Abstract
Mucopolysaccharidoses (MPS) are severe inherited metabolic disorders from the group of lysosomal storage diseases. They are caused by deficiency in the activity of enzymes involved in the degradation of glycosaminoglycans (GAGs) and resultant accumulation of these compounds in the cells of patients. Although enzyme replacement therapy has become available for some MPS types (MPS I, MPS II and MPS VI), this treatment is not efficient when neurological symptoms occur, especially in MPS III (Sanfilippo disease). Recent studies indicated that substrate reduction therapy (SRT) may be an effective option for the treatment of neurodegenerative lysosomal storage diseases, including MPS III. However, previous attempts to SRT for MPS III focused on the use of non-specific inhibitors of GAG synthesis. Thus, we aimed to use the small interfering RNA (siRNA) procedure to control expression of particular genes, whose products are involved in GAG synthesis. In this report we show that, in MPS IIIA fibroblasts, we were able to reduce mRNA levels of four genes, XYLT1, XYLT2, GALTI and GALTII, whose products are involved in GAG synthesis. This decrease in levels of transcripts corresponded to a decrease in levels of proteins encoded by them. Moreover, efficiency of GAG production in these fibroblasts was considerably reduced after treatment of the cells with siRNA. These results indicate that efficient reduction of GAG synthesis may be achieved by the use of siRNA.
Keywords: interference RNA, siRNA, Sanfilippo disease
Introduction
Mucopolysaccharidoses (MPS) are inherited metabolic diseases caused by mutations in genes coding for enzymes involved in the degradation of glycosaminoglycans (GAGs).1 Storage of GAGs in cells of patients suffering from MPS results in a progressive damage of the affected tissues, including the heart, respiratory system, bones, joints and, in some cases, the central nervous system (CNS).1 The disease is usually fatal, with average expected life span of one or two decades.1
Depending on the deficient enzyme, there are 11 types and subtypes of MPS.1 Currently, enzyme replacement therapy (ERT), based on an intravenous infusion of an active, recombinant form of a deficient enzyme, is available for treatment of only three of them, MPS I, MPS II and MPS VI.2 This therapy is helpful in managing many somatic symptoms. However, neurological symptoms developed because of GAG storage and secondary accumulation of gangliosides in the CNS cannot be managed by ERT owing to an inefficient delivery of proteins through the blood–brain barrier. The CNS dysfunction-related symptoms occur in some MPS I patients (subtype MPS IH), most of MPS II and MPS VII patients, and all MPS III patients,1 in which they are especially severe.
Sanfilippo disease (MPS III) is a group of four conditions (MPS IIIA, B, C and D) showing similar clinical symptoms and characterized by lysosomal storage of heparan sulfate (HS), one of GAGs.1 This condition is associated with severe learning difficulties and behavioral disturbances, and with milder somatic involvement relative to other MPS types. In most affected patients the progressive nature of the disease leads to death in the second (or rarely third) decade of life.1 As this disorder primarily affects the CNS, most attempts to cure the patients have not been successful and the best that could be offered was palliative or symptomatic care.
Recent studies showed that reduction in the efficiency of substrate (GAG) synthesis by small molecules, either genistein (5, 7-dihydroxy-3-(4-hydroxyphenyl)-4H-1-benzopyran-4-one) or rhodamine B ([9-(2-carboxyphenyl)-6-diethylamino-3-xanthenylidene]-diethylammonium chloride), has led to a substantial decrease in lysosomal storage in human and mouse cells.3, 4 Moreover, treatment of MPS IIIA mice with rhodamine B resulted in a behavioral improvement,5 and pilot clinical studies with the use of a genistein-rich extract for treatment of MPS IIIA and MPS IIIB human patients showed statistically significant improvement is all tested parameters, including cognitive functions.6 The improvement in the CNS functions in both mice and humans treated with inhibitors of GAG synthesis (either rhodamine B or genistein) were proposed to be due to a possibility of crossing the blood–brain barrier by these small molecules, and indicated that impairment of substrate production may be an effective therapeutic option for patients suffering from neuronopathic forms of lysosomal storage diseases.7 On the other hand, both genistein and rhodamine B are not specific inhibitors of GAG synthesis, and they have other biological functions.8, 9 Therefore, we aimed to develop a method for specific inhibition of GAG synthesis.
The RNA interference (RNAi) is a mechanism for selective silencing of expression of a particular gene by specific degradation of corresponding mRNA.10 The ability of synthetic small interfering RNA (siRNA) to silence genes in vivo, through the RNAi mechanism, has made them particularly well suited as drugs.11 In fact, development of therapeutics using siRNA is being advanced rapidly, including efforts to construct new selective drugs for cancer, allergic diseases, viral infections, neurological diseases and others.10 The major challenge in the development of real therapeutics based on siRNA seemed to consist of effective delivery of RNA molecules to the target tissue or organ. However, recent developments in this field provided a hope that this problem may be overcome.12 In fact, there are examples of several different clinical trials with the use of siRNA as therapeutics, and more clinical studies are planned for the near future.
GAG chain formation involves the following events: (i) chain initiation by the transfer of xylose residue to certain serine in the core protein, (ii) assembly of the tetrasaccharide linkage region, (iii) elongation by alternate addition of D-glucuronic acid (GlcUA) and N-acetyl-D hexosamine (GlcNAc for HS or GalNAc for chondroitin sulfate (CS) and dermatan sulfate (DS)) units to the linker tetrasaccharide. Finally, there are multiple sulfation and epimerization steps along the GAG chains.13
The assembly of CS/DS sulfate and HS/heparin GAG chains of proteoglycans (PGs) is initiated by the formation of a GlcA-β1,3-Gal-β1,3-Gal-β1,4-Xyl-β1-O-tetrasaccharide, which is linked to serine residues of a core protein. The assembly process involves the sequential transfer of monosaccharide residues from the corresponding nucleotide sugars starting at the reducing end. This step is initiated by β-D-xylosyltransferase (XYLT) that catalyzes transfer of a xylose (Xyl) to a serine residue. Two variants of O-xylosyltransferases (XYLT1 and XYLT2) are responsible for the initiation of GAG biosynthesis. XYLT1 and XYLT2 differ in terms of acceptor specificity and tissue distribution. In the next step, two galactose (Gal) residues are attached in reactions catalyzed by β1,4-galactosyltransferase 7 (GALTI) and β1,3-galactosyltransferase 6 (GALTII). GALTI (β4GALT7, a member of β1,4-galactosyltransferases family) catalyzes the transfer of galactose to the xylose residue (Galβ1,4Xyl). GALTII (β3GALT6) catalyzes the formation of Galβ1,3Gal linkage; therefore, it belongs to the family of β1,3-galactosyltransferases. The last step in the initiation process of GAG synthesis is catalyzed by the β1,3-glucuronosyltransferase (GLCATI), which transfers the glucuronic acid (GlcA) to the terminal Gal residue of the primer.13
After completion of the synthesis of the linkage tetrasaccharide sequence, polymerization of disaccharide-repeating units occurs by the alternate transfer of GlcA and GalNAc (for CS/DS) or GlcA and GlcNAc (for HS/heparan). The transfer of GalNAc or GlcNAc to the linkage oligosaccharide, by the action of β1,4-N-acetylgalactosaminyltransferase I (GALNACT I) or EXTL2, is the first step for mature CS/DS or HS/heparin formation. The elongation of HS/heparin chains are mediated by EXT1 and EXT2 glycosyltransferases. The polymerization reactions of CS chains are carried out by chondroitin-synthesizing enzymes, chondroitin synthase-1 (CHSY-1), chondroitin synthase-2 (CHSY-2), chondroitin sulfate synthase-3 (CSS-3) and chondroitin GalNAc transferases 1 and 2.13
The aim of this study was to impair the synthesis of GAGs by siRNA-mediated silencing of genes coding for enzymes involved in the initial steps of HS and DS synthesis. If successful, such a method could be potentially considered in the future for treatment of patients suffering from MPS, especially from those types for which no treatment is currently available and which involve the CNS dysfunction, such as MPS III (Sanfilippo disease).
Materials and methods
Cell cultures of MPS IIIA fibroblasts
Skin fibroblast lines were initiated from the forearm skin biopsies obtained from MPS IIIA patients. The patients were diagnosed with Sanfilippo A disease (MPS IIIA) on the basis of standard biochemical and enzymatic assays for levels of urinary GAGs and activity of lysosomal hydrolases. Fibroblasts were cultured in Dulbecco's modified Eagle's medium (DMEM) containing 10% fetal bovine serum (FBS), antibiotic antimycotic solution (Sigma-Aldrich, St Louis, MO, USA) at 37°C in a humidified atmosphere of 5% CO2.
Gene silencing with siRNA
Mucopolysaccharidoses IIIA cells were seeded into a 6-well tissue culture plate 24 h before transfection. Then, the cells were transfected with the siRNAs (25 nM of each) using the HiPerFect transfection reagent (Qiagen GmbH, Hilden, Germany) according to the manufacturer's instructions. Pre-designed siRNAs (chemically synthesized siRNAs that were designed for maximum potency and specificity by using an in silico algorithm) for the human XYLT1 gene (siRNA ID# 112165; 5′-GCAUCAUGCUACCAAUCUGtt-3′ (sense) and 5′-CAGAUUGGUAGCAUGAUGCtt-3′ (antisense), siRNA ID# 29873; 5′-GGAUGGCUACUUUUCUCAUtt-3′ (sense) and 5′-AUGAGAAAAGUAGCCAUCCtg-3′ (antisense)), XYLT2 gene (siRNA ID# 112168; 5′-CCUGUAUUUCUAUGACGACtt-3′ (sense) and 5′-GUCGUCAUAGAAAUACAGGtg-3′ (antisense), siRNA ID# 212974; 5′-CGAGCAGCACUUCUUUUACtt-3′ (sense) and 5′-GUAAAAGAAGUGCUGCUCGtg-3′ (antisense)), GALTI (B4GALT7) gene (siRNA ID# 19329; 5′-GGAGCUGGACUAUGGCUUUtt-3′ (sense) and 5′-AAAGCCAUAGUCCAGCUCCtc-3′ (antisense) , siRNA ID# 111770; 5′-GCAACAGCACGGACUACAUtt-3′ (sense) and AUGUAGUCCGUGCUGUUGCtg-3′ (antisense)) and GALTII (B3GALT6) gene (siRNA ID# 112321; 5′-GGAAAGACUUUUAUGUCAGtt-3′ (sense) and 5′-CUGACAUAAAAGUCUUUCCtt-3′ (antisense), siRNA ID# 112322; 5′-CCCAUUGUUUCUUUCUGAAtt-3′ (sense) and 5′-UUCAGAAAGAAACAAUGGGtt-3′ (antisense)) were purchased from Applied Biosystems/Ambion (Austin, TX, USA). AllStars Negative Control siRNA (Qiagen GmbH) served as a control for all siRNA transfections and mock control indicates the use of the transfection reagent only. Stability of siRNA was tested by treating the cells with labeled (fluorescent) oligonucleotide, non-complementary to any human gene (negative control), for 4, 24, 48 and 72 h, and measuring the fluorescence. The fluorescence level was stable for 48 h and decreased by about 30% at 72 h. The efficiency of transfection was estimated, and we found that the optimal siRNA concentration was 25 nM, with the range of effective transfection at concentrations between 10 and 50 nM. The expression level of XYLT1, XYLT2, GALTI (B4GALT7) and GALTII (B3GALT6) genes was analyzed by quantitative real-time PCR at 48 h after transfection. In some experiments, a second transfection was carried out at this time, and cells were cultured for next the 24 h before the analysis.
Quantitative real-time PCR
To examine siRNA-mediated knockdown of GALTI (B4GALT7) and GALTII (B3GALT6) expression, cDNA was prepared from transfected cells using the FastLane Cell cDNA kit (Qiagen GmbH), according to the manufacturer's instructions. To examine siRNA-mediated knockdown of XYLT1 and XYLT2 expression, total RNA was isolated from transfected cells using High Pure RNA Isolation Kit (Roche Applied Science, Indianapolis, IN, USA) according to the manufacturer's instructions. Total RNA (1 μg) was reverse transcribed into cDNA using Transcriptor First Strand cDNA Synthesis Kit (Roche Applied Science) according to the manufacturer's instructions. Quantification of mRNA expression was carried out using LightCycler FastStart DNA Master SYBR Green I (Roche Applied Science) with the LightCycler detection system (Roche Applied Science). Primers used for specific amplification of the target genes were as follows: RT2 qPCR Primer Assay for Human XYLT1 and XYLT2 (SABiosciences, Frederick, MD, USA), and QuantiTect Primer Assays for Human GALTI (B4GALT7), GALTII (B3GALT6) and GAPDH (Qiagen GmbH). The parameters of real-time PCR amplification were as follows: 15 min at 95°C, 45 cycles for 10 s at 95°C, 5 s at 55°C and 10 s at 72°C. The 2−ΔΔct method was used to determine the relative gene transcript levels after normalization to the reference gene coding for glyceraldehyde-3-phosphate dehydrogenase (GAPDH).14
Estimation of GAG synthesis efficiency
GAG synthesis was monitored by measuring incorporation of 35S from a radioactive sodium sulfate into PG. MPS IIIA cells, in passages from 2 to 4, were plated in a 24-well tissue culture plate at 24 h before transfection in DMEM, 10% FBS, at 37°C in a humidified atmosphere of 5% CO2. Then, the cells were transfected with the siRNAs (25 nM of each) using the HiPerFect transfection reagent (Qiagen GmbH) according to the manufacturer's protocol. The transfection was repeated after 48 h, and the cells were labeled with [35S]sodium sulfate (20 μCi/ml) for 24 h in DMEM, 10% FBS, at 37°C in a humidified atmosphere of 5% CO2. After this incubation, the cells were washed four times with Dulbecco's phosphate-buffered saline (PBS) and were subjected to papain digestion (0.03% papain in 0.1 M sodium acetate, pH 7.0). To evaluate [35S]sulfate incorporation, radioactivity of a portion (0.1 ml) of the sample was measured in a scintillation counter. Values of [35S]sulfate incorporation were normalized to the DNA content, measured spectrophotometrically using PicoGreen dsDNA Quantitation (Molecular Probes-Invitrogen, Carlsbad, CA, USA).
SDS-polyacrylamide gel electrophoresis and western blot analyses
Mucopolysaccharidoses IIIA cells were seeded into 6-well tissue cultured plates. After 24 h, the cells were transfected as described above. The cells were incubated for 48–72 h before western blotting analysis. After incubation, the cells were harvested by scraping in PBS, and protein lysates from cells were prepared in the lysis buffer (50 mM Tris–HCl pH 8.0, 150 mM NaCl, 1% TX-100, 0.5% sodium deoxycholate and 0.1% SDS), supplemented with protease inhibitor cocktail and phosphatase inhibitor cocktail (Roche Applied Science). In all, 60 μg of each sample were separated in a 4–10% polyacrylamide gel (MP Biomedicals, Solon, OH, USA) and transferred onto Immobilon-FL Transfer membrane (Millipore Corporation, Bedford, MA, USA). After the blocking reaction with TBS containing 5% nonfat dried milk and 0.1% Tween 20, for 90 min at room temperature, membranes were incubated with the following primary antibodies: rabbit polyclonal antibodies anti-B4GALT7 (Proteintech Group, Inc. Chicago, IL, USA), rabbit polyclonal antibodies anti-XYLT1 (Atlas Antibodies AB, Stockholm, Sweden), rabbit polyclonal antibodies anti-XYLT2 (Atlas Antibodies AB), mouse monoclonal antibodies anti-B3GALT6 (M06), clone 3E5 (Abnova, Taipei, Taiwan). The incubation was conducted at room temperature for 90 min or overnight at 4°C, and the following secondary antibodies, conjugated to horseradish peroxidase, were added: goat anti-rabbit IgG (Sigma-Aldrich) or goat anti-mouse IgG (H&L) (Abnova, Taipei, Taiwan). After incubation for 90 min at room temperature, membranes were developed with Western Lighting chemiluminescence reagent Plus (PerkinElmer Life Sciences, Waltham, MA, USA). The same blots were incubated with monoclonal anti-GAPDH–peroxidase antibodies (Sigma-Aldrich) to control protein load.
Statistical analysis
Comparison of data presented as mean ±SE was carried out by Student's t-test (unpaired). Percent reduction of individual protein expression was measured by densitometric analysis and is presented as a mean of three independent experiments. P<0.05 was considered significant, and P<0.001 was considered highly significant.
Results
siRNA-mediated decrease in mRNA levels
Mucopolysaccharidoses IIIA fibroblasts were cultured in vitro, and treated with either control siRNA (AllStars Negative Control siRNA, from Qiagen), which should not silence any human genes, or siRNAs specific for XYLT1, XYLT2, GALTI (B4GALT7) and GALTII (B3GALT6) genes. Following 48-h incubation after cell transfection, levels of mRNAs were quantified by real-time PCR and estimated relative to the GAPDH mRNA level (used as an internal standard).
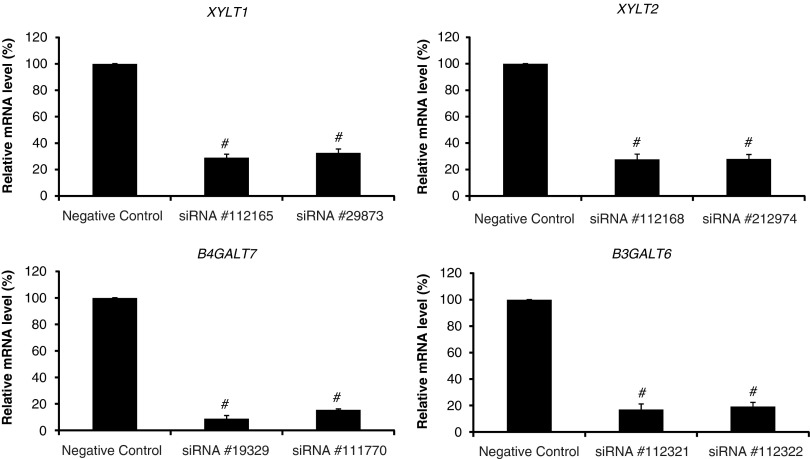
We found that siRNAs, specific for certain genes, caused a significant decrease in the mRNA levels of genes, XYLT1, XYLT2, GALTI (B4GALT7) and GALTII (B3GALT6), respectively (Figure 1). In all cases, two different siRNAs were effective in mRNA depletion and their efficiency was quite similar. Generally, mRNA amounts in cells treated with siRNAs were at the level of 20–40% of that measured in control samples, although siRNAs specific for GALTI (B4GALT7) were the most effective (Figure 1). These results indicate that by using the pre-designed siRNAs from Applied Biosystem/Ambion, we were able to downregulate the expression of human XYLT1, XYLT2, GALTI (B4GALT7) and GALTII (B3GALT6) genes at the level of mRNA amount.
Figure 1.
siRNA-mediated silencing of XYLT1, XYLT2, GALTI (B4GALT7) and GALTII (B3GALT6) genes. Fibroblasts were transfected with the indicated siRNAs as described in the experimental procedures. After cultivation for 48 h, levels of mRNAs were analyzed by quantitative real-time PCR. The presented values are the means of three independent experiments with corresponding SD. ‘Negative control' represents results with AllStars Negative Control siRNA (see experimental procedures for details). #P<0.0001 compared with negative-control siRNA.
Protein levels in siRNA-treated cells
The obvious question that must be asked in experiments with gene silencing by using siRNA is whether a decrease in mRNA level causes also a significant decrease in the amount of corresponding protein. The efficiency of protein synthesis may depend on various factors, such as efficiency of translation, protein stability and others. Therefore, we measured the levels of XYLT1, XYLT2, GALTI (B4GALT7) and GALTII (B3GALT6) proteins in cells transfected with siRNAs dedicated to silence corresponding genes. After protein isolation from tested cell cultures, amounts of proteins were estimated by western blotting, using specific antibodies for each protein, and the results were calculated relative to GAPDH level, which should be theoretically constant under these experimental conditions (in fact, differences in GAPDH levels were small, if any, in all experiments).
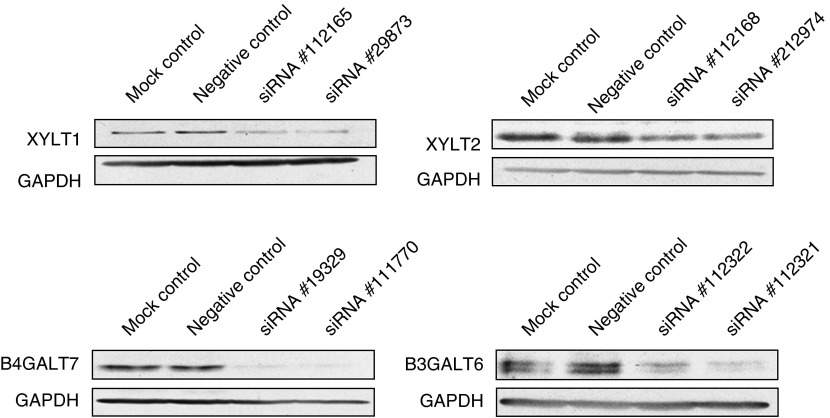
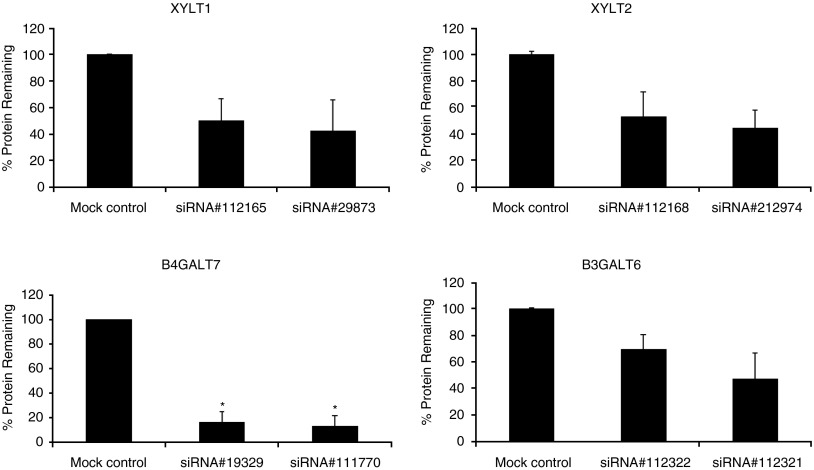
We found that levels of XYLT1, XYLT2, GALTI (B4GALT7) and GALTII (B3GALT6) proteins (estimated by western blotting) decreased significantly in cells treated with siRNAs specific for corresponding genes (Figure 2). The blots were quantified densitometrically, and the results indicated that protein levels dropped, depending on the protein, from 30 to 87% relative to the negative control (Figure 3). Therefore, we conclude that these siRNAs cause not only a decrease in the mRNA levels, but also impair syntheses of enzymes involved in the first stages of GAG production.
Figure 2.
Levels of XYLT1, XYLT2, GALTI (B4GALT7) and GALTII (B3GALT6) proteins, relative to the level of GAPDH (internal control) in fibroblasts transfected with indicated siRNAs. After transfection and further cultivation for 48 h, levels of mRNAs were analyzed by western blotting, as described in the experimental procedures. ‘Mock control' indicates conditions under which only a buffer was used in transfection-like experiments (without any siRNA), and ‘negative control' represents results with AllStars negative-control siRNA (see experimental procedures for details).
Figure 3.
Quantitation of levels of XYLT1, XYLT2, GALTI (B4GALT7) and GALTII (B3GALT6) proteins in cells treated with siRNA. Results of experiments exemplified in Figure 2 were quantified densitometrically using Quantity One software (Bio-Rad). Indicated values are means of three experiments ±SD. *P<0.05 compared with mock control.
Effects of gene silencing on GAG synthesis
As the aim of this study was to impair GAG synthesis by depleting enzymes involved in this process after transfection of cells with specific siRNAs, we have measured incorporation of [35S]sulfate into PG in siRNA-transfected MPS III fibroblasts. The results were normalized to the DNA content, which was assumed to be constant in normally growing cells.
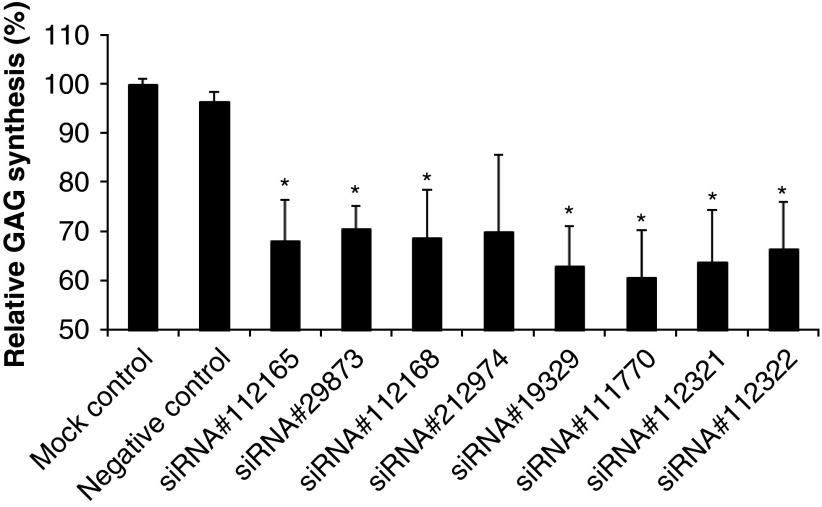
We found that transfection of the cells with siRNAs specific for genes coding for XYLT1, XYLT2, GALTI (B4GALT7) and GALTII (B3GALT6) proteins resulted in considerable impairment in GAG synthesis (Figure 4). Therefore, we conclude that by using siRNA that cause silencing of genes coding for proteins involved in the first stages of GAG synthesis, it is possible to reduce the efficiency of the latter process significantly.
Figure 4.
GAG synthesis in fibroblasts transfected with indicated siRNAs. Fibroblasts were transfected with indicated siRNAs, and after 48 h cultivation the transfection was repeated and cultivation was prolonged for another 24 h. GAG synthesis was estimated by measuring the incorporation of 35S as described in the experimental procedures. The presented values are the means of three independent experiments with corresponding SD. siRNAs for XYLT1 (siRNA ID# 112165, siRNA ID# 29873), XYLT2 (siRNA ID# 112168, siRNA ID# 212974), GALTI (B4GALT7) (siRNA ID# 19329, siRNA ID# 111770) and GALTII (B3GALT6) (siRNA ID# 112321, siRNA ID# 112322) genes were used. ‘Mock control' indicates conditions under which only a buffer was used in transfection-like experiments (without any siRNA), and ‘negative control' represents results with AllStars Negative Control siRNA (see experimental procedures for details). *P<0.05 compared with the no siRNA control.
Discussion
Although treatment of somatic symptoms of the three MPS types (MPS I, MPS II and MPS VI) by ERT has been established and introduced to medical practice,2 there is still a lack of efficient and approved treatment for the neuronopathic forms of MPS and other LSD.12 Sanfilippo disease (MPS III) is a special case of MPS as this disease manifests mostly by severe neurological symptoms, which currently cannot be managed, apart from a palliative care.1
Recent studies indicated that reduction of the substrate (GAG, or more precisely HS) synthesis may provide a way for treatment of MPS III patients.7 GAG synthesis could be reduced by the use of either genistein3 or rhodamine B,4, 5 compounds that were effective, but which have also other biological functions. Therefore, it seemed reasonable to look for a more specific method for reduction of GAG synthesis efficiency.
In this report, we show that siRNA-mediated silencing of genes coding for XYLT1, XYLT2, GALTI (B4GALT7) and GALTII (B3GALT6) enzymes resulted in a decrease in the levels of the gene products and impairment of GAG synthesis in transfected cells. It is worth noting that reductions in the levels of mRNAs, proteins and GAG synthesis were obtained in the experiments conducted, due to technical and methodological reasons, for relatively short times (48–72 h). This seems to be promising in the light of positive effects observed in in vitro studies,3, 4 in experiments on animals5 and in human clinical studies,6 when GAG production was decreased only a few times by prolonged treatment with either rhodamine B4, 5 or genistein.3, 6
The efficiency of GAG synthesis reduction determined in this study was efficient under conditions of partial silencing of any gene coding for an enzyme involved in the first stages of GAG production (Figure 3). Generally, more efficient reduction in the level of mRNA and particular protein corresponded to less effective GAG synthesis, which can be exemplified in experiments with silencing of GALTI (B4GALT7); the use of specific siRNAs resulted in the most effective decrease in the mRNA level, the protein level and GAG synthesis among all the tested genes and their products (Figures 1, 2, 3 and 4).
The main problem with the treatment of MPS III and other neuronopathic MSP types and LSDs is delivery of the specific drugs to the CNS, especially to the brain. This problem also concerns potential treatments with the use of the siRNA methodology. Moreover, for the use of any therapy based on RNA, one must consider a low stability of this nucleic acid in vivo. Nevertheless, recent achievements in the field of the use of this method in medicine are encouraging, and it seems that effective treatment of neurological diseases with siRNA-based drugs may be real in the near future.12, 15
In the accompanying paper, Kaidonis et al16 showed an effective RNAi-mediated silencing of EXTL2 and EXTL3 genes, whose products operate in the stages of HS synthesis downstream to those catalyzed by XYLT1, XYLT2, GALTI (B4GALT7) and GALTII (B3GALT6) enzymes. Impaired expression of EXTL2 and EXTL3 caused a reduction of GAG synthesis.16 These results corroborate our conclusions presented above.
If one considers the reduction in expression of gene(s) coding for enzyme(s) involved in HS synthesis as a therapeutic option for neuronopathic forms of MPS, it would be necessary to chose appropriate target(s) to avoid serious side effects. In fact, it was reported that mutations in exostosin 1 (EXT1), exostosin 2 (EXT2),17, 18, 19 N-deacetylase/N-sulfotransferase 1 (NDST1), N-deacetylase/N-sulfotransferase 2 (NDST2),20, 21, 22 heparan sulfate 2-O-sulfotransferase (HS2ST), heparan sulfate 3-O-sulfotransferase-1 (HS3ST1),23, 24, 25, 26 heparan sulfate 6-O-sulfotransferase-1 (HS6ST1)27 and heparan sulfate C5 epimerase (GLCE)28 genes caused serious defects in mice and/or humans. Therefore, a significant decrease in expression of one of these genes might be potentially dangerous for patients. To our knowledge, no reports indicating serious medical problems caused by dysfunction of genes coding for XYLT1, XYLT2, GALTI (B4GALT7) and GALTII (B3GALT6) were published to date. In this light, these genes might serve as attractive targets for a putative siRNA-mediated therapy of MPS. Furthermore, the corresponding enzymes catalyze the first steps in HS synthesis, thus, such a putative therapy might be potentially effective.
It is clear that further in vivo studies on animal (most probably mouse) models should be carried out before one can consider the siRNA-mediated treatment as a therapy for MPS. If such studies are planned, we assume that the doses and frequencies of siRNA application should be similar to those used in the experiments that are already conducted with mouse models of other diseases, for example, three injections of 50 μl of 2 μg/μl siRNA solution spaced over 1 week,29 single injection of 20–100 μg of siRNAs in 200 μl of the mixture,30 or three injections of 50 μg siRNA dissolved in 1 ml PBS in the intervals of 8 and 16 h.31
The question remains how the reduction of HS synthesis may influence clinical symptoms of MPS patients. This question may be partially answered by analysis of results of already published studies on the use of GAG synthesis inhibitor, genistein. In vitro experiments indicated that 10–30 μM genistein can reduce GAG synthesis three to four times.3, 32 The amounts of this compound administered to achieve similar levels in MPS III patients' plasma resulted in the improvement in all tested parameters, including cognitive functions, during an open-label pilot clinical study.6, 7 Therefore, we speculate that levels of the decrease in HS synthesis reported here (Figure 4) are encouraging in the light of possible further in vivo studies.
Acknowledgments
We are grateful to Xenia Kaidonis and Dr Sharon Byers for sharing their results before publication, and for discussions. This work was supported by the Ministry of Science and Higher Education of Poland (project Grant no. 3631/B/P01/2007/33 to JJB.). The authors express their special thanks to the Polish MPS Society for purchasing and providing the LightCycler detection system (Roche Applied Science).
References
- Neufeld EF, Muenzer J.The mucopolysaccharidosesIn: Scriver CR, Beaudet AL, Sly WS, Valle D (eds): The Metabolic and Molecular Bases of Inherited Disease New York: McGraw-Hill Co; 20013421–3452. [Google Scholar]
- Heese BA. Current strategies in the management of lysosomal storage diseases. Semin Pediatr Neurol. 2008;15:119–126. doi: 10.1016/j.spen.2008.05.005. [DOI] [PubMed] [Google Scholar]
- Piotrowska E, Jakóbkiewicz-Banecka J, Barańska S, et al. Genistein-mediated inhibition of glycosaminoglycan synthesis as a basis for gene expression-targeted isoflavone therapy for mucopolysaccharidoses. Eur J Hum Genet. 2006;14:846–852. doi: 10.1038/sj.ejhg.5201623. [DOI] [PubMed] [Google Scholar]
- Roberts AL, Thomas BJ, Wilkinson AS, Fletcher JM, Byers S. Inhibition of glycosaminoglycan synthesis using rhodamine B in a mouse model of mucopolysaccharidosis type IIIA. Pediatr Res. 2006;60:309–314. doi: 10.1203/01.pdr.0000233037.00707.da. [DOI] [PubMed] [Google Scholar]
- Roberts AL, Rees MH, Klebe S, Fletcher JM, Byers S. Improvement in behaviour after substrate deprivation therapy with rhodamine B in a mouse model of MPS IIIA. Mol Genet Metab. 2007;92:115–121. doi: 10.1016/j.ymgme.2007.06.016. [DOI] [PubMed] [Google Scholar]
- Piotrowska E, Jakóbkiewicz-Banecka J, Tylki-Szymańska A, et al. Genistin-rich isoflavone extract in substrate reduction therapy for Sanfilippo syndrome: an open-label, pilot study in 10 pediatric patients. Curr Ther Res Clin Exp. 2008;69:166–179. doi: 10.1016/j.curtheres.2008.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakóbkiewicz-Banecka A, Węgrzyn A, Węgrzyn G. Substrate deprivation therapy: a new hope for patients suffering from neuronopathic forms of inherited lysosomal storage diseases. J Appl Genet. 2007;48:383–388. doi: 10.1007/BF03195237. [DOI] [PubMed] [Google Scholar]
- Karstens T, Kobs K. Rhodamine B and rhodamine 101 as reference substances for fluorescence quantum yield measurements. J Phys Chem. 1980;84:1871–1872. [Google Scholar]
- Szkudelska K, Nogowski L. Genistein – a dietary compound inducing hormonal and metabolic changes. J Steroid Biochem Mol Biol. 2007;105:37–45. doi: 10.1016/j.jsbmb.2007.01.005. [DOI] [PubMed] [Google Scholar]
- Kim D, Rossi J. RNAi mechanisms and applications. Biotechniques. 2008;44:613–616. doi: 10.2144/000112792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Y. Advances in the development of siRNA-based therapeutics for cancer. Idrug. 2008;11:572–578. [PubMed] [Google Scholar]
- Juliano R, Alam MR, Dixit V, Kang H. Mechanisms and strategies for effective delivery of antisense and siRNA oligonucleotides. Nucleic Acids Res. 2008;36:4158–4171. doi: 10.1093/nar/gkn342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prydz K, Dalen K. Synthesis and sorting of proteoglycans. J Cell Sci. 2000;113:193–205. doi: 10.1242/jcs.113.2.193. [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Method. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Smith RA, Miller TM, Yamanaka K, et al. Antisense oligonucleotide therapy for neurodegenerative disease. J Clin Invest. 2006;116:2290–2296. doi: 10.1172/JCI25424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaidonis X, Liaw WC, Roberts AD, Ly M, Anson D, Byers S.Gene silencing of EXTL2 and EXTL3 as a substrate deprivation therapy for heparan sulphate storing mucopolysaccharidoses Eur J Hum Genet 2009. E-pub ahead of print; doi: 10.1038/ejhg.2009.143 [DOI] [PMC free article] [PubMed]
- Lin X, Wei G, Shi Z, et al. Disruption of gastrulation and heparan sulfate biosynthesis in EXT1-deficient mice. Dev Biol. 2000;224:299–311. doi: 10.1006/dbio.2000.9798. [DOI] [PubMed] [Google Scholar]
- Zak BM, Crawford BE, Esko JD. Hereditary multiple exostoses and heparan sulfate polymerization. Biochim Biophys Acta. 2002;1573:346–355. doi: 10.1016/s0304-4165(02)00402-6. [DOI] [PubMed] [Google Scholar]
- Forsberg E, Kjellen L. Heparan sulfate: lessons from knockout mice. J Clin Invest. 2001;108:175–180. doi: 10.1172/JCI13561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ringvall M, Ledin J, Holmborn K, et al. Defective heparan sulfate biosynthesis and neonatal lethality in mice lacking N-deacetylase/N-sulfotransferase-1. J Biol Chem. 2000;275:25926–25930. doi: 10.1074/jbc.C000359200. [DOI] [PubMed] [Google Scholar]
- Forsberg E, Pejler G, Ringvall M, et al. Abnormal mast cells in mice deficient in a heparin-synthesizing enzyme. Nature. 1999;400:773–776. doi: 10.1038/23488. [DOI] [PubMed] [Google Scholar]
- Grobe K, Inatani M, Pallerla SR, Castagnola J, Yamaguchi Y, Esko JD. Cerebral hypoplasia and craniofacial defects in mice lacking heparan sulfate Ndst1 gene function. Development. 2005;132:3777–3786. doi: 10.1242/dev.01935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merry CL, Bullock SL, Swan DC, et al. The molecular phenotype of heparan sulfate in the Hs2st-/- mutant mouse. J Biol Chem. 2001;276:35429–35434. doi: 10.1074/jbc.M100379200. [DOI] [PubMed] [Google Scholar]
- Wilson VA, Gallagher JT, Merry CL. Heparan sulfate 2-O-sulfotransferase (Hs2st) and mouse development. Glycoconj J. 2002;19:347–354. doi: 10.1023/A:1025325222530. [DOI] [PubMed] [Google Scholar]
- Merry CL, Wilson VA. Role of heparan sulfate-2-O-sulfotransferase in the mouse. Biochim Biophys Acta. 2002;1573:319–327. doi: 10.1016/s0304-4165(02)00399-9. [DOI] [PubMed] [Google Scholar]
- Shworak NW, HajMohammadi S, de Agostini AI, Rosenberg RD. Mice deficient in heparan sulfate 3-O-sulfotransferase-1: normal hemostasis with unexpected perinatal phenotypes. Glycoconj J. 2002;19:355–361. doi: 10.1023/A:1025377206600. [DOI] [PubMed] [Google Scholar]
- Habuchi H, Nagai N, Sugaya N, Atsumi F, Stevens RL, Kimata K. Mice deficient in heparan sulfate 6-O-sulfotransferase-1 exhibit defective heparan sulfate biosynthesis, abnormal placentation, and late embryonic lethality. J Biol Chem. 2007;282:15578–15588. doi: 10.1074/jbc.M607434200. [DOI] [PubMed] [Google Scholar]
- Li JP, Gong F, Hagner-McWhirter A, et al. Targeted disruption of a murine glucuronyl C5-epimerase gene results in heparan sulfate lacking L-iduronic acid and in neonatal lethality. J Biol Chem. 2003;278:28363–28366. doi: 10.1074/jbc.C300219200. [DOI] [PubMed] [Google Scholar]
- Hickerson RP, Vlassov AV, Wang Q, et al. Stability study of unmodified siRNA and relevance to clinical use. Oligonucleotides. 2008;18:345–354. doi: 10.1089/oli.2008.0149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sioud M, Sørensen DR. Cationic liposome-mediated delivery of siRNAs in adult mice. Biochem Biophys Res Commun. 2003;312:1220–1225. doi: 10.1016/j.bbrc.2003.11.057. [DOI] [PubMed] [Google Scholar]
- Song E, Lee SK, Wang J, et al. RNA interference targeting Fas protects mice from fulminant hepatitis. Nat Med. 2003;9:347–351. doi: 10.1038/nm828. [DOI] [PubMed] [Google Scholar]
- Jakóbkiewicz-Banecka J, Piotrowska E, Narajczyk M, Barańska S, Wegrzyn G. Genistein-mediated inhibition of glycosaminoglycan synthesis, which corrects storage in cells of patients suffering from mucopolysaccharidoses, acts by influencing an epidermal growth factor-dependent pathway. J Biomed Sci. 2009;16:26. doi: 10.1186/1423-0127-16-26. [DOI] [PMC free article] [PubMed] [Google Scholar]