et al. Losartan, an AT1 antagonist, prevents aortic aneurysm in a mouse model of Marfan syndrome. Science. 2006;312:117–125. doi: 10.1126/science.1124287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucarini L, Sticchi E, Sofi F, et al. ACE and TGFBR1 genes interact in influencing the susceptibility to abdominal aortic aneurysm. Atherosclerosis. 2009;202:205–210. doi: 10.1016/j.atherosclerosis.2008.04.038. [DOI] [PubMed] [Google Scholar]
- Golledge J, Clancy P, Jones GT. Possible association between genetic polymorphisms in transforming growth factor β receptors, serum transforming growth factor β1 and abdominal aortic aneurysm. BJS. 2009;96:628–632. doi: 10.1002/bjs.6633. [DOI] [PubMed] [Google Scholar]
- Santiago-Sim T, Mathew-Joseph S, Pannu H, et al. Sequencing of TGF-beta pathway genes in familial cases of intracranial aneurysm. Stroke. 2009;40:1604–1611. doi: 10.1161/STROKEAHA.108.540245. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.