Abstract
Neurological pathology is characteristic of the mucopolysaccharidoses (MPSs) that store heparan sulphate (HS) glycosaminoglycan (gag) and has been proven to be refractory to systemic therapies. Substrate deprivation therapy (SDT) using general inhibitors of gag synthesis improves neurological function in mouse models of MPS, but is not specific to an MPS type. We have investigated RNA interference (RNAi) as a method of targeting SDT to the HS synthesising enzymes, EXTL2 and EXTL3. Multiple shRNA molecules specific to EXTL2 or EXTL3 were designed and validated in a reporter gene assay, with four out of six shRNA constructs reducing expression by over 90%. The three EXTL2-specific shRNA constructs reduced endogenous target gene expression by 68, 32 and 65%, and decreased gag synthesis by 46, 50 and 27%. One EXTL3-specific shRNA construct reduced endogenous target gene expression by 14% and gag synthesis by 39%. Lysosomal gag levels in MPS IIIA and MPS I fibroblasts were also reduced by EXTL2 and EXTL3-specific shRNA. Incorporation of shRNAs into a lentiviral expression system reduced gene expression, and one EXTL2-specific shRNA reduced gag synthesis. These results indicate that deprivation therapy through shRNA-mediated RNAi has potential as a therapy for HS-storing MPSs.
Keywords: mucopolysaccharidosis IIIA, EXTL2, EXTL3, substrate deprivation therapy, heparan sulphate
Introduction
The mucopolysaccharidoses (MPSs) are a group of inherited metabolic disorders that have in common a reduction or complete absence of a lysosomal enzyme required for intracellular glycosaminoglycan (gag) degradation.1 A total of 11 different enzyme deficiencies have been characterised and 7 of these involve an enzyme required for the degradation of heparan sulphate (HS). The buildup of undegraded HS in brain cells of affected MPS children results in a progressive neurological deterioration. Aggressiveness and hyperactivity result, and clinical management of affected children is difficult. Treatment of neurological symptoms has remained essentially palliative, despite the development of enzyme replacement therapies for the non-neuronal symptoms of MPS.2, 3, 4 Although there is recent evidence that supraphysiological doses of recombinant enzyme may promote transfer across the blood–brain barrier (BBB),5 the balance of evidence suggests that the BBB remains a significant obstacle to successful enzyme replacement therapy in a majority of MPS children. Alternate approaches that target the CNS are required to provide effective multi-tissue treatments for these disorders.
One approach is substrate deprivation therapy (SDT), which acts by inhibiting the synthesis of the substrate for the missing enzyme.6 This has been used successfully to treat Gaucher disease,7, 8, 9 and has been clinically available since 2002. For the MPS disorders, a number of SDT agents have been shown to inhibit gag synthesis in vitro10, 11, 12 and gag storage in vivo.12, 13, 14 Importantly, SDT using a gag synthesis inhibitor reduced neurological pathology in the MPS IIIA mouse. Thus, SDT has the potential to treat all sites of pathology in MPS. However, all chemical SDT agents tested reduce the synthesis of all gag types and are, thus, general inhibitors. As such, they will also inhibit the production of gag types that are not involved in a specific MPS disorder. RNA interference (RNAi) technology is one way of specifically targeting the synthesis of an individual gag type by reducing the expression of one or more of the glycosyl transferases responsible for gag synthesis.
Heparan sulphate is one of five gag types, and is synthesised in the Golgi apparatus on a serine residue located in the core protein of the parent proteoglycan molecule. A linkage region consisting of Xyl–Gal–Gal–GlcA is common to chondroitin and dermatan sulphate (DS) gags, as well as to HS.15 The first committed step in HS synthesis is the addition of N-acetylglucosamine (GlcNAc), which is catalysed by the N-acetylglucosaminyltransferase-I (GlcNAcT-I) enzymes, EXTL2 and EXTL3.16, 17 This is then followed by the addition of alternating glucuronic acid (GlcUA) and GlcNAc sugar subunits catalysed by N-acetylglucosaminyltransferase-II (GlcNAcT-II)17 and GlcUA/GlcNAc transferase18 enzymes of the EXT/EXTL family, including EXTL3. Further modifications can be made to the HS chain and include epimerisation, sulphation and deacetylation, all of which contribute to the gag's final structure and specific function.19, 20
In this study, RNAi was used to specifically target EXTL2 and EXTL3 expression. Multiple shRNAs were designed with specificity towards EXTL2 and EXTL3 (shEXTL2 and shEXTL3, respectively). All shRNAs directed at EXTL2 significantly reduced target gene expression in a reporter gene assay, reduced endogenous target gene expression, significantly decreased gag synthesis and reduced lysosomal storage of gag in an MPS IIIA cell line. One of the three shEXTL3s decreased endogenous EXTL3 expression and subsequent gag synthesis. Incorporation of shRNAs into a stable lentiviral expression system also resulted in reduction of target gene expression in a reporter gene assay.
Materials and methods
Design of shRNA oligos
Top and bottom strand oligos specific to the EXTL2 or EXTL3 target gene were designed using the Invitrogen BLOCK-iT RNAi designer (Invitrogen, Melbourne, Australia). Each top strand was designed to include a gene homologous region, a loop, an antisense sequence and 5′ overhangs for cloning into the pENTR/U6 expression vector (Table 1). A blast search against the human genome confirmed no homology between the oligo sequences and any other genes, including other EXTL genes. A control sequence was also generated that had no homology to any known gene sequence.
Table 1. shRNA and target gene homologous sequences.
| shRNA | Target gene homologous sequence (5′–3′) | Site of target sequence | Accession number |
|---|---|---|---|
| shEXTL2.1 | GCAGCAATTTCCTGATCAAAT | 788–808 | AF000416 |
| shEXTL2.2 | GCAAGACTTCAGGGATATTTG | 1057–1077 | AF000416 |
| shEXTL2.3 | GGTGTTGCCACATCTGCAAAC | 292–312 | AF000416 |
| shEXTL3.1 | GCTTTCTCTGGGAGACTTACT | 2139–2159 | AF001690 |
| shEXTL3.2 | GGTGGTGATGTTGACTTATGA | 2593–2613 | AF001690 |
| shEXTL3.3 | GCTCAGCTTCACGCTCTTTGT | 694–714 | AF001690 |
Six shRNA oligos were designed, each with sequence homology to a different target sequence within the EXTL2 or EXTL3 gene. The position of each shRNA within the target gene is specified.
Cloning
To make shEXTL and ctrlEXTL pENTR/U6 constructs, top and bottom oligos were annealed at 95 °C and ligated directly into the pENTR/U6 vector (Invitrogen), according to the manufacturers instructions. The resultant constructs were termed shEXTL2.1, shEXTL2.2, shEXTL2.3, shEXTL3.1, shEXTL3.2, shEXTL3.3 and ctrlEXTL. EXTL2 and EXTL3 sequences were amplified from cDNA derived from human skin fibroblast RNA and ligated into pGEM-Teasy. Fragments containing the shRNAs were generated by digestion with NotI/SpeI (EXTL2) or NotI/XhoI (EXTL3), gel purified and cloned into the psiCHECK-2 vector (Promega Corp., Madison, WI, USA). The resultant vectors were termed psiCHECK-2EXTL2 and psiCHECK-2EXTL3.
Tissue culture
293T cells and human MPS skin fibroblasts were maintained in Dulbecco's modified Eagle's medium supplemented with 10% foetal calf serum, 50 U/ml penicillin and 50 μg/ml streptomycin, at 37 °C with 5% CO2 and 90% humidity. Transfections were performed using the lipofectamine 2000 reagent as per the manufacturers instructions (Invitrogen). Mock transfections used the lipofectamine 2000 reagent only (that is, no DNA).
Dual luciferase reporter (DLR) assay
293T cells were seeded in a 96-well plate at a density of 6000 cells per well and incubated at 37 °C for 24 h, before transfection with 20 ng psiCHECK-2 target gene and 120 ng of the shEXTL pENTR/U6 constructs. After a further 24-h incubation at 37 °C, the cells were washed with PBS and lysed as per the manufacturers instructions (Promega Corp.). Luminescence was measured on a Veritas luminometer 1-sec post-injection using the DLR protocol (Promega Corp.).
Real-time PCR of endogenous EXTL gene expression
293T cells seeded at 5000 cells/cm2 in a six-well plate were mock transfected or transfected with shEXTL2, shEXTL3 or ctrlEXTL pENTR/U6 constructs, and incubated at 37 °C for 72 h. Total RNA was extracted using TRIzol as per reagent instructions (Invitrogen) and reverse transcribed into cDNA using the QuantiTect reverse transcription kit (Qiagen, Hilden, Germany). EXTL2 (forward primer 5′-CCTGAACTGGAAACCAATGCAG-3′, reverse primer 5′-CAGGAAATTGCTGCCAAACTG-3′) or EXTL3 (forward primer 5′-CCCGAATGTCACTGGTGGATC-3′, reverse primer 5′-GCTGTCCTTGTTGGAAGCACC-3′) expression levels were determined by real-time PCR on an ABI 7300 system from cDNA equivalent to 100 ng of total RNA and normalised to cyclophilin A (forward primer 5′-GGTTGGATGGCAAGCATGTG-3′, reverse primer 5′-TGCTGGTCTTGCCATTCCTG-3′). The fold change in gene expression was calculated using the 2−ΔΔCt method.21
gag synthesis
293T cells were seeded at a concentration of 5000 cells/cm2 and were transfected with shEXTL2.1, shEXTL2.2, shEXTL2.3, shEXTL3.2 or ctrlEXTL constructs or mock transfected (no DNA). At 48 h post-transfection, cells were incubated with 5 μCi/ml 35SO4 for 4 h. The medium was collected and cell layer extracted into 4 M GuHCl, 50 mM Na acetate buffer, pH 5.8, containing 10 mM Na2EDTA, 0.1 M amino hexanoic acid and 0.1% (v/v) Triton-X-100. Unincorporated 35SO4 was removed by size exclusion chromatography on Sepharose G25 equilibrated in 2 M GuHCL, 0.1 M Na2SO4, 0.05 M Tris and 0.5% (v/v) Triton-X-100, pH 7.5. Sulphate incorporation was normalised to protein concentration that was determined using the Bradford method.22
gag storage
Human MPS IIIA or MPS I skin fibroblasts were mock transfected or transfected with one of the shEXTL2.1, shEXTL2.2, EXTL2.3 or shEXTL3.2 constructs. After 24 h, 5 μCi/ml 35SO4 was added as above, before harvesting with 0.012% (w/v) trypsin, 0.002% (w/v) EDTA and loading onto a 30% Percoll gradient.12 The lysosomal fractions were identified by β-hexosaminidase assay and the results were normalised to protein.22
Subcloning of shRNAs into a lentiviral vector and virus production
pENTR/U6 shEXTL2.1, 2.2, 2.3 and 3.2 constructs were digested with SalI/XbaI to liberate a fragment containing the U6 promoter, shRNA sequence and RNA polymerase III transcription terminator. This fragment was then cloned into the lentiviral vector pHIV-1SDmPGKeYFP23 immediately 5′ of the Rev-response element. The resultant plasmids (termed pHIV-1shEXTL2.1, etc., to distinguish them from the pENTR/U6 constructs) were either used directly in the DLR assay or used to generate virus for cell transduction. Virus was produced and purified as previously described.24 293T cells were transduced with virus in the presence of 4 μg/ml polybrene,23 and gag synthesis measured after 7 days by 35SO4 incorporation as described above.
Statistical analysis
Significant differences between means were determined using a one-way analysis of variance with Tukey's HSD post-hoc test where appropriate (SigmaStat 3.0, SPSS Inc., Chicago, IL, USA).
Results
Validation of shRNAs
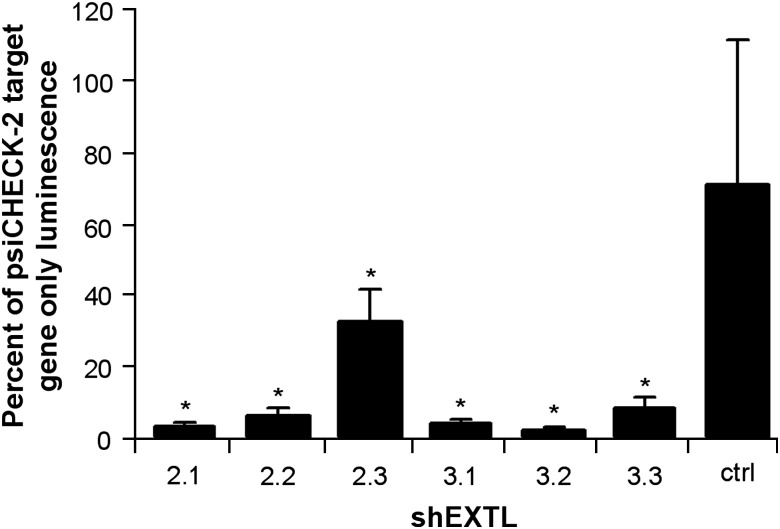
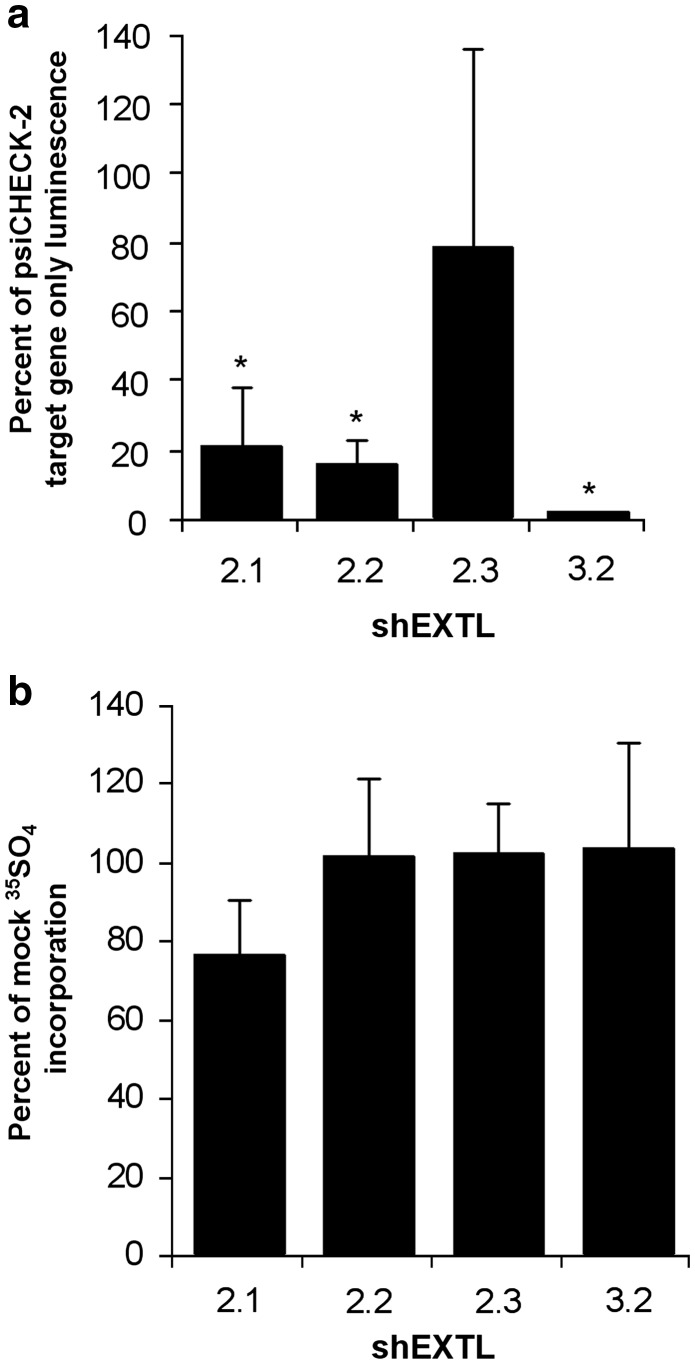
Before examining their effect on endogenous gene expression, the shEXTL constructs (Table 1) were tested for their ability to reduce target gene expression using a DLR assay. 293T cells were co-transfected with each of the pENTR/U6 shEXTL constructs and the corresponding psiCHECK-2EXTL2 or psiCHECK-2EXTL3 gene construct. A significant decrease in target gene expression by all six shEXTL constructs was observed compared with the psiCHECK-2 target gene only (Figure 1). Luminescence was reduced in the presence of shEXTL2.1, shEXTL2.2 and shEXTL2.3 by 97, 94 and 67%, respectively. Luminescence was also reduced in the presence of shEXTL3.1, shEXTL3.2 and EXTL3.3 by 90, 94 and 77%, respectively. The ctrlEXTL construct had no effect on target gene expression.
Figure 1.
Quantitative knockdown of EXTL2 and EXTL3. 293T cells were transfected with psiCHECK-2EXTL2 or psiCHECK-2EXTL3 only, or co-transfected with a psiCHECK-2 target gene construct and corresponding shEXTL or ctrlEXTL expressed from the pENTR/U6 vector. Control cells were mock transfected with lipofectamine 2000 reagent only. A DLR assay was conducted 24 h post-transfection, and results expressed as a percentage of psiCHECK-2 target gene expression. Results are the mean±standard deviation of n=10 replicates. *Significant difference from mock transfection P<0.001 (ANOVA, Tukey's HSD).
Knockdown of endogenous EXTL2 and EXTL3
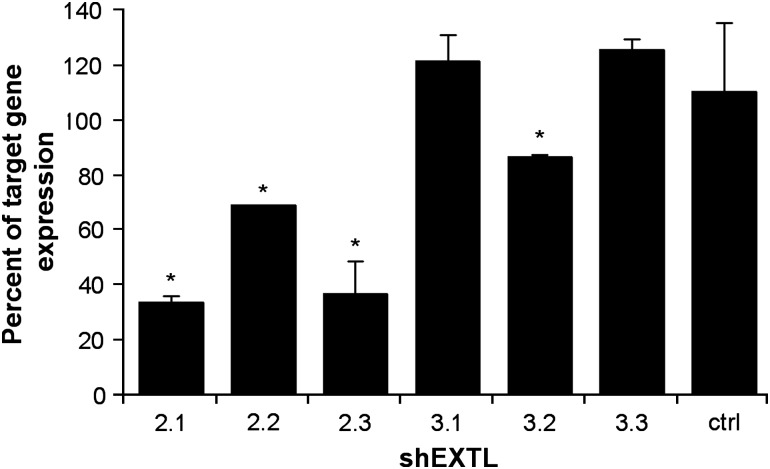
The ability of the shEXTL constructs to knockdown endogenous target gene expression was determined by real-time PCR 72 h after transfection of 293T cells. Only shEXTL2.1, shEXTL2.2, shEXTL2.3 and shEXTL3.2 reduced their target gene expression by 68, 32, 65 and 14%, respectively (Figure 2). The control shRNA construct did not alter target gene expression.
Figure 2.
Knockdown of endogenous EXTL2 and EXTL3. 293T cells were transfected with shEXTL2, shEXTL3 or ctrlEXTL pENTR/U6 constructs. After 72 h, total RNA was extracted and reverse transcribed to generate cDNA. EXTL2 or EXTL3 expression was determined by real-time PCR, and the fold change in expression was expressed as a percentage of the untransfected control and compared with a mock-transfected control (lipofectamine 2000 reagent only). Results are the mean±standard deviation of n=3 replicates. *Significant difference from mock transfection, P<0.001 (ANOVA, Tukey's HSD).
Reduction in gag synthesis and gag storage by EXTL2 and EXTL3 gene silencing
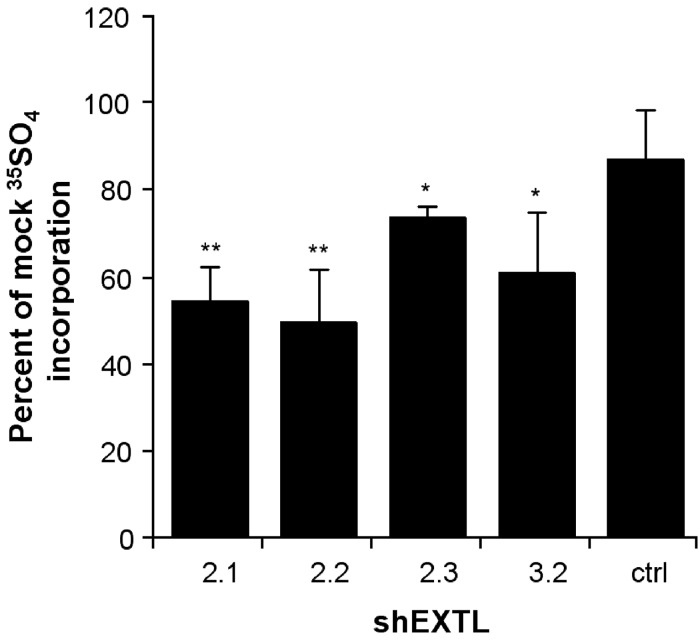
Gag synthesis was determined from the rate of 35SO4 incorporation in 293T cells 72 h after transfection with the shEXTL constructs that reduced endogenous gene expression. shEXTL2.1, shEXTL2.2, shEXTL2.3 and shEXTL3.2 constructs significantly reduced gag synthesis by 46, 50, 27 and 39%, respectively (Figure 3). Combined transfections of shEXTL2.1 with shEXTL2.2, or shEXTL2.1 with shEXTL3.2 did not result in any additional reduction in gag synthesis over that observed with shEXTL2.1 alone (data not shown).
Figure 3.
Reduction in gag synthesis. 293T cells were transfected with shEXTL2.1, shEXTL2.2, shEXTL2.3, shEXTL3.2 or ctrlEXTL expressed from the pENTR/U6 vector. A mock transfection consisting of lipofectamine 2000 reagent only served as a control. After 48 h, 5 μCi/ml 35SO4 was added and gag synthesis determined over 4 h. Results are the mean±standard deviation of three replicates, and are expressed as the percentage of the mock control. *Significant difference from mock transfection, P<0.05 (ANOVA, Tukey's HSD), **Significant difference from mock transfection, P<0.001 (ANOVA, Tukey's HSD).
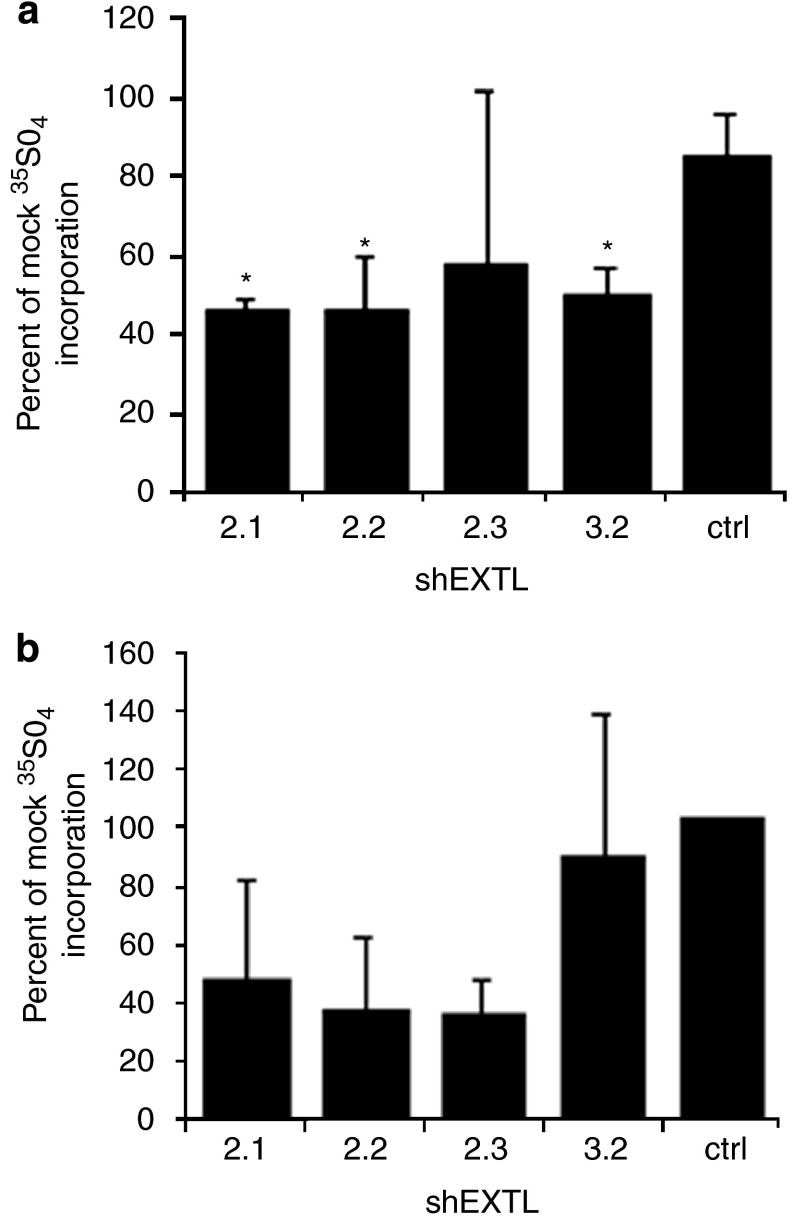
Lysosomal gag content was measured in human MPS IIIA or MPS I skin fibroblasts transfected with shEXTL2.1, shEXTL2.2, shEXTL2.3 or shEXTL3.2. A 55% reduction in gag storage was observed in MPS IIIA cells treated with shEXTL2.1, a 54% reduction in cells treated with shEXTL2.2 and a 50% reduction in cells treated with shEXTL3.2 (Figure 4a). A reduction in storage was also observed with shEXTL2.3, but this was not significant. Gag storage was also reduced in MPS I skin fibroblasts by shEXTL2.1, shEXTL2.2 and shEXTL3.2, but the reduction was not significant (Figure 4b).
Figure 4.
Reduction in gag storage. MPS IIIA (a) or MPS I (b) skin fibroblasts were transfected with shEXTL2.1, shEXTL2.2, shEXTL2.3 or shEXTL3.2. A mock transfection consisting of lipofectamine 2000 reagent only served as control. After 48 h, cells were harvested and the lysosomal fraction recovered after Percoll gradient centrifugation. Results are the mean±standard deviation of two experiments performed in duplicate and are expressed as the percentage of the mock control. *Significant difference from mock transfection, P<0.05 (ANOVA, Tukey's HSD).
Generation of a lentiviral construct encoding shRNAs
All of the above results were generated using transient expression of shRNAs from the pENTR/U6 vector. For stable expression of shRNAs suitable for in vivo testing, the shRNAs along with the U6 promoter and RNA polymerase III transcription terminator were subcloned into a lentiviral vector. Expression of the shRNA sequences from the lentiviral vector was confirmed in the DLR assay (Figure 5a). pHIV-1shEXTL2.1, pHIV-1shEXTL2.2 and pHIV-1shEXTL3.2 significantly reduced target gene expression by 80, 83 and 99%, respectively, a similar reduction to that observed after expression of the shRNAs from the pENTR/U6 vector (Figure 1). Although pHIV-1shEXTL2.3 also reduced EXTL2 expression, this was not significant (Figure 5a).
Figure 5.
Expression of shRNAs from a lentiviral vector. 293T cells were transfected with pHIV-1shEXTL2.1, pHIV-1shEXTL2.2, pHIV-1shEXTL2.3, pHIV-1shEXTL3.2 or pHIV-1ctrlEXTL, and a DLR assay was performed to determine whether shRNAs were expressed from the vector (a). 293T cells were also transduced with virus encoding the same shRNAs and the rate of gag synthesis measured 7 days after transduction (b). Results are the mean±standard deviation of three replicates and are expressed as the percentage of mock control. *Significant difference from mock transfection P<0.001 (ANOVA, Tukey's HSD).
Virus was produced as previously described24 and used to transfect 293T cells. The rate of gag synthesis was measured 7 days after transduction. Only cells transduced with virus expressing shEXTL2.1 displayed a reduction in gag synthesis; however, this was not significant (P=0.06) (Figure 5b).
Discussion
Substrate deprivation therapy is the first systemic therapy to clearly result in a measurable improvement in neurological function when administered to the MPS IIIA mouse.14 Improvements in speech, comprehension and general behaviour were also observed in MPS IIIA and MPS IIIB patients in a clinical trial of SDT.13 This type of therapeutic approach to the MPS disorders has the potential to address all major sites of pathology simultaneously. All of the SDT agents used to date10, 11, 12, 13, 14 are non-specific inhibitors of gag synthesis and are, therefore, not specific to a particular gag or MPS type. Although no side effects to SDT were observed in a 6-month in vivo trial in the MPS IIIA mouse,12, 14 there is still concern that reducing the synthesis of gags that are not involved in an MPS type, and whose metabolism is thus unaffected, may be detrimental. RNAi is a potential mechanism by which SDT can be focussed on an individual gag type by targeting the glycosyl synthases responsible for chain synthesis. In this study, we specifically targeted HS synthesis by inhibiting the GlcNAcT-1 activity encoded by EXTL2 and EXTL3, which is responsible for the initiation of HS chain synthesis, and the GlcUA/GlcNAc transferase activity encoded by EXTL3, which is responsible for HS chain elongation.16, 17
Multiple shRNAs with specificity towards EXTL2 or EXTL3 were designed, as knockdown efficacy is known to vary between molecules. The luciferase reporter gene assay showed that all shRNAs could be used by the cell to reduce expression of the EXTL2 or EXTL3 target genes. All three shRNA sequences designed towards EXTL2 resulted in a small decrease in endogenous gene expression, as did one of the shRNA sequences designed towards EXTL3. The larger size of the EXTL3 gene (2.76 kb compared with 0.99 kb for EXTL2), instability of the shRNA transcript, an inability of the cell's RNAi machinery to recognise and interact with the shRNAs, and difficulty recognising mRNA because of secondary structures in the complementary region of the transcript may all contribute to the differences observed between the expression levels of EXTL2 and EXTL3 in the presence of shRNAs.25
The four shRNA sequences (shEXTL2.1, shEXTL2.2, shEXTL2.3 and shEXTL3.2) that reduced endogenous target gene expression were investigated further for their ability to affect gag synthesis and storage. All resulted in a significant reduction in gag synthesis by 293T cells, whereas shEXTL2.1, shEXTL2.2 and shEXTL3.2 also significantly reduced HS gag storage in MPS IIIA fibroblasts. The same shRNAs were also tested in MPS I skin fibroblasts, which store DS gag in addition to HS gag. Reductions in overall gag storage were observed for MPS I fibroblasts treated with shEXTL2.1, shEXTL2.2 and shEXTL2.3; however, this was not significant. In addition, the efficacy of shEXTL2.3 and shEXTL3.2 differed from that observed in MPS IIIA fibroblasts. This difference in efficacy is not unexpected given that DS gag storage will not be affected by RNAi directed towards EXTL2 and EXTL3, which are specific for HS gag chain synthesis.
Inhibiting the expression of EXTL2 or EXTL3, thus, directs SDT towards those MPS that store HS gag. However, it will not distinguish between the HS-storing MPS. Absolute specificity of SDT using an RNAi approach, however, is possible for a number of the MPS disorders. For example, targeting the enzyme responsible for N-sulphation of HS (N-deacetylase/N-sulpho-transferase-1; NDST-1) would specifically reduce the storage material in MPS IIIA.1, 15, 26 Similarly, the other sulphatase deficiencies (MPS II, MPS IIID, MPS IVA and MPS VI) are amenable to this type of specific approach by targeting the individual sulpho-transferases involved.1, 15, 27, 28, 29, 30, 31, 32
The most effective shRNA was shEXTL2.1 followed by shEXTL2.2 and shEXTL3.2. Thus, these were used in combination to determine whether gag synthesis was further reduced by knocking down EXTL2 with two different shEXTL2 pENTR/U6 constructs to maximise the reduction in gene expression, or by targeting both EXTL2 and EXTL3, because of possible redundancy between the two genes. However, no further reduction in gag synthesis was achieved over that observed by shEXTL2.1 alone. The application of multiple shRNAs to a single gene, thus, has no additive effect, nor does targeting multiple genes with overlapping functions (EXTL2 and EXTL3). A more effective approach may be to use shEXTL2.1 in combination with RNAi to gene targets whose products have other roles in gag synthesis. Targeting of linkage region genes (XYLT1, XYLT2, GALTI, GALTII) with siRNA has been shown to be effective in decreasing gag synthesis in the accompanying paper.33 Linkage region genes are common to HS, DS and chondroitin sulphate synthesis, and act before those involved in HS initiation and elongation. Thus, these genes may be candidate targets for RNAi in combination with shEXTL2.1, as they act at different stages in the HS biosynthetic pathway.
The above results, particularly the reduction of lysosomal storage of gag in MPS IIIA cells, show the potential of SDT by the gene specific method of RNAi as a therapy for MPS IIIA. RNAi has been investigated in vivo in other diseases and shown to be non-toxic. siRNA administered by intravenous injection has been used to treat liver damage by targeting Fas in mice with autoimmune hepatitis.34 A mouse model of amyotrophic lateral sclerosis has been treated using an inducible shRNA knock-in.35 In addition, adenoassociated virus (AAV) shRNA constructs have been intrastriatially injected into Huntington's disease mice to target the multi-repeat disease gene and improve motor coordination.36 Lentiviral shRNA constructs have been administered through hippocampal injection in mice with prion disease to target PrP mRNA, resulting in improvements in burrowing and object recognition, and extending survival.37 As shRNAs are hairpin sequences, they lend themselves to AAV or lentiviral therapy from which they can incorporate into the host genome for long-term stable expression.
Preliminary experiments showed that shRNAs could be expressed from a lentiviral vector using a reporter gene assay. The production of virus encoding shRNAs and transduction of 293T cells resulted in a small decrease in gag synthesis in the presence of shEXTL2.1, but not in the presence of shEXTL2.2, shEXTL2.3 or shEXTL3.2. Expression from the lentiviral vector depends on uptake of the lentivirus into the cell, incorporation into the host cell genome and expression of the transgene. Although time parameters that we have previously shown to be effective for the production of lysosomal enzymes were used,23 they may not be optimal for shRNA production, and further refinement of our methodology is required. Of promise is the observation that a decrease in gag synthesis was achieved with shEXTL2.1. This shRNA construct consistently produced the greatest level of inhibition of gag synthesis when transiently expressed from the pENTR/U6 vector.
Overall, we have shown that RNAi methodologies can be employed to specifically reduce the expression of genes involved in HS gag synthesis, and that this results in a reduction in gag synthesis and storage in MPS IIIA and MPS I cells. These results have application in the treatment of CNS disease in MPS disorders and warrant trials, particularly of shEXTL2.1, in mouse models of MPS. Together with the accompanying paper,33 which shows the efficacy of inhibiting gag synthesis by targeting glycosyl transferases responsible for the addition of linkage region monosaccharides, this study shows the feasibility of using an RNAi approach in reducing the buildup of tissue gag characteristic of MPS disorders.
Acknowledgments
This study was supported by grants from the National Health and Medical Research Council of Australia, The National MPS Society and the Women's and Children's Hospital Research Foundation. The authors would like to thank Drs Dziedzic, Wegrzyn and Jakobkiewicz-Banecka for helpful discussions and access to their data before publication.
References
- Neufeld EF, Muenzer J.The mucopolysaccharidosesin Scriver CR, Beaudet AL, Sly WS, Valle D (eds): The Metabolic and Molecular Bases of Inherited Disease8th edn.USA: McGraw-Hill; 2001. vol 3, pp3421–3452. [Google Scholar]
- Harmatz P, Giugliani R, Schwartz I, et al. Enzyme replacement therapy for mucopolysaccharidosis type VI: a phase 3, randomized, double blind, placebo controlled multinational study of recombinant N-acetylgalactosamine-4-sulfatase (recombinant human arylsulfatase B or rhASB) and follow-on, open-label extension study. J Pediatr. 2006;148:533–539. doi: 10.1016/j.jpeds.2005.12.014. [DOI] [PubMed] [Google Scholar]
- Muenzer J, Gucsavas-Calikoglu M, McCandless SE, Schuetz TJ, Kimura A. A phase I/II clinical trial of enzyme replacement therapy in mucopolysaccharidosis II (Hunter syndrome) Mol Genet Metab. 2007;90:329–337. doi: 10.1016/j.ymgme.2006.09.001. [DOI] [PubMed] [Google Scholar]
- Sifuentes M, Doroshow R, Hoft R, et al. A follow-up study of MPS I patients treated with laronidase enzyme replacement therapy for 6 years. Mol Genet Metab. 2007;90:171–180. doi: 10.1016/j.ymgme.2006.08.007. [DOI] [PubMed] [Google Scholar]
- Vogler C, Levy B, Grubb JH, et al. Overcoming the blood–brain barrier with high-dose enzyme replacement therapy in murine mucopolysaccharidosis VII. Proc Natl Acad Sci USA. 2005;102:14777–14782. doi: 10.1073/pnas.0506892102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inokuchi J, Radin NS. Preparation of the active isomer of 1-phenyl-2-decanoylamino-3-morpholino-1-propanol inhibitor of murine glucocerebroside synthase. J Lipid Chem. 1987;28:565–571. [PubMed] [Google Scholar]
- Cox TM, Aerts JM, Andria G, et al. The role of the imino sugar N-butyldeoxynorjirimycin (miglustat) in the management of type I (non-neuronopathic) Gaucher disease: a position statement. J Inherit Metab Dis. 2003;26:513–526. doi: 10.1023/a:1025902113005. [DOI] [PubMed] [Google Scholar]
- Moyses C. Substrate reduction therapy: clinical evaluation in type I Gaucher disease. Phil Trans R Soc Lond B Biol Sci. 2003;358:955–960. doi: 10.1098/rstb.2003.1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimran A, Elstein D. Gaucher disease and the clinical experience with substrate reduction therapy. Phil Trans R Soc Lond B Biol Sci. 2003;358:961–966. doi: 10.1098/rstb.2003.1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berkin A, Szarek WA, Kisilevsky R. Synthesis of 4-deoxy-4-fluro analogues of 2-acetamido-2-deoxy-D-glucose and 2- acetamido-2-deoxy-D-galactose and their effects on cellular glycosaminoglycan biosynthesis. Carbohydr Res. 2000;30:250–263. doi: 10.1016/s0008-6215(00)00049-5. [DOI] [PubMed] [Google Scholar]
- Piotrowska E, Jakóbkiewicz-Banecka J, Barańska S, et al. Genistein-mediated inhibition of glycosaminoglycan synthesis as a basis for gene expression-targeted isoflavone therapy for mucopolysaccharidoses. Eur J Hum Genet. 2006;14:846–852. doi: 10.1038/sj.ejhg.5201623. [DOI] [PubMed] [Google Scholar]
- Roberts AL, Thomas BJ, Wilkinson AS, Fletcher JM, Byers S. Inhibition of glycosaminoglycan synthesis using rhodamine B in a mouse model of mucopolysaccaridosis type IIIA. Pediatr Res. 2006;60:309–314. doi: 10.1203/01.pdr.0000233037.00707.da. [DOI] [PubMed] [Google Scholar]
- Piotrowska E, Jakóbkiewicz-Banecka J, ki-Szymańska A, et al. Genistein-rich soy isoflavone extract in substrate reduction therapy for Sanfilippo syndrome: an open-label, pilot study in 10 pediatric patients. Curr Ther Res Clin Exp. 2008;69:166–179. doi: 10.1016/j.curtheres.2008.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts AL, Rees MH, Klebe S, Fletcher JM, Byers S. Improvement of behaviour after substrate deprivation therapy with rhodamine B in a mouse model of MPS IIIA. Mol Genet Metab. 2007;92:115–121. doi: 10.1016/j.ymgme.2007.06.016. [DOI] [PubMed] [Google Scholar]
- Sugahara K, Kitagawa H. Heparin and heparan sulphate biosynthesis. IUBMB Life. 2002;54:163–175. doi: 10.1080/15216540214928. [DOI] [PubMed] [Google Scholar]
- Kitagawa H, Shimakawa H, Sugahara K. The tumor suppressor EXT-like gene EXTL2 encodes an α1,4-N-acetylhexosaminyltransferase that transfers N-acetylgalactosamine and N-acetylglucosamine to the common glycosaminoglycan-protein linkage region. J Biol Chem. 1999;274:13933–13937. doi: 10.1074/jbc.274.20.13933. [DOI] [PubMed] [Google Scholar]
- Kim BT, Kitagawa H, Tamura J, et al. Human tumor suppressor EXT gene family members EXTL1 and EXTL3 encode α1,4-N-acetylglucosaminyltransferases that likely are involved in heparan sulphate/ heparin biosynthesis. Proc Natl Acad Sci USA. 2001;98:7176–7181. doi: 10.1073/pnas.131188498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lind T, Tufaro F, McCormick C, Lindahl U, Lidholt K. The putative tumor suppressors EXT1 and EXT2 are glycosyltransferases required for the biosynthesis of heparan sulphate. J Biol Chem. 1998;273:26265–26268. doi: 10.1074/jbc.273.41.26265. [DOI] [PubMed] [Google Scholar]
- Esko JD, Lindahl U. Molecular diversity of heparan sulphate. J Clin Invest. 2001;108:169–173. doi: 10.1172/JCI13530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardingham TE, Fosang AJ. Proteoglycans: many forms and many functions. FASEB J. 1992;6:861–870. [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCt method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Anson DS, McIntyre C, Thomas B, et al. Lentiviral-mediated gene therapy for mucopolysaccharidosis type IIIA. Genet Vaccines Ther. 2007;5:1–8. doi: 10.1186/1479-0556-5-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koldej R, Cmielewski P, Stocker A, Parsons DW, Anson DS. Optimisation of a multipartite human immunodeficiency virus based vector system; control of virus infectivity and large-scale production. J Gene Med. 2005;7:1390–1399. doi: 10.1002/jgm.803. [DOI] [PubMed] [Google Scholar]
- McMannus MT, Petersen CP, Haines BB, Chen J, Sharp PA. Gene silencing using micro-RNA designed hairpins. RNA. 2002;8:842–850. doi: 10.1017/s1355838202024032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusche-Gullberg M, Eriksson I, Pikas DS, Kjellén L. Identification and expression in mouse of two heparan sulfate glucosaminyl N-deacetylase/N-sulfotransferase genes. J Biol Chem. 1998;273:11902–11907. doi: 10.1074/jbc.273.19.11902. [DOI] [PubMed] [Google Scholar]
- Fukuta M, Kobayashi Y, Uchimura K, Kimata K, Habuchi O. Molecular cloning and expression of human chondroitin 6-sulfotransferase. Biochim Biophys Acta. 1998;1399:57–61. doi: 10.1016/s0167-4781(98)00089-x. [DOI] [PubMed] [Google Scholar]
- Habuchi O, Hirahara Y, Uchimura K, Fukuta M. Enzymatic sulfation of galactose residue of keratan sulfate by chondroitin 6-sulfotransferase. Glycobiology. 1996;6:51–57. doi: 10.1093/glycob/6.1.51. [DOI] [PubMed] [Google Scholar]
- Habuchi H, Kobayashi M, Kimata K. Molecular characterisation and expression of heparan-sulfate 6-sulfotransferase: complete cDNA cloning in human and partial cloning in Chinese hamster ovary cells. J Biol Chem. 1998;273:9208–9213. doi: 10.1074/jbc.273.15.9208. [DOI] [PubMed] [Google Scholar]
- Kobayashi M, Habuchi H, Habuchi O, Saito M, Kimata K. Purification and characterisation of heparan sulfate 2-sulfotransferase from cultured Chinese hamster ovary cells. J Biol Chem. 1996;271:7645–7653. doi: 10.1074/jbc.271.13.7645. [DOI] [PubMed] [Google Scholar]
- Kobayashi M, Sugumaran G, Liu J, Shworak NW, Silbert JE, Rosenberg RD. Molecular cloning and characterisation of a human uronyl 2-sulfotransferase that sulfates iduronyl and glucuronyl residues in dermatan/chondroitin sulfate. J Biol Chem. 1999;274:10474–10480. doi: 10.1074/jbc.274.15.10474. [DOI] [PubMed] [Google Scholar]
- Yamauchi S, Hirahara Y, Usui H, et al. Purification and characterisation of chondroitin 4-sulfotransferase from the culture medium of a rat chondrosarcoma cell line. J Biol Chem. 1999;274:2456–2463. doi: 10.1074/jbc.274.4.2456. [DOI] [PubMed] [Google Scholar]
- Dziedzic D, Wegrzyn G, Jakobkiewicz-BaneckaImpairment of glycosaminoglycan synthesis in MPS IIIA cells by using siRNA: a potential therapeutic approach for Sanfilippo disease Eur J Human GenetIn Press. [DOI] [PMC free article] [PubMed]
- Song E, Lee SK, Wang J, et al. RNA interference targeting Fas protects mice from fulminant hepatitis. Nat Med. 2003;9:347–351. doi: 10.1038/nm828. [DOI] [PubMed] [Google Scholar]
- Xia X, Zhou H, Huang Y, Xu Z. Allele-specific RNAi selectively silences mutant SOD1 and achieves significant therapeutics in vivo. Neurobiol Dis. 2006;23:578–586. doi: 10.1016/j.nbd.2006.04.019. [DOI] [PubMed] [Google Scholar]
- White MD, Farmer M, Mirabile I, Brandner S, Collinge J, Mallucci GR. Single treatment with RNAi against prion protein rescues early neuronal dysfunction and prolongs survival in mice with prion disease. Proc Natl Acad Sci USA. 2008;105:10238–10243. doi: 10.1073/pnas.0802759105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harper SQ, Staber PD, He X, et al. RNA interference improves motor and neuropathological abnormalities in a Huntington's disease mouse model. Proc Natl Acad Sci USA. 2005;102:5820–5825. doi: 10.1073/pnas.0501507102. [DOI] [PMC free article] [PubMed] [Google Scholar]