Abstract
Deletions in chromosome 17q12 encompassing the HNF1β gene cause cystic renal disease and maturity onset diabetes of the young, and have been recently described as the first recurrent genomic deletion leading to diabetes. Earlier reports of patients with this microdeletion syndrome have suggested an absence of cognitive impairment, differentiating it from most other contiguous gene deletion syndromes. The reciprocal duplication of 17q12 is rare and has been hypothesized to be associated with an increased risk of epilepsy and mental retardation. We conducted a detailed clinical and molecular characterization of four patients with a deletion and five patients with a reciprocal duplication of this region. Our patients with deletion of 17q12 presented with cognitive impairment, cystic renal disease, seizures, and structural abnormalities of the brain. Patients with reciprocal duplications manifest with cognitive impairment and behavioral abnormalities, but not with seizures. Our findings expand the phenotypic spectrum associated with rearrangements of 17q12 and show that cognitive impairment is a part of the phenotype of individuals with deletions of 17q12.
Keywords: 17q12, genomic rearrangements, cystic renal disease, cognitive impairment, LHX1, HNF1β
Introduction
Structural variation of the human genome is due to the occurrence of rearrangements such as deletions, duplications, insertions, and inversions. All of these genomic rearrangements, except for inversions, result in copy number variation (CNV) or deviation from the normal number of copies for a given genomic segment. Genomic rearrangements may convey phenotypes by changing the copy number of dosage-sensitive genes, disrupting genes, creating fusion genes, and other mechanisms.1 Recurrent genomic rearrangements of chromosome 17q12 are associated with varied clinical phenotypes. Deletions of 17q12, including the HNF1β (hepatocyte nuclear factor 1-beta also known as transcription factor 2, MIM 189907) gene, are associated with maturity onset diabetes of the young type 5 (MODY5), as well as with cystic renal disease, renal dilations, pancreatic atrophy, and liver abnormalities.2, 3, 4, 5, 6 Earlier reports of this contiguous gene deletion syndrome involving HNF1β have suggested that cognitive impairment is not part of the phenotype conveyed by these deletions. Mefford et al6 have reported that recurrent deletions in this region spanning 1.8 Mb are one of the few examples of contiguous gene deletion syndromes that present without mental retardation. The reciprocal duplication in this genomic region is rare in the normal population, and its frequency is higher in patients with mental retardation and epilepsy.6 To understand the phenotype associated with CNV in chromosome 17q12, we conducted a detailed clinical and molecular characterization of four patients with deletion and five patients with duplication of this region. Three of the four patients with deletions in 17q12 presented with developmental delay. Patients with a duplication in 17q12 presented with developmental delay, but seizures and other neurological findings were absent. Our findings increase the repertoire of characteristics associated with deletions and reciprocal duplications of chromosome 17q12. Importantly, contrary to previous reports, we show that developmental delay may be associated with the deletion of 17q12.
Clinical reports
Patient 1
This male patient was born at 34 weeks of gestation to a gravida 3, para 1 woman with no significant medical history. A prenatal ultrasound revealed the presence of multicystic dysplastic kidneys. His birth weight was 2.2 kg (50th centile), whereas measurements of his length and FOC were not available. During the first 2 years, it was noted that he had developmental delay, as evidenced by walking only by 2 years of age, and a vocabulary of 10 words at 3 years. Evaluation of speech was remarkable for significant expressive and receptive speech delays. At 5 years and 6 months, his weight (14.8 kg, z-score=−2.43) and height (96 cm, z-score=−3.41) were well below normal, whereas his FOC (49.6 cm, 10th centile) was normal. The physical examination revealed plagiocephaly, but no other craniofacial dysmorphisms were observed. His systemic examination was normal. MRI of the brain did not show any structural defects, whereas the imaging of kidneys showed multicystic renal dysplasia and a nonfunctioning right kidney. A peripheral blood karyotype revealed normal chromosome constitution, 46, XY.
Patient 2
The proband was born at 39 weeks of gestation. Prenatal ultrasounds were remarkable for cystic renal disease and oligohydramnios. His birth weight was 2179 gm (z-score=−2.8), length was 43 cm (z-score=−4), and head circumference was 34.5 cm (9th centile, z-score=−1.4). He had a large anterior fontanel, a shawl scrotum, and a significant coarctation of the aorta that required urgent surgical correction. He had significant failure to thrive and had not regained his birth weight by 7 weeks; however, his weight gain improved after 2 months of age and at the 4-month evaluation, his growth parameters were remarkable with a weight of 4100 gm (z-score=−4.1), length of 50 cm (z-score=−7.6), and a head circumference of 41.5 cm (54th centile, z-score=0.1). He did not have any facial dysmorphic features and his systemic examination was unremarkable. He developed a social smile at 3 months, but did not attain head control. An ultrasound of the head showed no structural abnormalities and renal imaging showed the presence of multiple small cysts.
Patient 3
This female patient was born to nonconsanguineous Caucasian parents. The maternal history was significant for generalized tonic–clonic seizures being treated with phenytoin, phenobarbitol, carbamazapine, and sodium valproate. Carbamazapine was discontinued during the latter half of the second trimester. A prenatal ultrasound showed the presence of multiple renal cysts. The proband was delivered by emergent cesarean section at 30 weeks because of maternal seizures. She developed complex partial seizures in the first year of life. Her attainment of motor milestones was initially delayed, as evident by rolling over at 9 months of age, but her subsequent motor development was normal and she walked by 15 months of age. She had difficulties in language development and showed learning difficulties, and is enrolled in special education classes. She was referred to a genetics clinic for the evaluation of learning difficulties and short stature at 13 years of age. Her growth parameters were remarkable for a height of 135 cm (z-score=−3.18), weight of 33.5 kg (z-score=−1.86), and a normal FOC of 52 cm (40th percentile). Her physical examination was unremarkable. Testing for fragile X syndrome and galactosemia were unremarkable. MRI of the brain showed hyperintensities in the hippocampi (Figure 1a), but no other structural abnormalities were observed. The results of evaluation for hormonal causes of short stature, including thyroid hormone, thyroid-stimulating hormone, follicle-stimulating hormone, luteinizing hormone, and insulin-like growth factor-1, were within normal limits.
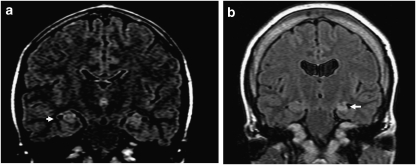
Figure 1.
MRI in two patients with a deletion of 17q12. (a) Patient 3 with hyperintensities in hippocampi (arrow). (b) Patient 4 with mild cerebral atrophy, substantial atrophy in the left hippocampus (arrow), and non-specific findings of increased FLAIR intensities in the subcortical white matter of the left superior frontal gyrus.
Patient 4
This female patient was born with a dysplastic right kidney and left renal agenesis. She developed complex partial seizures at 8 months of age and had been on treatment with various antiseizure medications. At the age of 3 years, she underwent her first cadaveric renal transplant that was rejected because of poor compliance with medications, and the patient was maintained on continuous cycling peritoneal dialysis for 7 years. At the age of 11 years, she underwent a second renal transplant and received long-term treatment with steroids that resulted in the development of diabetes mellitus. She was diagnosed with moderate mental retardation and had significant learning difficulties. She is enrolled in special education classes that train her in vocational skills. A physical examination at 20 years was remarkable for a height of 150 cm (z-score=−2.05) and a weight of 54.7 kg (35th centile, z-score=−0.39). The physical examination was remarkable for mild hirsutism and cushingoid habitus, and for bilateral punctuate cataracts of the lens. There were no facial dysmorphisms. Neurological examination showed normal muscle tone with no focal deficits. MRI of the brain revealed mild cerebral atrophy, substantial atrophy in the left hippocampus, and non-specific findings of increased FLAIR intensities in the subcortical white matter of the left superior frontal gyrus (Figure 1b)
Patient 5
This male patient was evaluated at 4 years and 6 months of age. His prenatal history was significant for maternal polyhydramnios. His birth weight was 2.66 kg (2nd centile, z-score=−2.1), whereas his length was 48.5 cm (5th centile, z-score=−1.7). Soon after birth, a tracheoesophageal fistula (TEF) with esophageal atresia, along with butterfly vertebra at the ninth thoracic vertebra, were noted. There were no associated renal, cardiac, or genitourinary abnormalities. He underwent a surgical correction of the TEF. When examined by a geneticist at 4 years and 6 months of age, his weight (16.4 kg, 32nd centile, z-score=−0.47), height (104 cm, 34th centile, z-score=−0.42), and FOC (50 cm, 10–25th centile) were normal. He had no craniofacial dysmorphisms and the systemic examination was unremarkable. His developmental parameters were age appropriate.
Patients 6 and 7
Patient 6 was born at 33 weeks of gestation by emergent cesarean section performed for placental abruption in the mother (Patient 7). He was large for gestational age, with a weight of 2.4 kg (97th centile) and length of 46.5 cm (97th centile). The immediate perinatal period was complicated by respiratory distress that required mechanical ventilation for 2 weeks. The first 3 years were characterized by delayed attainment of motor developmental milestones, as evidenced by rolling over at 9 months, sitting unsupported by 12 months, and walking by 32 months. His language development was delayed, and the proband spoke his first words by 2 years. He was noted to have significant behavioral abnormalities including aggressive and self-injurious behaviors. When examined at 4 years and 6 months of age, his weight (20.8 kg, 82nd centile, z-score=0.93), FOC (49.5 cm, 10–25th centile), and height (110 cm, 90th centile, z-score=1.32) were normal. His physical examination was remarkable for synophrys and mild syndactyly of the second and third fingers and toes bilaterally. A developmental evaluation revealed global developmental delay, receptive and expressive language deficits, and oppositional defiant behavior. MRI of the brain and echocardiography were normal.
The proband's mother (patient 7) was also found to have learning difficulties and required special education classes throughout her schooling. On examination at 45 years of age, she showed no craniofacial dysmorphisms, but shared the mild syndactyly of the second and third toes.
Patient 8
This female patient was referred for a genetics evaluation at 4 years and 4 months of age. The patient was in the care of foster parents because of social reasons. The details of her prenatal and birth history were unavailable, but there was concern for intrauterine exposure to alcohol and drugs. The mother of the proband was reported to have cognitive impairment. The patient was noted to have behavioral abnormalities including aggressive and self-injurious behaviors such as biting and hitting others, use of inappropriate language, and auditory hallucinations. Her developmental evaluation was remarkable for expressive speech delay. Her weight (15.56 kg, 11th centile) and height (98 cm, 32nd centile) were normal, whereas FOC (48 cm) measured below the 2nd centile. Her physical examination revealed no significant facial dysmorphisms, except for small palpebral fissures and a thin upper lip. Her systemic examination was normal.
Patient 9
This 3-year-old male patient, half-sibling of patient 8, was evaluated by genetics for developmental delay. The mother apparently had cognitive impairment, and abused alcohol and drugs during pregnancy. When transferred to the care of foster parents at 2 years of age, the proband was nonverbal. At 3 years, he had developed a vocabulary of around 50 words and spoke in several word phrases with around 50 percent intelligibility. His weight (13.4 kg, 25th centile), height (91 cm, 13th centile), and FOC (48.6 cm, ∼10th centile) were normal. His physical examination showed minor facial dysmorphic features including a triangular face, small palpebral fissures, epicanthal folds, and small mouth. His systemic examination was unremarkable.
Methods
Human subjects
After the initial result of the clinical diagnostic testing using array comparative genomic hybridization (aCGH), clinical information was obtained through the primary health provider. Consent was obtained from patient 3 for further analysis by a high-density whole-genome aCGH. The protocol was approved by the institutional review board for human subjects research at the Baylor College of Medicine (BCM).
FISH analysis
FISH analyses with bacterial artificial chromosome clones were performed using standard procedures.7 The BAC clone of interest was grown in TB media with 20 μg/ml chloramphenicol. DNA was extracted from BAC clones (Eppendorf Plasmid Mini Prep kit, Hamburg, Germany) and directly labeled with SpectrumOrangeTM dUTP by nick translation (Vysis, Downer Grove, IL, USA) according to the manufacturer's instructions. Digital FISH images were captured by a Power Macintosh G3 System using MacProbe software version 4.4 (Applied Imaging, San Jose, CA, USA).
Clinical chromosome microarray analysis
Chromosome Microarray Analysis (CMA) was performed on all patients. The microarrays were designed in the Medical Genetics Laboratory of BCM. The oligonucleotide array, V7.2OLIGO, is a custom made array with ∼105 000 oligonucleotides of 60 base pairs manufactured by Agilent Technologies Inc (Santa Clara, CA, USA). This array selects the best performing oligonucleotides from the electronic library of Agilent, and contains probes for virtually all the known microdeletion or microduplication syndromes, as well as the pericentromeric and subtelomeric regions with an average resolution of 10–20 kb. In addition to these targeted regions, the entire genome (between disease regions) is covered with an average resolution of 30 kb, excluding the repetitive sequences through a combination of bioinformatics and computation. The array also includes a few regions of known polymorphic variants, such as the AMY1A (amylase) gene cluster in 1p21.2, UGT2B17 (UDP glucuronosyltransferase 2 family) genes in 4q13.2–q13.3, CCL3L1 (chemokine C–C motif) in 17q12, and DEFB107B (defensin gene family) in 8p23.1, to serve as internal control for hybridization efficiency. The procedures for DNA digestion, labeling, and hybridization, as well as data analysis, were performed as described.8
High-resolution whole-genome oligonucleotide array
To determine the minimal extent of deletions in this region, the DNA sample from patient 3 was interrogated using the Agilent 244k Whole Genome Oligo Microarray Kit (Agilent Technologies Inc). This array contains 238 459 arrayed 60-mer oligonucleotides, representing a compiled view of the human genome at an average resolution of 6.4 kb. The procedures for DNA digestion, labeling, and hybridization were performed according to the manufacturer's instructions with some modifications.9
Results
Clinical CMA
We performed aCGH analysis on the clinical microarray platform routinely used in our institution. Four patients had a deletion, whereas five had a duplication involving chromosome 17q12. The deletions and duplications in all patients encompassed at a minimum a 1.06 Mb region that mapped from LHX1 to LOC28400 genes (32 221 569–33 288 139) (Figures 2 and 3). The region between coordinates 3154257–32221567 and 33288139–3410803 represents flanking low copy repeats and is not covered by oligonucleotide probes. The positions of the adjacent proximal and distal oligonucleotides at 31 548 257 bp and 34 010 803 bp, respectively, suggested that the maximal possible size of the deletions or duplications is 2.46 Mb, extending from CCL3L3 to SNIP (Figure 4). All the deletions and duplications were confirmed by FISH using BAC clones RP11-87D17 or RP11-115K3 (Figures 2c and 3b). The deletion in patient 2 was confirmed to be a de novo event. The duplication in patient 6 is inherited from the mother (patient 7), whereas both patients 8 and 9 presumably inherited the duplication from the mother. Parental studies were not available for patients 1, 4, 5, 8, and 9. In patient 3, the mother does not have the deletion, whereas the father was not available for testing.
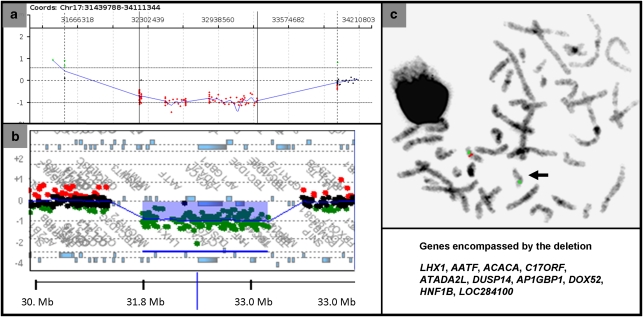
Figure 2.
(a) Results of aCGH with oligonucleotide array V7.2OLIGO in patient 3 with a deletion of 17q12. The results depicted are representative of the deletions in all patients. Each point represents an oligonucleotide probe. The normalized data for each probe are represented along a horizontal line that indicates its relative position on 17q. Loss of copy number is indicated by deviation below a mean log2 ratio of −0.2 (depicted in red). The regions between genomic coordinates 3154257–32221567 and 33288139−3410803 have many copy number polymorphisms and are not represented by oligonucleotide probes. (b) Results of high-resolution whole-genome array in patient 3. The deletion encompassed a region of 1.4 Mb (31889297–33323037 bp) mapping from ZNHIT3 to LOC28400 genes. (c) FISH analysis showing the deletion of 17q12 detected by probe RP11-87D17 (red signal). The green signal is a centromeric marker on chromosome 17.
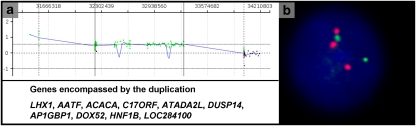
Figure 3.
(a) Results of aCGH with oligonucleotide array V7.2OLIGO in patient 6 with a duplication of 17q12. The results depicted are representative of the duplications in all patients. Each point represents an oligonucleotide probe. The normalized data for each probe are represented along a horizontal line that indicates its relative position on 17q. Gain of copy number is indicated by deviation above a mean log2 ratio of 0.2 (depicted in green).(b) FISH analysis showing the duplication of 17q12 detected by probe RP11-115K3 (red signal). The green signal is a centromeric marker on chromosome 17.
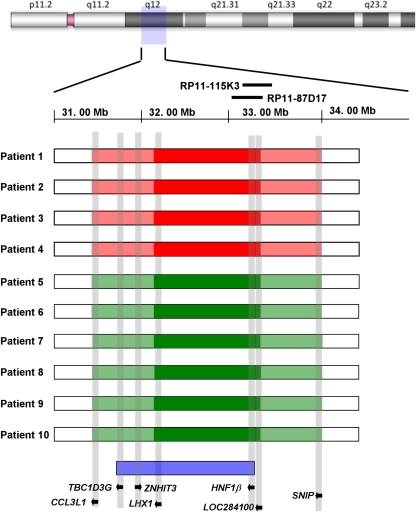
Figure 4.
The size, extent, and genomic content of the deletions and duplications of 17q12 in our cohort of patients. The cases with deletions are depicted in red, whereas those with duplications are shown in green. The dark colored blocks represent the regions of minimal deletion or duplication, whereas the extended areas shaded in lighter color represent the maximal possible extent of the rearrangements. The previously mapped minimal critical region for renal malformations and/or diabetes due to recurrent deletions of 17q12 is depicted at the bottom.6
High-resolution whole-genome oligonucleotide array
As there was a gap in coverage on the clinical array in this region, a high-resolution whole-genome array was used to further delineate the end points in patient 3.The high-resolution array showed that the minimal deletion in patient 3 encompassed a region of 1.4 Mb (31889297–33323037 bp) mapping from ZNHIT3 to LOC28400 genes. However, the regions between genomic coordinates 31504564–31889297 and 33323031–33708879 represent many copy number polymorphisms and are not covered by this whole-genome array (Figure 2b). Hence, the higher resolution array did not allow a further fine mapping of deletions as compared with the clinical array.
Clinical features
Phenotype associated with deletion of 17q12 involving HNF1β
The patients with deletions of chromosome 17q12 involving HNF1β generally presented with renal disease (Table 1). The renal manifestations included cystic renal disease, multicystic renal dysplasia, and renal agenesis. Two patients (2 and 3) had preserved renal function, whereas one had a nonfunctioning right kidney (patient 1) and one had end-stage renal disease (patient 4) treated with a renal transplant. All four patients also had evidence for growth restriction, with the height in all being less than the 3rd centile. Interestingly, three patients (1, 3, and 4) had features suggestive of central nervous system involvement. Patient 1 had significant receptive and expressive speech problems, patient 3 had speech delay and complex partial seizures, and patient 4 had moderate-to-severe mental retardation and complex partial seizures.
Table 1. Clinical features of patients with deletions in chromosome 17q12.
| Patient 1 | Patient 2 | Patient 3 | Patient 4 | |
|---|---|---|---|---|
| Age at evaluation | 5 years 6 months | 4 months | 13 years | 18 Years |
| Sex | M | M | F | F |
| Prenatal renal abnormalities | + | + | + | NA |
| Dysmorphic features | − | − | − | − |
| Nervous system | ||||
| Developmental delay/cognitive impairment | + | − | + | + |
| Seizures | − | − | + | + |
| Hypotonia | − | − | − | − |
| Hypertonia | − | − | − | − |
| Structural brain abnormalities | − | − | Hyperintensities in hippocampus | Left mesial temporal sclerosis, mild cortical atrophy |
| Hearing loss | − | NA | − | Bilateral sensorineural loss |
| Renal | ||||
| Cysts | + | + | − | + |
| Renal function | Nonfunctioning right kidney | Normal | Normal | ESRD, renal transplant |
| Cystic renal disease | + | + | Normal | + |
| Renal agenesis | − | − | − | + |
| Diabetes | NA | NA | NA | Drug-induced DM after renal transplant |
Abbreviations: +, a characteristic is present; –, a characteristic is absent; DM, diabetes mellitus; ESRD, end-stage renal disease; F, female; M, male; NA, datum was not available.
Phenotype associated with duplication of 17q12 involving HNF1β
Patients with a reciprocal duplication of the region generally presented with cognitive impairment (Table 2). Patient 5 hadnonsyndromic esophageal atresia. His development was age appropriate. The other four patients had cognitive impairment. Patient 6 had global developmental delay and behavioral abnormalities, including aggression and self-injurious behaviors, whereas his mother (patient 7) had significant learning difficulties. Both patients 8 and 9 (half-sibling pair) presented with developmental delay, with patient 8 also showing behavioral abnormalities. None of the patients with a duplication in our cohort had seizures.
Table 2. Clinical features of patients with duplications in chromosome 17q12.
| Patient 5 | Patient 6 | Patient 7 | Patient 8 | Patient 9 | |
|---|---|---|---|---|---|
| Age at evaluation | 4 years 6 months | 4 years 5 months | 45 years | 4 years 4 months | 3 years 4 months |
| Sex | M | M | F | F | M |
| Dysmorphic features | − | − | − | − | + |
| Nervous system | |||||
| Developmental delay/cognitive impairment | − | + | + | + | + |
| Seizures | − | − | − | − | − |
| Hypotonia | − | − | − | − | − |
| Hypertonia | − | − | − | − | − |
| Structural brain abnormalities | − | − | NA | NA | NA |
| Hearing loss | − | − | − | NA | NA |
| Renal | |||||
| Cysts | − | NA | NA | NA | NA |
| Renal function | Normal | NA | NA | NA | NA |
| Renal imaging | Normal | NA | NA | NA | NA |
| Diabetes | − | NA | NA | NA | NA |
Abbreviations: +, a characteristic is present; –, a characteristic is absent; F, female; M, male; NA, datum was not available.
Discussion
Microdeletions of chromosome 17q12, including HNF1β, are associated with MODY5, cystic renal disease, pancreatic atrophy, and liver abnormalities.2, 3, 4, 5, 6 Earlier reports of individuals with the same or similar microdeletions have emphasized that these individuals do not have cognitive impairment or a neurological phenotype.6 However, three of the patients with deletions in 17q12 in our cohort had features of neurological involvement: two patients had speech delay and one patient had moderate-to-severe mental retardation. Two patients also presented with complex partial seizures and structural abnormalities in the hippocampi or frontal cortex. Defining a critical region for neurological involvement is hampered by the extensive and variable copy number polymorphisms in the region flanking the deletions. Mefford et al have suggested previously that the minimal critical region of these recurrent microdeletions (1.52 Mb) is between 31 835 000 and 33 357 000, and maps from TBC1D3G to HNF1β. In our patients, the minimal possible deletion mapping from LHX1 to LOC284100 spans 1.06 Mb, whereas the maximal possible deletion mapping from CCL3L1 to SNIP spans 2.46 Mb. Many of the genes that map outside the minimal critical region, but within the maximal possible deletions, such as chemokine (C–C motif) ligand receptors (CCL3L3, CCL4L1, CCL3L1, and CCL4L2) and TBC1 domain family, member 3 (TBC1D3C, TBC1D3G, TBC1D3, TBC1D3E, and TBC1D3D), have multiple copies as a normal population variant and are not likely to be dosage sensitive. Thus, the deletion of genes or regulatory elements in the common minimal critical region may be responsible for the neurological phenotype. This phenotype is clearly incompletely penetrant, as other patients with the deletion do not have evidence of neurological/cognitive problems.6
The reciprocal duplication in 17q12 is rare, and few cases with a duplication of this region have been reported.6, 10 The phenotypes associated with duplication include focal cortical dysplasia, seizures, and mental retardation.6 One case with sex reversal, Peters' anomaly, microphthalmia, glaucoma, cleft soft palate, atrial septal defect, and severe mental retardation has also been reported with a duplication of this region.10 Four of the five patients with a duplication of 17q12 in our cohort had cognitive impairment and behavioral abnormalities. Although it has been suggested that this duplication is enriched in patients with epilepsy,6 none of the patients described in this study had seizures. This may be because of an incomplete penetrance of the seizure phenotype, or because four of the five patients in our cohort were young and seizures may not manifest until a later age.
A candidate gene that may be pertinent to the neurological phenotype seen with both deletions and duplications in 17q12 is LIM homeobox 1 (LHX1).6 LHX1 encodes a member of a large protein family that contains the LIM domain, a unique cysteine-rich zinc-binding domain. LHX1 is expressed in the brain and has been implicated in Purkinje cell development in the cerebellum11 and in the migration of motor axons to the limbs.12 A mouse model with a targeted mutation of LHX1 had a lack of head structures anterior to rhombomere 3 in the hindbrain.13 These data suggest that LHX1 may be involved in the normal development of the brain.
We have shown that contrary to previous reports, individuals with deletions of 17q12 involving HNF1β can present with cognitive impairment, seizures, and structural abnormalities of the brain, in addition to the previously established features of cystic renal disease and diabetes. The reciprocal duplication in this region can be associated with cognitive impairment and behavioral abnormalities. On the basis of our genotype–phenotype correlation in our cohort, and expression and animal model studies of LHX1, we hypothesize that it is a dosage-sensitive gene and that it is a candidate gene responsible for the neurocognitive phenotype associated with rearrangements in this region.
Acknowledgments
We thank the participating families for their kind cooperation. This work was supported in part by fellowship grants from the Osteogenesis Imperfecta Foundation (SNSC), DK081735-01A1, NIH/NIGMS T32 contract grant number GM07526 and by the National Urea Cycle Foundation Research Fellowship (AE), and grant R13-0005-04/2008 from the Polish Ministry of Science and Higher Education (PS).
References
- Lupski JR, Stankiewicz P. Genomic disorders: molecular mechanisms for rearrangements and conveyed phenotypes. PLoS Genet. 2005;1:e49. doi: 10.1371/journal.pgen.0010049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellanne-Chantelot C, Clauin S, Chauveau D, et al. Large genomic rearrangements in the hepatocyte nuclear factor-1beta (TCF2) gene are the most frequent cause of maturity-onset diabetes of the young type 5. Diabetes. 2005;54:3126–3132. doi: 10.2337/diabetes.54.11.3126. [DOI] [PubMed] [Google Scholar]
- Ulinski T, Lescure S, Beaufils S, et al. Renal phenotypes related to hepatocyte nuclear factor-1beta (TCF2) mutations in a pediatric cohort. J Am Soc Nephrol. 2006;17:497–503. doi: 10.1681/ASN.2005101040. [DOI] [PubMed] [Google Scholar]
- Decramer S, Parant O, Beaufils S, et al. Anomalies of the TCF2 gene are the main cause of fetal bilateral hyperechogenic kidneys. J Am Soc Nephrol. 2007;18:923–933. doi: 10.1681/ASN.2006091057. [DOI] [PubMed] [Google Scholar]
- Faguer S, Bouissou F, Dumazer P, et al. Massively enlarged polycystic kidneys in monozygotic twins with TCF2/HNF-1beta (hepatocyte nuclear factor-1beta) heterozygous whole-gene deletion. Am J Kidney Dis. 2007;50:1023–1027. doi: 10.1053/j.ajkd.2007.06.016. [DOI] [PubMed] [Google Scholar]
- Mefford HC, Clauin S, Sharp AJ, et al. Recurrent reciprocal genomic rearrangements of 17q12 are associated with renal disease, diabetes, and epilepsy. Am J Hum Genet. 2007;81:1057–1069. doi: 10.1086/522591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu X, Shaw CA, Patel A, et al. Clinical implementation of chromosomal microarray analysis: summary of 2513 postnatal cases. PLoS ONE. 2007;2:e327. doi: 10.1371/journal.pone.0000327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ou Z, Kang SH, Shaw CA, et al. Bacterial artificial chromosome-emulation oligonucleotide arrays for targeted clinical array-comparative genomic hybridization analyses. Genet Med. 2008;10:278–289. doi: 10.1097/GIM.0b013e31816b4420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Probst FJ, Roeder ER, Enciso VB. Chromosomal microarray analysis (CMA) detects a large X chromosome deletion including FMR1, FMR2, and IDS in a female patient with mental retardation. Am J Med Genet A. 2007;143A:1358–1365. doi: 10.1002/ajmg.a.31781. [DOI] [PubMed] [Google Scholar]
- Mencarelli MA, Katzaki E, Papa FT, et al. Private inherited microdeletion/microduplications: implications in clinical practice. Eur J Med Genet. 2008;51:409–416. doi: 10.1016/j.ejmg.2008.06.003. [DOI] [PubMed] [Google Scholar]
- Zhao Y, Kwan KM, Mailloux CM, et al. LIM-homeodomain proteins Lhx1 and Lhx5, and their cofactor Ldb1, control Purkinje cell differentiation in the developing cerebellum. Proc Natl Acad Sci USA. 2007;104:13182–13186. doi: 10.1073/pnas.0705464104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kania A, Johnson RL, Jessell TM. Coordinate roles for LIM homeobox genes in directing the dorsoventral trajectory of motor axons in the vertebrate limb. Cell. 2000;102:161–173. doi: 10.1016/s0092-8674(00)00022-2. [DOI] [PubMed] [Google Scholar]
- Shawlot W, Behringer RR. Requirement for Lim1 in head-organizer function. Nature. 1995;374:425–430. doi: 10.1038/374425a0. [DOI] [PubMed] [Google Scholar]