Abstract
The basis for vesicoureteral reflux (VUR) is considered to be primarily genetic, with a 30–50% incidence of VUR in first-degree relatives of patients. The search for the causative gene or genes has been elusive, likely because of VUR being genetically heterogeneous with complex inheritance patterns. In this study, a genome-wide analysis of VUR with high-density single nucleotide polymorphisms was conducted with the aim of identifying susceptibility loci for VUR in 98 families with two or more affected children. Using the affected sib-pair method of analysis in 150 sib-pairs, we identified a genome-wide statistically significant linkage peak with an LOD score greater than 4 on chromosome 5 and two linkage peaks with LOD scores greater than 3.6 on chromosomes 13 and 18 were identified in these 98 families. These results suggested that multiple genes are likely to contribute to the formation of VUR phenotype. Further mapping of these linkage peaks may help identify the causative genes.
Keywords: vesicoureteral reflux, linkage analysis, sib-pair analysis, chromosome
Introduction
Vesicoureteral reflux (VUR) is a common congenital disease in which urine in the bladder refluxes back into the ureters and kidneys. When complicated by urinary tract infection (UTI), the kidneys may become directly infected resulting in renal injury with permanent scarring (reflux nephropathy). Substantial kidney scarring can lead to the possibility of hypertension, compromised renal function and complications during pregnancy. In fact, VUR is a significant contributor to kidney failure and the subsequent need for kidney transplantation. Given its major heath consequences, early detection of VUR is essential to limit the sequelae of renal injury.
It is estimated that the overall incidence of VUR is approximately 10% in the general population. VUR is usually diagnosed following investigation for other urinary tract problems, the most common being UTI and antenatal hydronephrosis. It is diagnosed in 40% of children who present with a febrile UTI and in 15–20% of children with a history of antenatal hydronephrosis. Consequently, there is a bimodal distribution in the age of VUR presentation, with one being less than 3 months of age and the other at approximately 2–4 years of age. There appears to be a gender difference in those affected with VUR; infants with antenatally detected reflux are more likely to be boys, whereas children with reflux diagnosed following a febrile UTI are more likely to be girls. In addition, there appears to be a racial difference in those affected with VUR; Caucasian girls are 10 times more likely to have reflux than their African-American counterparts.1
Over the past 30 years, clinical observations have suggested a genetic basis for VUR.2 There is a 30–50% incidence of VUR in first-degree relatives of probands,3, 4 a 100% concordance of VUR among monozygotic twins and 50% among dizygotic twins.5 Genetic linkage to loci on chromosomes 1, 6, 10 and 13 has been previously observed.6 Together, these findings suggest an autosomal dominant mode of inheritance with variable penetrance in a subset of families studied. However, the genetic factors in VUR may be heterogeneous and may also fit other genetic models. Reflux has been found to be associated with several loci in the HLA antigen complex.7, 8, 9 Several groups have sought to identify associations with VUR and various candidate genes, including PAX-2,10, 11 G proteins,12 angiotensin II, type 2 receptor13 and components of the renin–angiotensin system.14, 15, 16, 17 Unfortunately, none of these have been proven to be reliable markers of the reflux trait. There is a lack of replication of previously reported linkages.
Single nucleotide polymorphism (SNP) genotyping technologies have been used in many studies to identify genetic loci containing disease causative gene or genes. Polygenetic diseases and their susceptibility variants can be mapped with genome-wide linkage analysis of familial samples rather than just evaluating specific candidate genes.18, 19 The purpose of this study was to assess the susceptibility loci for VUR in families who have two or more affected children, using high-density SNPs for affected sib-pair analysis.
Patients and methods
Patient population
Enrollment of children with VUR started in 1998 using an IRB-approved Reflux Registry. Approximately 600–700 new patients were seen per year at Children's Hospital, Boston. Currently, 4151 patients are enrolled in this registry. VUR was confirmed radiologically and graded with either a voiding cystourethrogram (VCUG) or radionuclide cystogram (RNC). A total of 2456 children have been followed exclusively at Children's, whereas the remainder have been referred for subsequent follow-up after their initial diagnosis and management at another facility. The total number followed for more than 2 and 4 years exceed 2400 and 800 patients, respectively. Our total VUR population consisted of 988 boys and 3163 girls of which 82% were Caucasians, 8% African Americans, 6% Hispanics and 4% Asians. Total 73% of the patients presented with a UTI, whereas 11% had a history of prenatally diagnosed hydronephrosis (PNH) (Table 1). 47% of the patients were younger than 1 year of age, whereas 30% were between 1 and 5 years at time of presentation. 34% of the patients had low-grade reflux (VUR International Classification Grade 1–2), 51% had grade 3 VUR and only 15% had high-grade reflux (grade 4–5). Bilateral reflux was observed in 57%.
Table 1. Demographics and VUR characteristics of the total VUR population and study group.
| Total VUR population (N=4151) | Study group proband (N=98) | Study group sibling (N=123) | |
|---|---|---|---|
| Females | 3113 (75%) | 78 (80%) | 74 (60%) |
| Ethnicity | |||
| Caucasians | 3404 (82%) | 98 (100%) | 123 (100%) |
| African American | 332 (8%) | — | — |
| Hispanics | 249 (6%) | — | — |
| Asian | 166 (4%) | — | — |
| Mode of presentation | |||
| UTI | 3030 (73%) | 73 (75%) | 10 (8%) |
| PNH | 457 (11%) | 13 (13%) | 4 (3%) |
| Sibling screen | 498 (12%) | — | 109 (89%) |
| Parental history | 83 (2%) | 8 (8%) | — |
| Othersa | 83 (2%) | 4 (4%) | — |
| Age at presentation | |||
| <1 year | 1951 (47%) | 44 (45%) | 43 (35%) |
| 1–5 years | 1245 (30%) | 32 (33%) | 47 (38%) |
| 5–10 years | 706 (17%) | 20 (20%) | 29 (24%) |
| >10 years | 249 (6%) | 2 (2%) | 4 (3%) |
| Grade of VURb | |||
| 1–2 | 1411 (34%) | 30 (31%) | 64 (52%) |
| 3 | 2117 (51%) | 51 (52%) | 54 (44%) |
| 4–5 | 623 (15%) | 17 (17%) | 5 (4%) |
| Laterality | |||
| Bilateral | 2366 (57%) | 54 (55%) | 41 (33%) |
| Unilateral | 1785 (43%) | 44 (45%) | 82 (67%) |
Abbreviations: PNH, prenatally diagnosed hydronephrosis; VUR, vesicoureteral reflux; UTI, urinary tract infection.
Others mode of presentation include voiding problems and abdominal pain.
In patients with bilateral reflux, the highest grade noted between the two sides was selected.
All nuclear families (135) with two or more offspring affected with VUR were recruited for the study. Five families declined participation at the initial time of recruitment, and 15 failed to provide all the required specimens. Consequently, the overall participation rate was 85%. Upon genotyping analysis, a fairly constant Mendelian error rate of less than 1% was observed across each of four sets of 1500 SNPs assayed. Progeny software identified five families with Mendelian inconsistencies, and those families were excluded from the analysis. For the remaining families, approximately 30 000 Mendelian errors were detected and corrected within the Progeny database (South Bend, IN, USA). In cases where Mendelian errors were detected that could not be solved by repeated genotyping, the markers were set to unknown. Families with at least one family member having more than 30% missing genotypes were excluded (12 families), leaving 450 subjects (out of the original 499) consisting of 98 nuclear families with 150 affected sib-pairs to be included in the analysis. Unaffected siblings were included in the study.
Among the 98 nuclear families, 75 families had two affected offspring, whereas 21 families had three and 2 families had four. All nuclear families had complete parental genotypes. All were Caucasians; in the overall registry, there were no families with two or more children with VUR from any ethnic minority groups. Of the affected offspring included in the analysis, there were 151 girls and 70 boys. All children were healthy without any clinical syndrome (such as spina bifida, neurological disorders and diabetes). Among all children 2% had asthma/reactive airway disease and 5% were diagnosed with attention-deficit disorder. No other medical conditions were seen in both the proband and affected siblings.
Of the 98 probands, UTI was the most common mode of presentation, occurring predominately in girls (90%). Prenatal hydronephrosis was the mode of presentation in 13% of the probands, occurring more commonly in boys (69%). Total 45% of the probands were less than 1 year of age, whereas 33% were between 1 and 5 years. The probands had a similar distribution for the grade of VUR and laterality when compared to the total VUR population (Table 1).
A total of 221 affected offspring were included in the analysis. Although all had a sibling with reflux, 8% of the siblings were diagnosed for VUR only when they presented with a UTI and 3% had a concurrent history of PNH. 35% of the patients were younger than 1 year of age at time of diagnosis, whereas 38% were between 1 and 5 years of age (Table 1). Compared to the probands, the grade of VUR was low (1–2) in 52% of the children and was high (4–5) in only 4%. In the sibling study group, unilateral VUR was more common than bilateral VUR (67% compared to 33%).
SNP genotyping
Genomic DNA was extracted from blood or saliva from all the patients. DNA was quantified by optical density or by PicoGreen Assay per protocol (Invitrogen, Carlsbad, CA, USA). Genotype analysis was performed with approximately 6000 SNPs using the Illumina GoldenGate Assay (Illumina, San Diego, CA, USA) per the manufacturer's protocol. Genotype data were manually reviewed within Illumina's BeadStudio genotype calling software for genotype assay success and Mendelian inconsistencies as an indicator of the quality of each sample and of SNP assay quality. Poorly performing SNPs were removed from the analysis and samples with poor or inconsistent quality were genotyped in a second experiment. For cases in which only one offspring or parent exhibited Mendelian inconsistencies for more than 10% of markers in the raw data derived by the BeadStudio software (before any error correction), a new aliquot of the sample was re-screened. If the second attempt genotyping and analysis did not reduce the Mendelian error rate to less than 10%, the inconsistent subjects were removed.
The proofread data sets were then formatted with BeadStudio software and uploaded into a Progeny database (http://www.progenygenetics.com). This is a comprehensive database that stored all pedigree and genotype information for this study and ran a variety of error-checking programs, allowing one to review and correct each Mendelian inconsistency individually. It also provided a flexible file-formatting tool for exporting the data for further analysis using other software. Following importation of each data set into the Progeny database, Mendelian errors were tallied and displayed.
The data, once cleaned of Mendelian errors, were formatted through the Progeny software into source files for analysis with the PedWipe software (New Berlin, WI, USA). This software detects non-Mendelian errors in a sliding-window scan of SNP haplotypes across the genome in each family. Non-Mendelian errors are likely because of genotype errors that did not cause Mendelian inconsistency but are classified as errors because they would have required unlikely recombination events in a given family. An SNP exhibiting such non-Mendelian errors was flagged only in the affected family/families, thus preserving the data for a given SNP in every other family in the study. The nonaffected siblings were used to assess Mendelian consistency but were not included in the analysis.
Linkage analysis
Nonparametric linkage (NPL) analysis was implemented to obtain an ‘unbiased' result, because a complete parental genotype was obtained. The multipoint (MERLIN) analyses20 were assessed using the nuclear families with affected sib-pairs. Multiple sib-ships within a family were entered in MERLIN as one family (www.sph.umich.edu/csg/abecasis/Merlin/download). The easyLINKAGE software21 was adopted for the genome-wide linkage analysis. The Illumina 5K deCODE genetic map (http://compbio.charite.de/genetik/hoffmann/easyLINKAGE/; www.sourceforge.net/projects/easylinkage) was used in the linkage analysis. In an effort to avoid the inflated type I error rate using highly correlated SNP data, linkage disequilibrium (LD) was limited by removing SNPs with r2 values greater than 0.1 in the founders. The call rate, defined as the percentage of successful genotype calls among subjects, was used as a measure of data quality. A wide range of call rates for markers were examined ranging from 80 to 98%, and the same regions with linkage signals were identified. Therefore, the results based on the 95% call rate were reported.
In MERLIN, allele frequencies were estimated from founders only. Both the standard NPL Z score and nonparametric LOD score were calculated using the Kong–Cox exponential and linear model.22 The results were similar using each of the models, and the scores were used to generate graphic plots of the whole-genome scan results. This model is designed to identify small increases in allele-sharing spread across a large number of families, which is to be expected in a complex disease. The 1-LOD drop support interval was used as an estimate for the 90% confidence interval (CI) of the putative loci.23
Results
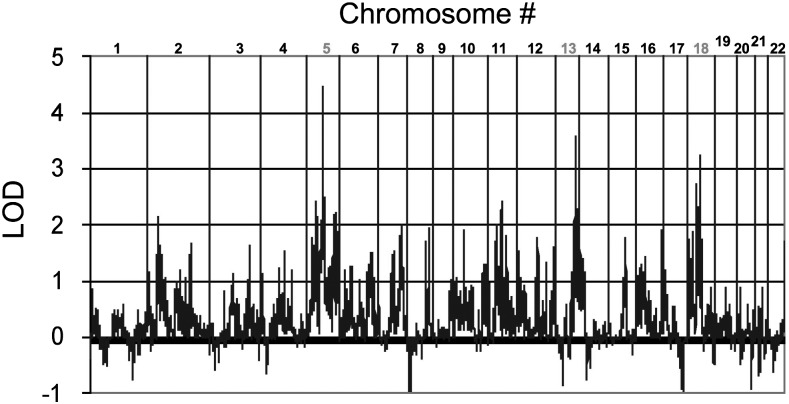
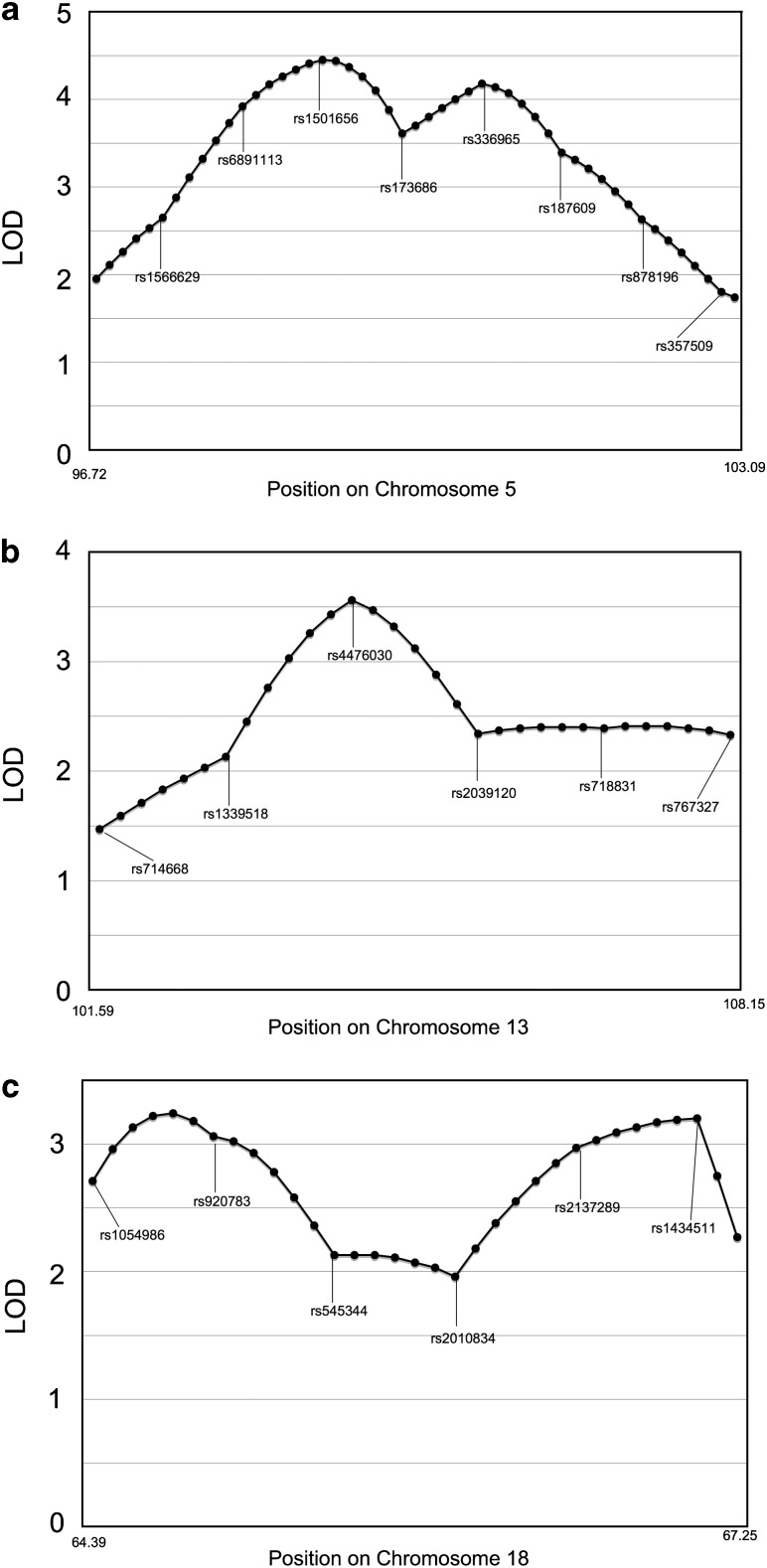
Using the MERLIN software for the affected sib-pair method, we detected linkage peaks with LOD scores greater than 3.6 on three different chromosomes (Figure 1). The highly significant linkage peak with a multipoint LOD score 4.46 was identified for VUR on chromosome 5 at 98.58 cM (nearest marker RS1501656, NPL Z=4.15 P=0.00002; LOD=4.46, P=0.00001). A secondary peak with an LOD score of 3.7 was found on chromosome 13 at 104.36 cM (nearest marker RS4476030, NPL Z=4.09, P=0.00002; LOD=3.7, P<0.00001). In addition to chromosomes 5 and 13, an interval on chromosome 18 with an LOD score of 3.71 at 65.28 cM was discovered (nearest marker RS1054986, NPL Z=3.71, P=0.0001; LOD=3.71, P=0.00002). The number of informative SNPs for chromosomes 5, 13 and 18 are listed in Table 2. To assess the quality of LOD peaks, the LOD scores surrounding those specific loci were evaluated (Figure 2a–c). The linkage information content (LIC) value, which is measure of marker informativeness specifically for model-free linkage analysis of a particular type of relative pair, at the linkage peak on chromosome 5 is 0.92. For chromosomes 13 and 18, the LICs are 0.87 and 0.85, respectively. The quality of the linkage peak is assured by: (1) complete parental genotypes for all nuclear families, (2) and LD was properly removed by r2 greater than 0.1 and (3) high LIC values for the linkage peaks. The loci exhibiting the highest linear LOD scores on chromosome 5 had sharp drop offs peripheral to the associated SNPs, whereas this was less so with the loci on chromosomes 13 and 18.
Figure 1.
Multipoint nonparametric LOD score of genome-wide scan for sibling VUR.
Table 2. Chromosome regions suggestive of linkage in sibling VUR.
| Chromosome | 5 | 8 | 13 |
|---|---|---|---|
| Total no. of SNPs | 324 | 191 | 178 |
| No. of SNPs with call rate ≤80% | 35.8% | 19.4% | 7.9% |
| Final no. of SNPs | 208 | 154 | 164 |
Figure 2.
LOD scores around loci exhibiting the highest linear LOD scores on chromosomes 5 (a), 13 (b) and 18 (c).
Discussion
A review of the literature by Sargent24 estimated the prevalence of VUR to be approximately 9% (95% CI=6–12%) in normal children. The incidence increased to 31% (95% CI=27–35%) in children with UTI and 21% (95% CI=19–22%) in children with a history of prenatal hydronephrosis. The predominace of girls, the frequencies of the modes of presentation, the distribution of the grade of VUR and laterality in the study total VUR population and in the probands of the study group are similar to those previously reported (reviewed by Godley25), suggesting that the study's total VUR population and the proband study group is representative of the general VUR population. A review of the literature by Chertin and Puri26 and Hollowell and Greenfield27 estimated the incidence of sibling reflux to be approximately 32%. The ratio of girls/boys were reported to be approximately 1.1:126 and 1.4:1.27 The incidence of low-grade VUR (grade 1–2) was found to be 48–60%, compared to high-grade VUR (grade 4–5) with an incidence of approximately 2%. Unilateral reflux was found to be more common than bilateral reflux (53–60%). The demographics and VUR characteristics of our sibling population reflect well those previously reported, validating that our sibling study group was also representative of the general sibling VUR population.
This affected sib-pair linkage study used the allele-sharing method for identifying loci associated with urinary reflux. The advantage of this approach is that linkage can be detected in the absence of any common ancestral allele. Most algorithms used to detect linkage are designed to detect an ancestral allele common to all or many affected individuals. This approach would not be successful for genetic disorders in which many pathogenic variants have arisen such that every family has a different mutation. Complex genetic disorders by definition do not show classic Mendelian inheritance and are hypothesized to result from a combination of genetic and environmental factors that exert separate effects on the phenotype. The allele-sharing method applied in this study detects an increase in the phenomenon of allele sharing (at a putatively causative locus) rather than an increase in sharing of a particular allele. Therefore, this method can be applied to a large number of small families, most of which may have the disorder due to different mutations in the same gene.
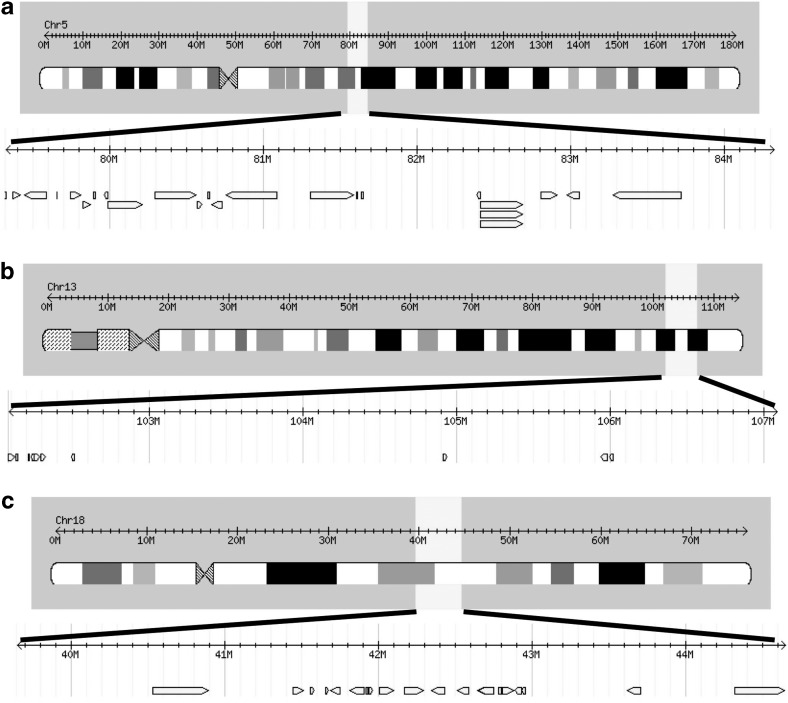
The allele-sharing algorithm identified a linkage peak on chromosome 5 well surpassing an LOD score of 3.6, which is considered to be a threshold for statistical significance.28 Thus, the linkage peak on chromosome 5 with an LOD score of 4.5 identified in this study would be considered highly significant. Using the International HapMap Project (www.hapmap.org/index.html.en), we analyzed potential genes in the region of the suspected loci. Given the 1-LOD drop position is approximately 5 Mbp, we evaluated potential genes within 5 Mbp of the suspected loci on chromosome 5 (Figure 3a). In addition to the candidate genes shown in Figure 3a, there are also several hypothetical protein encoding sequences on chromosome 5. These sequences represent as yet unconfirmed gene coding sequences that may be considered candidate genes in the future. Preliminary review of these genes did not demonstrate any obvious correlation with other established renal diseases or anomalies.
Figure 3.
Genes located within 5 Mbp of the suspected loci for sibling VUR: (a) chromosome 5, (b) chromosome 13 and (c) chromosome 18.
Two additional linkage peaks with LOD scores of greater than 3.6 were also identified on chromosomes 13 and 18. These linkage peaks may also contain candidate genes. Because the genetics of VUR is likely to involve multiple genes, additional peaks are expected. Candidate VUR genes within these loci are shown in Figure 3b and c. In addition, there is an open-reading frame in the region of interest on chromosome 18. More detailed analysis with additional SNPs and mutational analysis of these putative loci on chromosomes 5, 13 and 18 will help identify the causative genes. One apparent limitation of this study is that linkage analysis of the X chromosome was not performed; we initially wanted to focus on autosomal linkages. A higher incidence of VUR among female members in the pedigrees studied has been observed, which may indicate a modifying gene on the X chromosome.6, 29
Feather et al6 first performed a genome-wide linkage analysis on seven European families with nonsyndromic VUR. Using the model of autosomal dominant inheritance with reduced penetrance, they observed linkage within a 20-cM interval on chromosome 1p13, obtaining an LOD score of 5.4 in 78% of families. They also reported a linkage peak on chromosome 13 (distance=90–117 cM) with a parametric LOD score of 2.03 (NPL=2.34, P=0.03). This is similar to the peak we identified on chromosome 5 (distance=104 cM). Sanna-Cherchi et al30 subsequently performed linkage analysis of 35 markers on chromosomes 1p13, 3p12, 6p21, 10q26 and 19q13 in seven additional families, using models of locus heterogeneity and nonparametric, allele sharing. Unfortunately, they did not find any evidence of linkage at any of the loci tested. The authors concluded that their findings showed substantial genetic heterogeneity and suggested that future mapping strategies should rely on a large number of family members. More recently, Conte et al31 reported a nonparametric, affected-only linkage analysis in 24 families and identified four genomic areas on chromosomes 1, 3, 4 and 22 with the best result corresponded to the D3S3681-D3S1569 interval on chromosome 3 (NPL=2.75, P=0.008). Kelly et al32 performed a genome-wide linkage scan using 4710 SNPs in 609 individuals from 129 Irish families with at least two affected members. They observed a moderately suggestive linkage at chromosome 2q37 (NPLmax=2.67, P=0.001). Only with the analysis of a subset of patients did this linkage peak reach levels of genome-wide statistical significance (NPL=4.1, P=0.001). The authors also observed suggestive linkage at 10q26 and 6q27. Our data implicate new VUR disease loci but do not confirm those mentioned previously except for chromosome 13. This provides further evidence of marked genetic heterogeneity associated with VUR. Further analysis of the linkage peak on chromosome 13 would be of importance, because this linkage peak was the only to be suggested by two separate studies.
Genotype analysis for VUR will not only help identify causative genes but may help develop future diagnostic and prognostic tests. Currently, VUR is diagnosed by performing a VCUG or an RNC. These tests are invasive, which require placement of a urethral catheter and exposure to radiation. They often create more anxiety in both parents and children than the diagnosis of VUR itself. The findings noted in the current study could obviate these issues as the advent of diagnostic markers would identify patients for evaluation. Moreover, VUR is diverse in its natural history. The majority of cases resolve spontaneously, whereas others persist with the potential for complications such as recurrent UTI, renal scarring, hypertension and compromised renal function. Genotype screening of the specific subgroups may identify genetic markers for predicting patients in which VUR will or will not spontaneously resolve and those at risk for renal scarring and its sequelae.
Conclusion
Linkage peaks on chromosomes 5, 13 and 18 were identified from a large cohort, which represents the general population of patients affected with VUR. This study identified an LOD score demonstrating a high degree of statistically significant evidence of VUR linkage to any chromosomal region. Sequence analysis of the genes within the linkage intervals will lead to mutations that are causative for VUR and the causative gene can be assayed in additional patients to better understand the underlying biological pathways leading to VUR. VUR is likely to be genetically heterogeneous. Consequently, additional genotype-based approaches to identifying genetic factors in VUR, such as the affected sib-pair analysis conducted for this study, will likely be of great importance in the development of improved diagnostic and prognostic tests for VUR.
References
- Elder JS, Peters CA, Arant BS, Jr, et al. Pediatric Vesicoureteral Reflux Guidelines Panel summary report on the management of primary vesicoureteral reflux in children. J Urol. 1997;157:1846–1851. [PubMed] [Google Scholar]
- Zel G, Retik AB. Familial vesicoureteral reflux. Urology. 1973;2:249–251. doi: 10.1016/0090-4295(73)90457-3. [DOI] [PubMed] [Google Scholar]
- Noe HN. The long-term results of prospective sibling reflux screening. J Urol. 1992;148:1739–1742. doi: 10.1016/s0022-5347(17)37017-9. [DOI] [PubMed] [Google Scholar]
- Noe HN, Wyatt RJ, Peeden JN, Jr, Rivas ML. The transmission of vesicoureteral reflux from parent to child. J Urol. 1992;148:1869–1871. doi: 10.1016/s0022-5347(17)37053-2. [DOI] [PubMed] [Google Scholar]
- Kaefer M, Curran M, Treves ST, et al. Sibling vesicoureteral reflux in multiple gestation births. Pediatrics. 2000;105:800–804. doi: 10.1542/peds.105.4.800. [DOI] [PubMed] [Google Scholar]
- Feather SA, Malcolm S, Woolf AS, et al. Primary, nonsyndromic vesicoureteric reflux and its nephropathy is genetically heterogeneous, with a locus on chromosome 1. Am J Hum Genet. 2000;66:1420–1425. doi: 10.1086/302864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey RR, Janus E, McLoughlin K, Lynn KL, Abbott GD. Familial and genetic data in reflux nephropathy. Contrib Nephrol. 1984;39:40–51. doi: 10.1159/000409234. [DOI] [PubMed] [Google Scholar]
- Torres VE, Moore SB, Kurtz SB, Offord KP, Kelalis PP. In search of marker for genetic susceptibility to reflux nephropathy. Clin Nephrol. 1980;14:217–222. [PubMed] [Google Scholar]
- Kawauchi A, Takahara S, Sada M, Goto R, Nakatani T, Miki T. Susceptibility to vesicoureteral reflux in Japanese is linked to HLA-DR antigen. Urology. 2001;58:1036–1040. doi: 10.1016/s0090-4295(01)01459-5. [DOI] [PubMed] [Google Scholar]
- Choi KL, McNoe LA, French MC, Guilford PJ, Eccles MR. Absence of PAX2 gene mutations in patients with primary familial vesicoureteric reflux. J Med Genet. 1998;35:338–339. doi: 10.1136/jmg.35.4.338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lama G, Esposito Salsano M. [Molecular basis of vesicoureteral reflux] Minerva Pediatr. 2002;54:415–422. [PubMed] [Google Scholar]
- Zagradisnik B, Bracic K, Varda NM, Kokalj Vokac N, Gregoric A. G-protein beta3 subunit gene C825T polymorphism in patients with vesico-ureteric reflux. Ann Genet. 2004;47:209–216. doi: 10.1016/j.anngen.2004.04.003. [DOI] [PubMed] [Google Scholar]
- Hohenfellner K, Hunley TE, Yerkes E, Habermehl P, Hohenfellner R, Kon V. Angiotensin II, type 2 receptor in the development of vesico-ureteric reflux. BJU Int. 1999;83:318–322. doi: 10.1046/j.1464-410x.1999.00923.x. [DOI] [PubMed] [Google Scholar]
- Haszon I, Friedman AL, Papp F, et al. ACE gene polymorphism and renal scarring in primary vesicoureteric reflux. Pediatr Nephrol. 2002;17:1027–1031. doi: 10.1007/s00467-002-0968-1. [DOI] [PubMed] [Google Scholar]
- Ozen S, Alikasifoglu M, Saatci U, et al. Implications of certain genetic polymorphisms in scarring in vesicoureteric reflux: importance of ACE polymorphism. Am J Kidney Dis. 1999;34:140–145. doi: 10.1016/s0272-6386(99)70120-4. [DOI] [PubMed] [Google Scholar]
- Pardo R, Malaga S, Coto E, et al. Renin–angiotensin system polymorphisms and renal scarring. Pediatr Nephrol. 2003;18:110–114. doi: 10.1007/s00467-002-1031-y. [DOI] [PubMed] [Google Scholar]
- Yim HE, Jung MJ, Choi BM, et al. Genetic polymorphism of the rennin–angiotensin system on the development of primary vesicoureteral reflux. Am J Nephrol. 2004;24:178–187. doi: 10.1159/000076620. [DOI] [PubMed] [Google Scholar]
- Kim S, Misra A. SNP genotyping: technologies and biomedical applications. Annu Rev Biomed Eng. 2007;9:289–320. doi: 10.1146/annurev.bioeng.9.060906.152037. [DOI] [PubMed] [Google Scholar]
- Nickerson DA, Taylor SL, Weiss KM, et al. DNA sequence diversity in a 9.7-kb region of the human lipoprotein lipase gene. Nat Genet. 1998;19:233–240. doi: 10.1038/907. [DOI] [PubMed] [Google Scholar]
- Abecasis GR, Cherny SS, Cookson WO, Cardon LR. Merlin – rapid analysis of dense genetic maps using sparse gene flow trees. Nat Genet. 2002;30:97–101. doi: 10.1038/ng786. [DOI] [PubMed] [Google Scholar]
- Hoffmann K, Lindner TH. easyLINKAGE-Plus – automated linkage analyses using large-scale SNP data. Bioinformatics. 2005;21:3565–3567. doi: 10.1093/bioinformatics/bti571. [DOI] [PubMed] [Google Scholar]
- Kong A, Cox NJ. Allele-sharing models: LOD scores and accurate linkage tests. Am J Hum Genet. 1997;61:1179–1188. doi: 10.1086/301592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupuis J, Siegmund D. Statistical methods for mapping quantitative trait loci from a dense set of markers. Genetics. 1999;151:373–386. doi: 10.1093/genetics/151.1.373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sargent MA. What is the normal prevalence of vesicoureteral reflux. Pediatr Radiol. 2000;30:587–593. doi: 10.1007/s002470000263. [DOI] [PubMed] [Google Scholar]
- Godley ML.Vesicoureteral reflux: pathophysiology and experimental studiesin Gearhart JP, Rink RC, Mouriquand PD (eds):: Pediatric Urology Philadelphia: WB Saunders; 2001. 1st edn, pp359–381. [Google Scholar]
- Chertin B, Puri P. Familial vesicoureteral reflux. J Urol. 2003;169:1804–1808. doi: 10.1097/01.ju.0000058428.00284.d5. [DOI] [PubMed] [Google Scholar]
- Hollowell JG, Greenfield SP. Screening siblings for vesicoureteral reflux. J Urol. 2002;168:2138–2141. doi: 10.1016/S0022-5347(05)64337-6. [DOI] [PubMed] [Google Scholar]
- Lander E, Kruglyak L. Genetic dissection of complex traits: guidelines for interpreting and reporting linkage results. Nat Genet. 1995;11:241–247. doi: 10.1038/ng1195-241. [DOI] [PubMed] [Google Scholar]
- MacGregor ME, Freeman P. Childhood urinary infection associated with vesico-ureteric reflux. Q J Med. 1975;44:481–489. [PubMed] [Google Scholar]
- Sanna-Cherchi S, Reese A, Hensle T, et al. Familial vesicoureteral reflux: testing replication of linkage in seven new multigenerational kindreds. J Am Soc Nephrol. 2005;16:1781–1787. doi: 10.1681/ASN.2004121034. [DOI] [PubMed] [Google Scholar]
- Conte ML, Bertoli-Avella AM, de Graaf BM, et al. A genome search for primary vesicoureteral reflux shows further evidence for genetic heterogeneity. Pediatr Nephrol. 2008;23:587–595. doi: 10.1007/s00467-007-0675-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly H, Molony CM, Darlow JM, et al. A genome-wide scan for genes involved in primary vesicoureteric reflux. J Med Genet. 2007;44:710–717. doi: 10.1136/jmg.2007.051086. [DOI] [PMC free article] [PubMed] [Google Scholar]