Abstract
We are developing a method to identify cellular resistance to carboplatin by using accelerator mass spectrometry to measure carboplatin-DNA adducts formed from drug microdoses (~1/100th the therapeutic dose). Such an approach would be particularly useful if it is still valid in combination chemotherapy. We examined whether the addition of gemcitabine, another chemotherapeutic drug, could influence carboplatin-DNA adduct levels. There were no substantial differences in the levels of carboplatin-DNA adducts in cells upon exposure to the carboplatin/gemcitabine combination at various doses and schedules. These data demonstrate that microdosing is feasible for characterization of carboplatin resistance when given in combination with gemcitabine.
Carboplatin (cis-diammine(cyclobutane-1,1-dicarboxylate-O,O’)platinum(II)) is among the most commonly used anti-cancer chemotherapeutic drugs. Chemoresistance is among the most frequent causes of treatment failure. For example, the tumor response rate for non-small cell lung cancer (NSCLC), the most common cause of cancer death, is typically less than 30% following platinum (Pt)-based combination chemotherapy. It is approximately 50% for advanced bladder cancer. Carboplatin kills cells mainly through induction of carboplatin-DNA adducts. It is likely that low levels of adducts and/or rapid DNA repair result in cellular resistance to carboplatin. Several clinical showed that low levels of DNA adducts correlated with poor chemotherapy outcomes (1-11). However, the reported Pt-DNA measurement methods, such as atomic absorption spectroscopy and immunohistochemistry, lack the sensitivity required for clinical use, since patients have to receive high-dose toxic chemotherapy before chemoresistance can be identified.
We report here the use of accelerator mass spectrometry (AMS) to detect carboplatin-DNA monoadducts induced in cells by carboplatin “microdoses”, defined as 1/100th of the therapeutic dose (12). AMS measures 14C at the attomole (10-18) level or less in milligram-size specimens (13). After treatment with 14C-labeled carboplatin, AMS can measure 14C bound to genomic DNA, and allow the calculation of carboplatin-DNA monoadducts, the precursors of all other forms of carboplatin-DNA damage (Figure 1). Because of the high sensitivity of AMS, we are able to measure carboplatin-DNA adducts after treatment with subtoxic microdoses of [14C]carboplatin, and can perform repeated measurements with small clinical specimens. We hypothesize that carboplatin-DNA monoadducts induced by microdoses of carboplatin are useful as biomarkers of chemoresistance. However, carboplatin is almost always combined with other drugs in standard clinical practice. For example, the combination of carboplatin and gemcitabine (4-amino-1-(2-deoxy-2,2-difluoro-β-D-erythro-pentofuranosyl)pyrimidin-2(1H)-on 2′,2′-difluoro-2′-deoxycytidine) is widely accepted as a first-line regimen for patients with bladder cancer, NSCLC and other cancer types. The goal of the current study was to determine if addition of gemcitabine affects the formation of carboplatin-DNA monoadducts and, therefore, potentially confound our approach to identifying chemoresistance using the microdose strategy.
Figure 1. Mechanism of carboplatin-DNA monoadduct and crosslink formation.
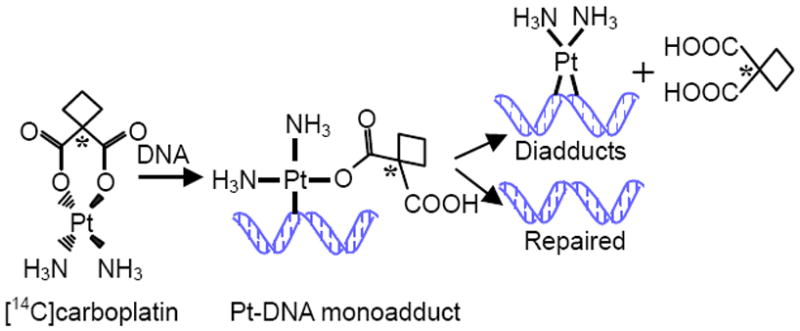
AMS can measure the 14C tag on the cyclobutane dicarboxylate ligand present in carboplatin-DNA monoadducts. Upon diadduct formation or DNA repair, the 14C falls off and cannot be detected in the DNA with AMS. (An asterisk represents the location of the 14C atom).
We compared the carboplatin-DNA monoadduct formation and repair to IC50 values for the 5637, T24 and TCCSUP bladder cancer cell lines. The carboplatin IC50 values, as determined with the MTT assay (14), were 10.2±1.2, 21.8±1.5 and 35.0±6.6 μM, respectively, for 5637, T24 and TCCSUP cells. For DNA monoadduct measurements, the cells were dosed for 4h with [14C]carboplatin at 1 μM (microdose) or 100 μM (therapeutic dose), washed with PBS, and incubated in carboplatin-free cell culture media for 20h. DNA was extracted from the cells at 0, 2, 4, 8 and 24 h time points after the initial exposure to carboplatin (in triplicate for each time point). The 14C content of the purified genomic DNA was measured by AMS and the carboplatin-DNA monoadduct concentrations were calculated from the data as previously described (15, 16). As shown in Figure 2A-B, each cell line had a different amount of carboplatin-DNA damage over 24h. The damage ranged from ~1-10 monoadducts per 108 nucleotides (nt) for the microdose, and ~100-1000 monoadducts per 108 nt for the therapeutic dose--an approximate 100-fold difference. The concentration of monoadducts induced by microdoses was linearly proportional to that produced by therapeutic carboplatin (Figure 2C). The maximum damage occurred between the 4h and 8h time points post dose, and was in the order of TCCSUP>T24>5637. This order of monoadduct levels is contrary to that expected to support the hypothesis that carboplatin-DNA adducts correlate positively with chemosensitivity. However, the DNA repair rates for each cell line were in the same order as would be expected if the persistence of DNA damage influences resistance. The average repair rate for each microdosed cell line was TCCSUP>T24>5637 (3.4, 3.9 and 4.1 % per hour, with p =0.019 and 0.006 for comparison of the T24 and TCCSUP to 5637). The average repair rate for each therapeutically dosed cell line was same order as above (1.6, 3.5 and 4.1 % per hour, respectively, with p =0.037 and 0.012 for comparison of the T24 and TCCSUP to 5637). In conclusion, the overall DNA damage and repair rate was unique for each cell line; carboplatin-DNA monoadduct levels were proportional to dose for each cell line; and DNA repair rates correlated positively with chemoresistance that are consistent with previous reports (17-19).
Figure 2. Carboplatin-DNA monoadducts induced by [14C]carboplatin without or with gemcitabine.
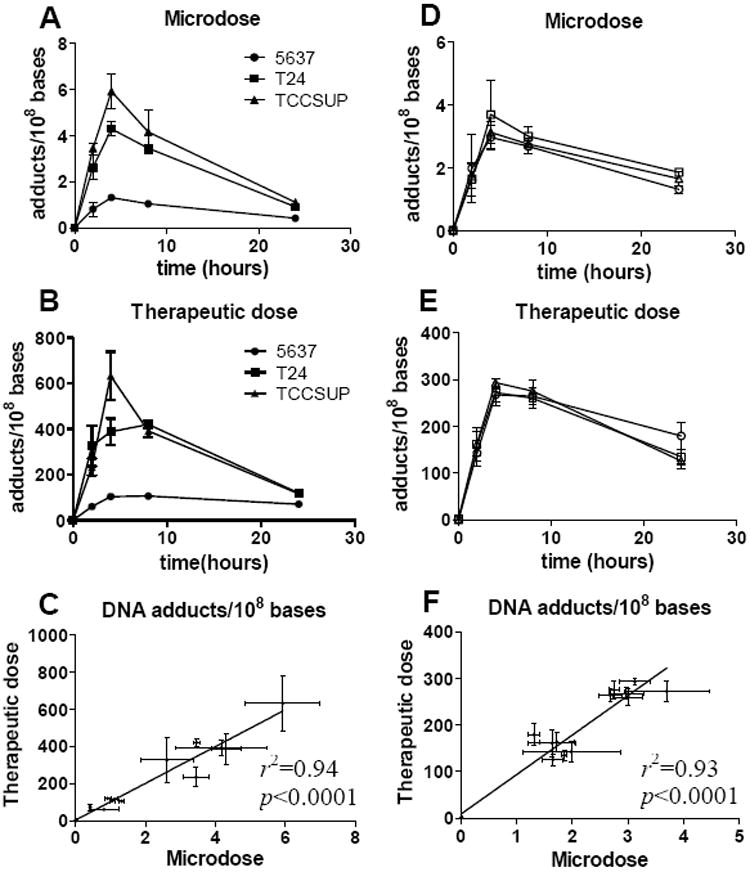
A and B: The levels of carboplatin-DNA monoadducts in three bladder cancer cell lines 5637, T24 and TCCSUP cells. C. Linear regression analysis of the DNA adduct data from A and B. The concentration of carboplatin-DNA monoadducts formed by [14C]carboplatin microdoses are linearly proportional to those caused by therapeutic carboplatin. D and E: The levels of carboplatin-DNA monoadducts as a result of gemcitabine exposure at 0.3, 3 and 30 μM, are represented by squares, circles and triangles, respectively, as the mean ± s.d.. Cells were exposed to gemcitabine for 4 h followed by carboplatin exposure at 1 μM (D), or 100 μM for 4 hours (E). F. Linear regression analysis of the data from D and E. The concentration of carboplatin-DNA monoadducts formed by [14C]carboplatin microdoses are linearly proportional to those caused by therapeutic carboplatin, regardless of gemcitabine exposure
We determined whether addition of different doses of gemcitabine before carboplatin treatment could modulate carboplatin-DNA monoadduct levels. We used the 5637 bladder cancer cell line for all experiments due to the clinical relevance of gemcitabine and carboplatin for bladder cancer treatment. Cells were treated with gemcitabine at 0.3 μM (microdose), 3 μM (intermediate dose) and 30 μM for 4h, washed and subsequently incubated for 4h with [14C]carboplatin at 1 μM (microdose) or 100 μM (therapeutic dose). The highest gemcitabine and carboplatin drug concentrations used in this study were based upon the reported Cmax values of 30 μM and 100 μM, respectively, in human plasma during chemotherapy (20, 21). Addition of gemcitabine did not change the kinetics of carboplatin-DNA monoadduct formation and disappearance when corrected for carboplatin dose. Microdose, intermediate, or therapeutic doses of gemcitabine did not significantly change carboplatin-DNA monoadduct levels caused by micro- (Figure 2D) or therapeutic doses of carboplatin (Figure 2E). The three curves in Figure 2D or E overlapped, indicating little or no influence of gemcitabine on monoadduct levels. Compared to the 0.3 μM gemcitabine group, the p values were 0.098 and 0.128, respectively, for the 3 and 30 μM groups in the microdose carboplatin treatment (Figure 2D); and 0.753 and 0.229, respectively, in the therapeutic carboplatin treatment (Figure 2E), not statistically significant. Regression analysis showed a strong linear relationship between the DNA adduct levels induced both doses of carboplatin in the absence or presence of different concentrations of gemcitabine (p<0.0001, Figure 2F).
We then determined if different doses and sequences of carboplatin and gemcitabine exposure would influence carboplatin-DNA monoadduct levels at the 4 h time point, the time of peak carboplatin-DNA monoadduct formation. We compared the adduct levels in 8 different dose-schedule combinations: gemcitabine followed by carboplatin versus carboplatin followed by carboplatin/gemcitabine, microdose carboplatin versus therapeutic dose carboplatin, and microdose gemcitabine versus therapeutic dose gemcitabine (2 × 2 × 2 =8 combinations) (Figure 3). Compared the adduct levels in cells treated with carboplatin alone, there was a slight trend toward increased carboplatin-DNA monoadducts in cells treated with microdose or therapeutic gemcitabine before exposure to carboplatin, but the trend did not reach statistical significance (p = 0.19 and 0.095, respectively). Addition of gemcitabine after carboplatin exposure, either at microdose or therapeutic dose concentrations, did not significantly change the levels of carboplatin-DNA monoadducts (Figure 3A, p=0.91 and 0.83, respectively). Only upon treatment with gemcitabine at 30 μM for 4 hours followed by carboplatin treatment at 100 μM for 4 more hours did carboplatin-DNA monoadduct levels significantly increase (21 % compared to therapeutic dose carboplatin alone, p=0.037, denoted by an asterisk in Figure 3B). It is possible that the increase in carboplatin-DNA adduct levels might be related to gemcitabine toxicity (22), as 30 μM of gemcitabine is about 350 times the IC50 (0.086 μM) for this cell line. There was no statistical difference between the two groups when cells were treated with microdose (0.3 μM) or therapeutic (30 μM) concentrations of gemcitabine followed by carboplatin (p = 0.10), or in any other treatment groups.
Figure 3. Carboplatin-DNA monoadducts at 4h after dosing with carboplatin and gemcitabine at different doses and schedules.
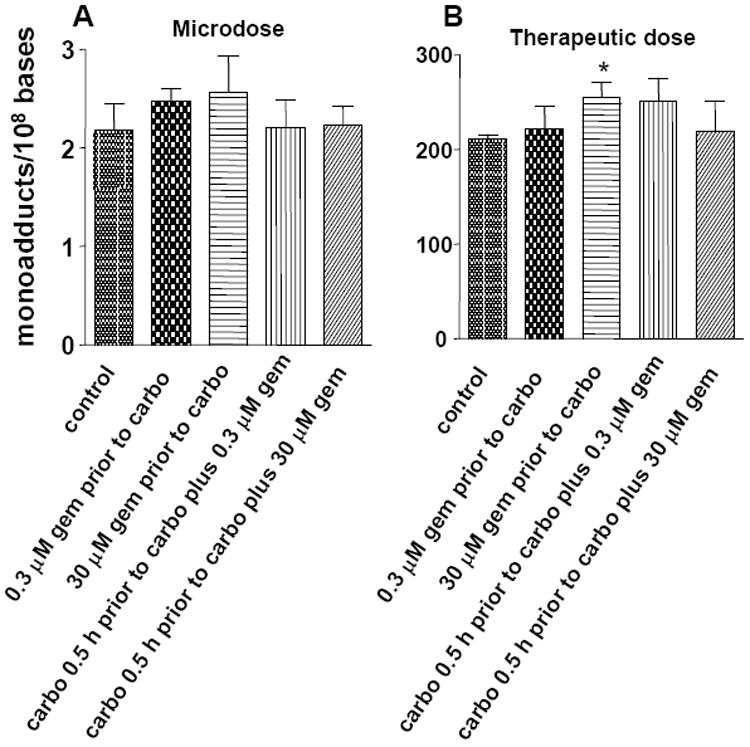
DNA adducts are shown as the mean ± s.d. of three independent experiments. The dose and treatment schedules are outlined below the bar graph. *p=0.037 when compared with the control.
We are developing a highly sensitive methodology using AMS to determine if chemoresistance to carboplatin can be characterized and identified by measuring monoadduct formation and repair, the critical step of carboplatin cytotoxicity. Clinically, carboplatin is usually combined at least one other chemotherapeutic drug. This study addressed a critical issue of our microdosing approach--whether the addition of another chemotherapy drug affects the formation and repair of carboplatin-DNA monoadducts. Among all of the doses and administration schedules we tested in this study, there was only one instance where a statistically significant difference was found after adding gemcitabine to the cells. However, this difference is not clinically relevant as it involved treating the cells with gemcitabine at 30 μM for 4 hours. This would not occur clinically as the in vivo peak concentration (Cmax) of gemcitabine is around 30 μM (26), and it is rapidly metabolized in plasma with a half life of less than 10 minutes (27). Therefore, the total exposure of the cells to gemcitabine in this study was well above what cancer cells are exposed in vivo under physiological conditions.
In conclusion, addition of gemcitabine does not significantly affect the levels of carboplatin-induced DNA monoadducts, except when super-physiologic exposures to gemcitabine are used prior to therapeutic carboplatin dosing. These observations are important for the extension of the carboplatin microdosing approach to animal and human studies of drug resistance.
Acknowledgments
We thank Ms. Miaoling He for technical support. This work was supported by the Susan and Gerry Knapp Foundation (PTH) and National Institutes of Health Grant RR13461 (KWT), and by the American Cancer Society Institutional Research Grant (CXP).
References
- 1.Parker RJ, Gill I, Tarone R, Vionnet JA, Grunberg S, Muggia FM, Reed E. Platinum-DNA damage in leukocyte DNA of patients receiving carboplatin and cisplatin chemotherapy, measured by atomic absorption spectrometry. Carcinogenesis. 1991;12:1253–1258. doi: 10.1093/carcin/12.7.1253. [DOI] [PubMed] [Google Scholar]
- 2.Bonetti A, Apostoli P, Zaninelli M, Pavanel F, Colombatti M, Cetto GL, Franceschi T, Sperotto L, Leone R. Inductively coupled plasma mass spectroscopy quantitation of platinum-DNA adducts in peripheral blood leukocytes of patients receiving cisplatin- or carboplatin-based chemotherapy. Clin Cancer Res. 1996;2:1829–1835. [PubMed] [Google Scholar]
- 3.Fichtinger-Schepman AM, van der Velde-Visser SD, van Dijk-Knijnenburg HC, van Oosterom AT, Baan RA, Berends F. Kinetics of the formation and removal of cisplatin-DNA adducts in blood cells and tumor tissue of cancer patients receiving chemotherapy: comparison with in vitro adduct formation. Cancer Res. 1990;50:7887–7894. [PubMed] [Google Scholar]
- 4.Fisch MJ, Howard KL, Einhorn LH, Sledge GW. Relationship between platinum-DNA adducts in leukocytes of patients with advanced germ cell cancer and survival. Clin Cancer Res. 1996;2:1063–1066. [PubMed] [Google Scholar]
- 5.Hoebers FJ, Pluim D, Hart AA, Verheij M, Balm AJ, Fons G, Rasch CR, Schellens JH, Stalpers LJ, Bartelink H, Begg AC. Cisplatin-DNA adduct formation in patients treated with cisplatin-based chemoradiation: lack of correlation between normal tissues and primary tumor. Cancer Chemother Pharmacol. 2008;61:1075–1081. doi: 10.1007/s00280-007-0545-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Motzer RJ, Reed E, Perera F, Tang D, Shamkhani H, Poirier MC, Tsai WY, Parker RJ, Bosl GJ. Platinum-DNA adducts assayed in leukocytes of patients with germ cell tumors measured by atomic absorbance spectrometry and enzyme-linked immunosorbent assay. Cancer. 1994;73:2843–2852. doi: 10.1002/1097-0142(19940601)73:11<2843::aid-cncr2820731130>3.0.co;2-d. [DOI] [PubMed] [Google Scholar]
- 7.Reed E, Ozols RF, Tarone R, Yuspa SH, Poirier MC. Platinum-DNA adducts in leukocyte DNA correlate with disease response in ovarian cancer patients receiving platinum-based chemotherapy. Proc Natl Acad Sci U S A. 1987;84:5024–5028. doi: 10.1073/pnas.84.14.5024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Reed E, Ozols RF, Tarone R, Yuspa SH, Poirier MC. The measurement of cisplatin-DNA adduct levels in testicular cancer patients. Carcinogenesis. 1988;9:1909–1911. doi: 10.1093/carcin/9.10.1909. [DOI] [PubMed] [Google Scholar]
- 9.Reed E, Parker RJ, Gill I, Bicher A, Dabholkar M, Vionnet JA, Bostick-Bruton F, Tarone R, Muggia FM. Platinum-DNA adduct in leukocyte DNA of a cohort of 49 patients with 24 different types of malignancies. Cancer Res. 1993;53:3694–3699. [PubMed] [Google Scholar]
- 10.Schellens JH, Ma J, Planting AS, van der Burg ME, van Meerten E, de Boer-Dennert M, Schmitz PI, Stoter G, Verweij J. Relationship between the exposure to cisplatin, DNA-adduct formation in leucocytes and tumour response in patients with solid tumours. Br J Cancer. 1996;73:1569–1575. doi: 10.1038/bjc.1996.296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.van de Vaart PJ, Belderbos J, de Jong D, Sneeuw KC, Majoor D, Bartelink H, Begg AC. DNA-adduct levels as a predictor of outcome for NSCLC patients receiving daily cisplatin and radiotherapy. Int J Cancer. 2000;89:160–166. [PubMed] [Google Scholar]
- 12.Henderson PT, Pan C-x. Human microdosing for the prediction of patient response. Bioanalysis. 2010;2:373–376. doi: 10.4155/bio.10.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Turteltaub KW, Dingley KH. Application of accelerated mass spectrometry (AMS) in DNA adduct quantification and identification. Toxicol Lett. 1998;102-103:435–439. doi: 10.1016/s0378-4274(98)00344-0. [DOI] [PubMed] [Google Scholar]
- 14.Mosmann T. Rapid colorimetric assay for cellular growth and survival: application to proliferation and cytotoxicity assays. J Immunol Methods. 1983;65:55–63. doi: 10.1016/0022-1759(83)90303-4. [DOI] [PubMed] [Google Scholar]
- 15.Dingley KH, Ubick EA, Vogel JS, Haack KW. DNA isolation and sample preparation for quantification of adduct levels by accelerator mass spectrometry. Methods Mol Biol. 2005;291:21–27. doi: 10.1385/1-59259-840-4:021. [DOI] [PubMed] [Google Scholar]
- 16.Hah SS, Stivers KM, de Vere White RW, Henderson PT. Kinetics of carboplatin-DNA binding in genomic DNA and bladder cancer cells as determined by accelerator mass spectrometry. Chem Res Toxicol. 2006;19:622–626. doi: 10.1021/tx060058c. [DOI] [PubMed] [Google Scholar]
- 17.Parker RJ, Eastman A, Bostick-Bruton F, Reed E. Acquired cisplatin resistance in human ovarian cancer cells is associated with enhanced repair of cisplatin-DNA lesions and reduced drug accumulation. J Clin Invest. 1991;87:772–777. doi: 10.1172/JCI115080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dabholkar M, Parker R, Reed E. Determinants of cisplatin sensitivity in non-malignant non-drug-selected human T cell lines. Mutat Res. 1992;274:45–56. doi: 10.1016/0921-8777(92)90042-2. [DOI] [PubMed] [Google Scholar]
- 19.Dabholkar M, Bradshaw L, Parker RJ, Gill I, Bostick-Bruton F, Muggia FM, Reed E. Cisplatin-DNA damage and repair in peripheral blood leukocytes in vivo and in vitro. Environmental Health Prospectives. 1992;98:53–59. doi: 10.1289/ehp.929853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Soo RA, W LZ, Tham LS. A multicentre randomised phase II study of carboplatin in combination with gemcitabine at standard rate or fixed dose rate infusion in patients with advanced stage non-small-cell lung cancer. Annals of Oncology. 2006;17:1128–1133. doi: 10.1093/annonc/mdl084. [DOI] [PubMed] [Google Scholar]
- 21.Oguri S, Sakakibara T, Mase H, Shimizu T, Ishikawa K, Kimura K, Smyth RD. Clinical pharmacokinetics of carboplatin. J Clin Pharmacol. 1988;28:208–215. doi: 10.1002/j.1552-4604.1988.tb03134.x. [DOI] [PubMed] [Google Scholar]
- 22.Yang LY, Li L, Jiang H, Shen Y, Plunkett W. Expression of ERCC1 antisense RNA abrogates gemicitabine-mediated cytotoxic synergism with cisplatin in human colon tumor cells defective in mismatch repair but proficient in nucleotide excision repair. Clin Cancer Res. 2000;6:773–781. [PubMed] [Google Scholar]
- 23.Stewart DJ. Mechanisms of resistance to cisplatin and carboplatin. Crit Rev Oncol Hematol. 2007;63:12–31. doi: 10.1016/j.critrevonc.2007.02.001. [DOI] [PubMed] [Google Scholar]
- 24.Matsuoka S, Ballif BA, Smogorzewska A, McDonald ER, 3rd, Hurov KE, Luo J, Bakalarski CE, Zhao Z, Solimini N, Lerenthal Y, Shiloh Y, Gygi SP, Elledge SJ. ATM and ATR substrate analysis reveals extensive protein networks responsive to DNA damage. Science. 2007;316:1160–1166. doi: 10.1126/science.1140321. [DOI] [PubMed] [Google Scholar]
- 25.Roberti A, La Sala D, Cinti C. Multiple genetic and epigenetic interacting mechanisms contribute to clonally selection of drug-resistant tumors: current views and new therapeutic prospective. J Cell Physiol. 2006;207:571–581. doi: 10.1002/jcp.20515. [DOI] [PubMed] [Google Scholar]
- 26.Soo RA, Wang LZ, Tham LS, Yong WP, Boyer M, Lim HL, Lee HS, Millward M, Liang S, Beale P, Lee SC, Goh BC. A multicentre randomised phase II study of carboplatin in combination with gemcitabine at standard rate or fixed dose rate infusion in patients with advanced stage non-small-cell lung cancer. Ann Oncol. 2006;17:1128–1133. doi: 10.1093/annonc/mdl084. [DOI] [PubMed] [Google Scholar]
- 27.Abbruzzese JL, Grunewald R, Weeks EA, Gravel D, Adams T, Nowak B, Mineishi S, Tarassoff P, Satterlee W, Raber MN, et al. A phase I clinical, plasma, and cellular pharmacology study of gemcitabine. J Clin Oncol. 1991;9:491–498. doi: 10.1200/JCO.1991.9.3.491. [DOI] [PubMed] [Google Scholar]


