Abstract
In several individuals with a Charcot–Marie–Tooth (CMT) phenotype, we found a copy number variation (CNV) on chromosome 17p12 in the direct vicinity of the peripheral myelin protein 22 (PMP22) gene. The exact borders and size of this CNV were determined by Southern blot analysis, MLPA, vectorette PCR, and microarray hybridization analyses. All patients from six apparently unrelated families carried an identical 186-kb duplication different from the commonly reported 1.5-Mb duplication associated with CMT1A. This ancestral mutation that was not reported in the human structural variation database was only detected in affected individuals and family members. It was absent in 2124 control chromosomes and 40 patients with a chronic inflammatory demyelinating polyneuropathy (CIDP) and therefore should be regarded as causative for the disease. This variant escapes most routine diagnostic screens for CMT1A, because copy numbers of PMP22 probes were all normal. No indications were found for the involvement of the genes that are located within this duplication. A possible association of this duplication with a mutation in the PMP22 coding regions was also excluded. We suggest that this CNV proximal of the PMP22 gene leads to CMT through an unknown mechanism affecting PMP22 expression.
Keywords: CMT, HMSN, PMP22, TEKT3, CNV
Introduction
Segmental duplications or low-copy repeats that typically share a high rate of homology (>90%) are considered pivotal for evolution of the human genome. The duplicated regions provide a substrate for non-allelic homologous recombination and as such represent recombination hotspots resulting in duplication, deletion, or inversion of the intervening sequences. Frequently, they are associated with structural variations or copy number variations (CNVs), many of which include functional genes and a relatively large part also contributes to human disease.1
One of the best studied examples is the reciprocal duplication and deletion of a 1.5-Mb region on chromosomal segment 17p12 caused by unequal crossing over because of misalignment of the highly homologous flanking 24-kb Charcot–Marie–Tooth (CMT) repeat regions that results in the most prevalent form of demyelinating CMT type 1A, and hereditary neuropathy with pressure palsies (HNPP), respectively.2, 3, 4
The peripheral myelin protein 22(PMP22) gene that is located within this large genomic region was identified as the disease causing gene5, 6, 7, 8 as is supported by the occurrence of natural mouse Pmp22 mutants Trembler and Trembler-J,9, 10 CMT patients with point mutations in PMP2211, 12 and several mouse and rat CMT models with similar phenotypes that harbor more copies of PMP22.13, 14, 15 PMP22 is an integral membrane protein that contributes to compact myelin of the peripheral nervous system. It was originally isolated as a growth arrest-specific gene (Gas3) in mouse fibroblasts16 and is highly expressed in myelinating Schwann cells using an alternative promoter.17 Both abnormal localization and expression have been described in nerve biopsies of CMT patients18, 19 and whereas altered gene dosage is the generally accepted mechanism through which the disease develops, further details of this mechanism are still largely unknown.
A few alternatively sized duplications or deletions on 17p11.2 have been reported to be associated with CMT in patients, but in all cases PMP22 was located within the genomic aberration.20, 21, 22 In this study, we describe an identical duplication of 186 kb containing the TEKT3 gene proximal to PMP22 in 11 patients with CMT from six apparently unrelated families that cosegregates with the disease in two families studied and is absent in more than 2000 control chromosomes. We postulate that this duplication also leads to CMT through an as yet unidentified mechanism possibly affecting the expression of PMP22.
Materials and methods
Patients
Clinical information of the patients is given in Table 1. For two patients, SD4 and SD9, several family members could be tested (Figure 1). The phenotype is variable but seems rather mild in most cases with a relatively late age of onset in some cases and normal to brisk reflexes in most patients and therefore, clinically more resembled an axonal polyneuropathy. For three patients, other diagnoses than CMT1 were considered. However, EMG findings showed the demyelinating nature of the disease although some NCVs were also only mildly reduced. Chronic inflammatory demyelinating polyneuropathy (CIDP) patients were diagnosed according to ENMC guidelines.23
Table 1. Clinical information of patients from separate families.
| SD1 | SD2 | SD3 | SD4 (family II) | SD9 (family I) | SD14 |
|---|---|---|---|---|---|
| Gender: Male | Male | Male | Female | Male | Male |
| Age of onset: <10y | 22–23y | 58y | 58y | 36y | 54y |
| Foot abnormalities: Pes cavus | Normal feet | Foot drop | Foot drop | Pes cavus | Normal feet |
| Family history: parents neurologically examined and found normal | Unclear; parents and siblings healthy; possibly affected brother of grandfather | Unclear; footdrop present in family of healthy mother | Affected childrena and two other family members | Affected mother and childrena | Affected sistera |
| Characteristics: Weakness and sensory abnormalities arms and legs Tremor hands Disturbed sense of vibration, touch, and pain in feet and legs Course slowly progressive; patient examined several times between ages of 32–44 | Weakness arms and hands, slight reduction of strength of legs Disturbed sense of vibration and position of hands and foot Reduced sense of vibration and pain in fingers and feet Course progressive | Absence of sensory abnormalities Severe atrophy hands and lower legs History of back pains | Progressing atrophy in muscles of hands and legs; 4 years after presentation disturbed sensitivity lower legs; sense of vibration and pain intact; 7 years after presentation spontaneous activity hands and legs | Discrete weakness lower legs, no sensory disturbances HMSN found by coincidence due to complaints of pain in back and legs Hip surgeries at age 14 | No weakness in upper arms or legs, no atrophy hands, distal weakness and sensory abnormalities: reduced sense of pain legs, reduced sense of vibration feet |
| Reflexes at first low, later described as brisk. Pyramidal signs (Babinski) | Reflexes normal, 1 year later brisk | Reflexes normal | Reflexes normal, 7 years after presentation low or non-responsive | Reflexes: normal | Reflexes normal except for Achilles tendon |
| EMG: nerve conduction uniformly decreased MNCVs median nerve 31–39 m/s Sural nerve biopsy shows de- and remyelination (onion bulbs) | EMG: multifocal motor and sensory demyelinating PNP that also fulfill criteria CIDP; NCV legs: 23–30 m/s, arms normal except for ulnar nerve: 40 m/s | EMG: NCVs 35–40 m/s; in legs 19–26 m/s; SNAP radial and sural nerve NR, decreased SNAP amplitude of ulnar and median nerve with reduced NCVs; MNCV ulnar nerve severely reduced | EMG: MNCV right ulnar nerve 45.6 m/s, left sensory ulnar nerve 48.8 m/s, MNCV right tibial nerve 18.5 m/s, left: DML 15.0 ms, MNCV peroneal nerve, sural nerve NR | EMG: peroneal nerve NR, normal NCVs for ulnar nerve with extended DML (3.8 ms), MNCV tibial nerve 24 m/s with decreased amplitude and extended DML (5.5 ms) | EMG of arms: motor and sensory NCVs uniformly severely decreased, extended DML, reduced SNAP median and ulnar nerve |
| Other diagnoses considered: HMSN5 | CIDP | Distal spinal muscle atrophy, CIDP, HMSN2; EMG demonstrated HSMN1 |
Abbreviations: NR, non-responsive; y, year.
Information on family members: SD5, SD6 are two affected children of SD4 (index patient family II); one female with pes cavus and mild motor and sensory abnormalities, normal reflexes, reduced MNCVs of tibial and sural nerve; normal median nerve MNCV with DML slightly extended, and one male with history of back complaints, temporary loss of sensitivity but otherwise no evident atrophies and normal reflexes. For related family members D7 and D8 no detailed information was available. SD9 is the index patient of family I, EMG data of ulnar, peroneal, and tibial nerve of the affected children showed a mildly reduced MNCV (39 m/s) in the tibial nerve of one affected child (SD13) and a mildly reduced (38 m/s), and reduced (31 m/s) MNCV with lower amplitude (2.1 mV), of the ulnar and tibial nerve with extended DML (4.3 and 5 ms, respectively) in the other affected child (SD10). The mother of SD9, SD11, was examined for the presence of carpal tunnel syndrome only (extended DML of 8.3 ms and low amplitude (1.1 mV) of median nerve). The affected sister of SD14 showed axonal NCVs on EMG.
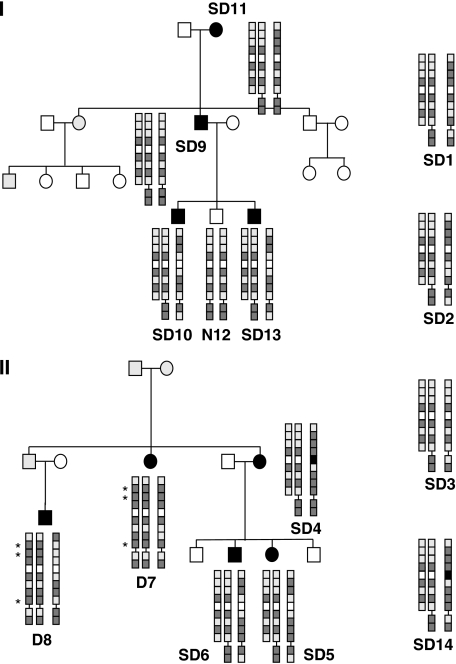
Figure 1.
Pedigrees of Charcot–Marie–Tooth (CMT) families I and II and haplotype of patients. Family trees of both families are depicted with affected members with filled symbols. Gray symbols indicate that the phenotype could not be certainly assessed or was not known. All affected members and unaffected member N12 from family I were screened for the presence of duplications. Patients D7 and D8 from family II carried the conventional 1.5-Mb duplication instead of the 186-kb duplication present in patients SD4–6. TEKT3 and PMP22 polymorphisms are represented as squares in the order as they appear on the coding/-strand of the chromosome from centromere to telomere (nine TEKT3 and two PMP22 SNPs). For reasons of clarity the (inferred) haplotypes of the tandem duplication are depicted next to each other. Dark shaded squares represent the most frequently occurring allele, and the light shaded squares the minor allele. In one case, for rs11411664, frequencies were unknown and the alleles were represented by black and white boxes, respectively. Represented SNPs are rs396445, rs7226363, rs2305959, rs230901, rs11411664, rs230898, rs230897, rs2286516, and rs13961 of the TEKT3 gene and rs231020 and rs3744333 of PMP22. Known frequencies of the TEKT3 alleles associated with the small duplication of represented SNPs are 0.14–0.18, 0.135–0.217, 0.2, 0.9, 0.47–0.54, 0.475, 0.7, 0.217–0.25, respectively. The haplotypes of single cases SD1–3 and SD14 are also provided in the same manner.
Southern blot analysis and MLPA
DNA isolation of blood samples was performed according to standard methods. After digestion of 5–10 μg of DNA with appropriate restriction enzymes and size fractionation on agarose gels, DNA was blotted onto Hybond N-Plus membranes (Amersham, Diegem, Belgium). Hybridization was performed according to the method of Church and Gilbert (1984)24 and 32P-α-dCTP radiolabeled probes of small PCR fragments (312–1197 bp) located between PMP22 and TEKT3 were made by random prime labeling. For normalization, control probe E3.9, located on chromosome 2225 was added to the hybridization mixture. To suppress background signals because of repetitive sequences, 5–10 μl of Hybridime (10 mg/ml) was preannealed to the probe mixture. A VAW409/exon 6 TEKT3 probe was included as a duplication control, DNA of patients with or without the 1.5-Mb duplication and reciprocal deletion were added as control samples. Signals were visualized by phosphor imaging and analyzed using the AIDAv3.45 software (Raytest, Straubenhardt, Germany). The average of signals of three different normal DNAs was used as a reference. Relative normalized intensities of <0.7 and >1.2 were considered indicative for a deletion and duplication, respectively.
MLPA was performed using the MLPA kit (P033B; MRC Holland, Amsterdam, the Netherlands) according to the protocol of the manufacturer. Data were analyzed by the ABI Genescan programs (Applied Biosystems, Foster City, CA, USA). Average peak areas of three different normal DNAs were taken as a reference. The total peak area of probes outside of the 17p11.2 region was used for normalization. Cutoff values for duplication and deletion were >1.2 and <0.7 respectively.
Vectorette and long-range PCRs
Vectorette PCR was performed using 1 μg of purified 5–10-kb genomic XbaI fragments using the Universal Vectorette System UVS-1 (Sigma, Zwijndrecht, the Netherlands). Additional long-range PCR reactions were performed using ExTaq (Takara, Bio Inc., Otsu, Japan). Briefly, genomic XbaI fragments 5–7.5 and 7.5–10 kb in size were ligated to an Xba vectorette cassette that was made using the method described by Riley et al. (1990).26 PCRs were primed with specific primers from the duplicated region inv1 or n2 and a vectorette primer using a touchdown protocol (3 min at 98°C, 7 cycles of 5 s 94°C; 9 min 72°C, 32 cycles 10 s 94°C; 9 min 68°C, final extension 9 min 68°C). Second or third round PCRs were performed on 1:100–1:1000 diluted PCR products of the previous round with several nested primers within the junction region (n1–n4; see Figure 2; 3 min 98°C, 7 × 5 s 98°C; 9 min 72°C, 2 × 5 s 98°C; 9 min 70°C, 31 cycles of 10 s 98°C; 9 min 68°C). PCRs were performed in buffer supplied by the manufacturer, 500 μ dNTPs, 2.5 m MgCl2, 0.5 μ of nested vector primer, and 1 μ of specific primer with or without betain 1 M as an additive. Junction PCRs on 20 ng of genomic DNA were performed using Hotfire Taq (Solis Biodyne, Tartu, Estonia) in the buffer supplied by the manufacturer, and 4 m of MgCl2, 0.25 m dNTPs, 500 n of primers (j1, j5) at an annealing temperature of 52°C. Primer sequences are given in Supplementary Table S1.
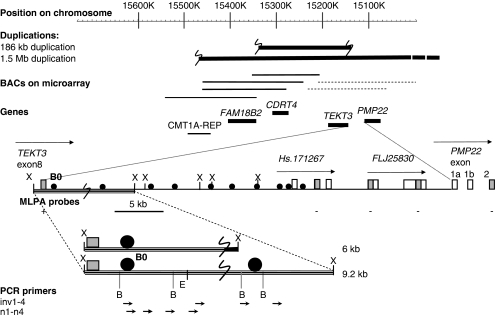
Figure 2.
Schematic overview of junction region: genes and duplicated regions on chromosome 17. The 1.5-Mb duplication is only partially drawn as indicated by the dotted line on the end because it is larger than the depicted region. BACs with a duplicated signal in the microarrays are represented by a solid thick line, the BAC with a normal signal by a thin line and the two BACs that showed partial duplication by a dotted line. The region between TEKT3 and PMP22 is shown in detail; E, B, and X represent EcoRI, BamH1, and XbaI restriction sites, filled circles represent probes used for Southern blot analysis, gray squares represent the positions of the MLPA probes. The normal 9.2-kb XbaI fragment and aberrant 6-kb XbaI fragment that were detected by Southern blot analysis by probe Bo and contain the location of the junction (large S), are depicted at the bottom. Vectorette PCR primed with inv1 or n2 and nested primers n1–n4 gave expected products of 3–4 kb in size all containing the duplication junction.
Microarray CGH
A custom-made chromosome 17 tiling path array covering the 17p13.3–17p11.2 region was made as described before.27 Shortly, clones were selected (Welcome Trust Sanger Institute, Hinxton, Cambridge, UK, http://www.ensembl.org), grown, amplified using a routine DOP–PCR protocol, and spotted in triplicate. The genomic microarray was hybridized with a combination of male and female patient control DNA mixed together with Cot DNA, scanned and the resulting images were analyzed using Genepix Pro 6.0 (Molecular Devices, Sunnydale, CA, USA). Cutoff value for duplication was a tester to reference ratio of 1.2.
Sequence analysis
After amplification, PCR products were treated using shrimp alkaline phosphatase and exonuclease I and analyzed by direct sequencing using the ABI Big Dye Terminator cycle sequencing kit and an ABI3730 sequencer (Applied Biosystems). Sequence traces were compared with the reference genomic clone sequences AC005517 and AC005703 or refseq sequences (NM_000304.2, NM_153321.1, and NM_031898.1) for PMP22 and TEKT3 using the Codon Code Aligner software (Dedham, MA, USA). Primer sequences are supplied in Supplementary Table 1.
Results
Identification of TEKT3 copy number alterations in CMT patients
A group of 3578 patients suspected of a genetic cause of CMT1 were screened for the presence of the commonly found duplication of the 1.5-Mb region on 17p12 containing the PMP22 and TEKT3 genes using Southern blot analysis or MLPA. In 20.4% of patients, the conventional 1.5-Mb duplication was found and in 9.5% of patients, a deletion of the same area was encountered. Out of 950 patients without any duplication or deletion, 44 patients carried a mutation in the coding region of PMP22. Six CMT patients showed normal copy numbers for PMP22 probes but duplication of the TEKT3 probes. For two cases, screening of family members could be performed and this resulted in the identification of five more affected individuals with the same pattern of duplication. Three additional MLPA probes located in the genomic region between PMP22 and TEKT3 (Figure 2) also showed normal copy numbers for all these patients.
Mapping of the duplication and identification of a junction fragment
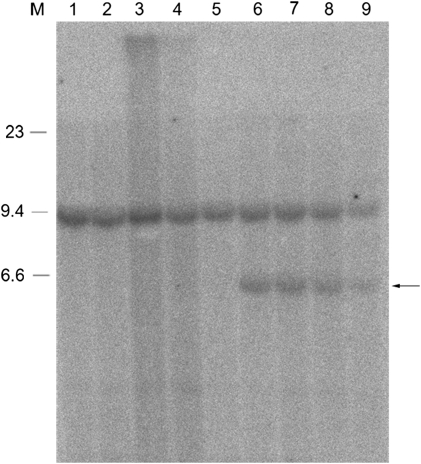
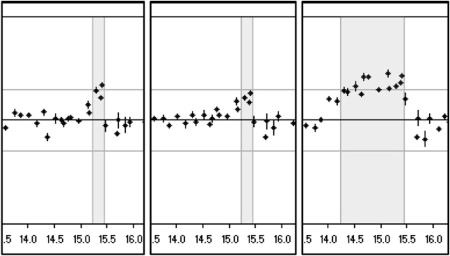
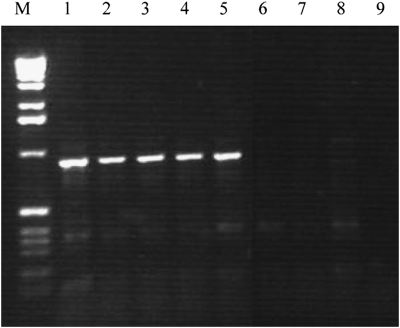
To determine the exact size of the duplicated region, several PCR probes were designed between PMP22 and TEKT3 that mapped proximal to the MLPA probes with normal copy numbers, and used for hybridization on Southern blots that contained DNAs from these patients as well as from patients with the conventional 1.5-Mb duplication, reciprocal deletion, or normal persons. Relative normalized intensities of the signals were calculated and compared with normal and duplicated DNA to determine whether the PCR probes were located within or outside of the duplication. All probes examined turned out to have normal copy numbers. Finally, a small 357-bp probe within 5-kb downstream of the TEKT3 gene (B0; Figure 2) yielded an aberrantly sized XbaI restriction fragment of approximately 6 kb in patients with the aberrant duplication in addition to the normally hybridizing 9.2-kb XbaI fragment that was present in all individuals tested (Figure 3). To obtain the sequence attached to the junction breakpoint as present in the aberrant 6-kb fragment (Figure 3), a vectorette PCR was set up using specific primers within the known duplicated sequence approximately 300-bp downstream of TEKT3. After one or more rounds of amplification, we identified several products of the expected size using nested primers on purified XbaI digested genomic DNA 5–10 kb in size of two patients with this specific duplication. Sequence analysis of these overlapping fragments showed that the junction of the duplicated region was located 4.7-kb downstream of TEKT3 (large S in Figure 2). The sequence attached resides in a repeat-rich region of more than 10 kb of continuous repeats interspersed only by four short sequences of unique sequence of 30–250 bp and maps 90-kb distal to the proximal CMT repeat region in which the breakpoints of the conventional 1.5-Mb duplication are mapped. To confirm these data, microarray CGH analysis on a custom-made chromosome 17 BAC microarray using DNA from two male patients with this specific duplication and two normal females as reference probes was performed. In both cases, three different BACs (RP11-726O12, RP11-378O18, and RP11-765E8 17 located 15.23–15.41 Mb from the telomere) clearly showed duplication and the two adjacent BACs (RP11-686G16 and RP11-655L10 15.07–15.17 Mb from telomere) were partially duplicated while the surrounding BACs showed normal signal ratios (Figure 4). The aberrantly hybridizing BACs all mapped outside of the CMT 24 kb repeats again showing this duplication to be different at both ends from the conventional 1.5-Mb duplication. To exclude the possibility that the junction products were PCR artifacts, we developed a PCR on genomic DNA of all patients with the aberrant duplication and controls using primers on both sides of the junction yielding a junction fragment of 1 kb in size. All patients with the aberrant duplication showed the 1-kb junction fragment (Figure 5) that was absent in all controls indicating that this fragment indeed represents the genuine junction fragment. To determine whether this duplication was not a rare CNV, 2124 chromosomes from healthy controls were screened, as well as DNA of 40 patients with CIDP. The junction fragment was not found in any of the control or CIDP cases screened nor was this specific duplication present in the human structural variation database.
Figure 3.
Southern blot analysis reveals the junction fragment. Southern blot analysis using B0 as a probe and XbaI-digested DNA from patients with the 1.5-Mb duplication (lanes 1, 3) without duplication from family I (lane 2) or from unrelated individuals (lanes 4 and 5), and from patients with the smaller duplication (lanes 6–9). In addition to the normal XbaI fragment of 9.2 kb that is detected with this probe, a junction fragment can also be seen of approximately 6 kb in patients carrying the smaller duplication (arrow). On the left, three bands of the lambda-HindIII marker are shown.
Figure 4.
Microarray analysis of duplicated region. Microarray CGH analysis of the same region on chromosome 17 from 17p13.3 to 17p11.2 (position in Mb on X-axis; ratio on Y-axis; upper horizontal line depicts the 1.2 cutoff) for two patients with the small duplication (left two panels) and one with the conventional 1.5-Mb duplication (right panel). The duplicated region (shaded) clearly shows three duplicated BAC signals in the middle of this box for the two patients with the small duplication (RP11-726O12, RP11-378O18, RP11-765E8). The two BACs on the distal side (RP11-686G16, RP11-655L10) still show a partial duplication (see also dotted lines in Figure 2).
Figure 5.
Detection of genomic junction fragments. PCR performed directly on genomic DNA from five patients with the 186-kb duplication (lanes 1–5), a patient with the 1.5-Mb deletion (lane 6) or 1.5-Mb duplication (lane 7) or on normal DNA (lane 8) using primers located at 258 and 593 bp from the opposite sites of the junction breakpoints, respectively. Lane 9 contains the water PCR control. The molecular marker shown is the 1-kb ladder (Invitrogen Life Science, Breda, The Netherlands).
Cosegregation with CMT in two families and identification of a founder haplotype
For two patients, additional family members were available for testing (Figure 1). In family I, the junction fragment could be detected in all members with CMT whereas it was lacking in the unaffected sibling. In family II, the junction fragment was also present in the affected children (SD5, SD6) of the index patient (SD4). In addition, the conventional 1.5-Mb duplication was found in two other family members with CMT (D7, D8) in another branch of this family.
Sequence analysis of the junction fragment revealed the sequence to be identical in all patients examined, which raised the question whether these persons were distally related to one another. Microsatellite analysis of three markers (D17S793, D17S918, and D17S261 located 48-kb proximal and 46-kb distal to the duplication and within the duplication, respectively) indicated that all patients with the specific duplication shared the most prevalent alleles of these markers. Analysis of nine TEKT3 polymorphisms in these patients, the unaffected person from family I without any duplication and two persons with the conventional duplication from family II proved more informative and revealed an identical haplotype in all patients with the TEKT3 duplication that was present in a homozygous state in patients SD3 and SD9. For three of these nine SNPs, the patients shared a more frequently occurring allele with frequencies of 50–90%, but for five other SNPs they shared the minor allele with described frequencies of 13–47%. In one case no frequency data were available. This identical haplotype of nine TEKT3 SNPs in patients with the TEKT3 duplication in combination with identical junction breakpoints is indicative of a founder mutation. Chances that this is due to coincidence are highly improbable. We studied family II in which another branch carried the conventional duplication of 1.5 Mb, which would suggest that recombination events could lead to the alternative TEKT3 duplication, in more detail. Relative peak ratios of the examined SNPs in a heterozygous state could be used to deduce, which allele was contained within the duplication. Inferred haplotypes are shown in Figure 1. For three TEKT3 SNPs (asterisks D7, D8), the two patients carrying the conventional duplication showed duplication of a different allele than that found in the smaller duplication indicating that recombination events of the conventional duplication cannot explain the occurrence of the TEKT3 duplication in this family.
To exclude the possibility that the 186-kb duplication would be associated with an as yet unidentified mutation in PMP22 and would not be responsible for the CMT phenotype itself, all coding exons and the non-coding alternative exons 1A and 1B, including exon–intron boundaries with at least 20 nt of adjacent sequence were screened for mutations. However, except for two known intronic polymorphisms (rs231020 and rs3744333), no mutations were found. We also excluded mutations in a recently described region in the PMP22 3′UTR targeted by miR-29a that was shown to regulate expression of PMP22.28 The TEKT3 gene that is located within the duplication was additionally examined for mutations but again, only polymorphisms were encountered (Figure 1).
Discussion
In 11 patients from six apparently unrelated families, an identical duplication was found of 186 kb with the junction breakpoints located in a repeat-rich region, located at a 90-kb distance of the proximal CMT repeat region on one side, and 3-kb upstream of PMP22 in the genomic region between PMP22 and TEKT3 on the other side. As this duplication was neither detected in 2124 control chromosomes nor present in 40 CIDP patients, and in addition, not described in the structural variation database, it is improbable that this is a neutral CNV. Moreover, in the two families for which we had more members available for research the presence of the 186-kb duplication correlated with the disease. We postulate that this CNV is associated with the disease.
The junction created by this duplication is located outside of any known genes or open reading frames and as such does not disturb any gene. In addition, no predicted new binding sites for transcription factors are created. The PMP22 gene, known to cause CMT1A and the obvious candidate in this region, is located just outside of this smaller duplication. It harbors no associated mutations in its coding exons, alternative first exons 1A or 1B or adjacent sequences nor in a recently described microRNA binding site in its non-coding 3′ tail.28
A search for mutations in TEKT3 that is located within the duplication also did not yield any abnormalities suggesting that this 186-kb duplication is in fact responsible for the disease. TEKT3 is primarily expressed in the male germ lineage in which it is believed to be involved in spermatozoa transport29 with a much lower expression in brain. In some tumors, and some other tissues expression is also detected. It encodes tektin3 that is a member of a filament forming family and like some proteins known to be involved in CMT such as LMNA (CMT2B1),30 periaxin (CMT4F),31, 32 and NEFL (CMT1F/CMT2E)33, 34 a cytoskeletal protein.
The region between PMP22 and the TEKT3 gene spans 38 kb and contains two uncharacterized transcripts that are described in public databases, FLJ25830 and Hs171267, both of which are noncoding RNAs that are represented by two ESTs each only with expression found in testis and in just one cDNA library made of equal amounts of mRNAs from fetal cells, testis, and B cells, respectively. Although they may be regulatory RNAs, they lie outside of the duplicated region and MLPA probes located in the corresponding genomic sequences show normal copy numbers. Regulatory sequences important for endogenous expression of Pmp22 in mice, especially during late myelination were reported within 10 kb of the start codon of Pmp22 that is located in exon 2,35, 36 also map outside of the duplicated region.
Except for TEKT3 only one other gene, CDRT4 (CMT duplicated region transcript 4), has been identified within the duplicated region, in addition to several non-characterized transcripts. For CDRT4 little is known; it is ubiquitously expressed, has no known conserved domains and is predicted to be a nuclear protein (PSORTII). It is highly represented (1.1%) in a uterus tumor cDNA library indicating that it may be a structural protein. In Affymetrix microarray experiments (NCBI Geoprofiles) expression has been described for PMP22, as well as for TEKT3 and CDRT4 in heregulin and forskolin mitogenically stimulated cultured Schwann cells of four different persons. In this experimental setup, TEKT3 gave unreliable signals and CDRT4 expression was unaffected whereas PMP22 expression decreased in three of the four cell lines on further passaging. The uncharacterized transcripts (Hs.677286, 667666, 697356, 690540, LOC729004, Hs. 528883, and Hs. 398012) within the duplication are all single exon transcribed sequences represented by only a few ESTs at most without described expression in nerves, with repeats present in many cases as well as A-stretches at the 3′-end in the genome in three cases indicating that these clones may have originated by priming on genomic DNA instead of mRNA and therefore raising doubt whether these really represent genuine transcripts. One of them, LOC729004 represents a pseudogene similar to ribosomal protein L9.
Recently, it has become clear that a much larger part of the genome is transcribed outside of gene annotations37, 38 and that chimeric transcripts may exist between genes that are possibly important for the regulation of gene expression. On the centromeric side of the duplication, FAM18B2 is located. FAM18B2-CDRT4 chimeric transcripts do exist as is supported by the presence of several chimeric ESTs and these genes are described in Unigene as parts of one transcription unit. The FAM18B2-CDRT4 region is interrupted by this duplication although it does not affect CDRT4 or FAM18B2 as separate units and may thus also deregulate expression of neighboring genes such as PMP22 through changes in chromatin structure. Chimeric ESTs containing CDRT4 and TEKT3 sequences or TEKT3 and PMP22 sequences were not found in the public databases. Our attempts to show chimeric transcripts by RT-PCR in fibroblasts of patients with the 186-kb duplication in the TEKT3–PMP22 region also yielded no specific products (data not shown). Alternatively, unknown regulatory sequences may be duplicated that influence expression of PMP22. Some examples of aberrations outside a dosage sensitive gene associated with disease have been described in literature. A duplication downstream of the dosage-sensitive PLP1 gene, that causes Pelizaeus–Merzbacher disease when duplicated or mutated, was associated with a spastic paraplegia phenotype39 by virtue of a position effect that resulted in gene silencing. More recently, some cases of Pierre Robin sequence, a subgroup of cleft palate, were reported to result from developmental misexpression of SOX9 because of disruption of very long-range cis-regulatory elements by translocation (breakpoints 1- to 1.2-Mb upstream of SOX9) or microdeletion (both approximately 1.5-Mb centromeric and approximately 1.5-Mb telomeric of SOX9).40
In an attempt to analyze whether PMP22 expression was affected, quantitative RT-PCR experiments were performed on skin fibroblasts of two patients with the TEKT3 duplication. We observed a relatively high Schwann cell-specific PMP22 expression (transcript 1; NM_000304) in these patients as compared with two normal unrelated persons, however, this specific transcript was also relatively high in an unaffected family member of one of the TEKT3 duplication patients (results not shown). No differences were seen for the ubiquitously expressed PMP22 transcript or TEKT3 that was hardly expressed at all. Possibly, a higher Schwann cell-specific PMP22 expression may also be present in these patients in other cell types than fibroblasts and as such contribute to the CMT phenotype.
Remarkably, the junction breakpoints of all patients analyzed were shown to be identical indicative of a founder mutation. This was an unexpected finding because these patients resided in different parts of the country and were not suspected to be related in any way. Polymorphisms in the TEKT3 gene clearly showed an identical haplotype shared by all patients, which is very unlikely to be caused by chance. This ancestral mutation may have arisen because of the presence of over 10 kb of clustered repeats on the proximal site of the duplicated region and an Alu repeat just downstream of the distal junction breakpoint. The co-occurrence of both a small and large duplication in one family could imply that the smaller duplication had arisen from the larger one because of a rare recombination event, however, we could exclude that this was the case because the larger duplication contained another haplotype than the smaller one (asterisks Figure 1).
No specific clinical features were observed in this small group of patients, phenotypes were variable as is the case for CMT patients with the conventional PMP22 duplication although the phenotype was rather mild and in some cases clinically more resembled an axonal neuropathy. EMG and/or nerve biopsy did reveal the demyelinating nature of the disease in most patients. We did not find any indications for the involvement of other genes than PMP22. In conclusion, we identified a 186-kb ancestral CNV, proximal of the PMP22 gene that is different from the frequently occurring 1.5-Mb CMT1A duplication, which does not represent a rare polymorphism but is associated with the disease. Although PMP22 is not directly affected at the genomic level, we postulate that also this duplication affects PMP22 expression levels through an as yet unidentified mechanism. Finally, it is important to realize that this mutation remains undetected in most clinical and diagnostic assays whereas it is the cause of the CMT phenotype in these patients.
Acknowledgments
The authors thank Dr Jessica Hoogendijk (UMCU Utrecht) and Anneke Zorgdrager (Winschoten) for reviewing of clinical data of patients and Hai-yen Man and Dr Jan Hoovers (Clinical Genetics, AMC) for their expert technical assistance of microarray experiments.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies the paper on European Journal of Human Genetics website (http://www.nature.com/ejhg)
Supplementary Material
References
- Bailey JA, Gu Z, Clark RA, et al. Recent segmental duplications in the human genome. Science. 2002;297:1003–1007. doi: 10.1126/science.1072047. [DOI] [PubMed] [Google Scholar]
- Chance PF, Alderson MK, Leppig KA, et al. DNA deletion associated with hereditary neuropathy with liability to pressure palsies. Cell. 1993;72:143–151. doi: 10.1016/0092-8674(93)90058-x. [DOI] [PubMed] [Google Scholar]
- Lupski JR, Oca-Luna RM, Slaugenhaupt S, et al. DNA duplication associated with Charcot-Marie-Tooth disease type 1A. Cell. 1991;66:219–232. doi: 10.1016/0092-8674(91)90613-4. [DOI] [PubMed] [Google Scholar]
- Pentao L, Wise CA, Chinault AC, Patel PI, Lupski JR. Charcot-Marie-Tooth type 1A duplication appears to arise from recombination at repeat sequences flanking the 1.5 Mb monomer unit. Nat Genet. 1992;2:292–300. doi: 10.1038/ng1292-292. [DOI] [PubMed] [Google Scholar]
- Matsunami N, Smith B, Ballard L, et al. Peripheral myelin protein-22 gene maps in the duplication in chromosome 17p11.2 associated with Charcot-Marie-Tooth 1A. Nat Genet. 1992;1:176–179. doi: 10.1038/ng0692-176. [DOI] [PubMed] [Google Scholar]
- Patel PI, Roa BB, Welcher AA, et al. The gene for the peripheral myelin protein PMP-22 is a candidate for Charcot-Marie-Tooth disease type 1A. Nat Genet. 1992;1:159–165. doi: 10.1038/ng0692-159. [DOI] [PubMed] [Google Scholar]
- Timmerman V, Nelis E, Van Hul W, et al. The peripheral myelin protein gene PMP-22 is contained within the Charcot-Marie-Tooth disease type 1A duplication. Nat Genet. 1992;1:171–175. doi: 10.1038/ng0692-171. [DOI] [PubMed] [Google Scholar]
- Valentijn LJ, Bolhuis PA, Zorn I, et al. The peripheral myelin gene PMP-22/GAS-3 is duplicated in Charcot-Marie-Tooth disease type 1A. Nat Genet. 1992;1:166–170. doi: 10.1038/ng0692-166. [DOI] [PubMed] [Google Scholar]
- Suter U, Welcher AA, Ozcelik T, et al. Trembler mouse carries a point mutation in a myelin gene. Nature. 1992;356:241–244. doi: 10.1038/356241a0. [DOI] [PubMed] [Google Scholar]
- Suter U, Moskow JJ, Welcher AA, et al. A leucine-to-proline mutation in the putative first transmembrane domain of the 22-kDa peripheral myelin protein in the trembler-J mouse. Proc Natl Acad Sci USA. 1992;89:4382–4386. doi: 10.1073/pnas.89.10.4382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valentijn LJ, Baas F, Wolterman RA, et al. Identical point mutations of PMP-22 in Trembler-J mouse and Charcot-Marie-Tooth disease type 1A. Nat Genet. 1992;2:288–291. doi: 10.1038/ng1292-288. [DOI] [PubMed] [Google Scholar]
- Roa BB, Dyck PJ, Marks HG, Chance PF, Lupski JR. Dejerine-Sottas syndrome associated with point mutation in the peripheral myelin protein 22 (PMP22) gene. Nat Genet. 1993;5:269–273. doi: 10.1038/ng1193-269. [DOI] [PubMed] [Google Scholar]
- Adlkofer K, Martini R, Aguzzi A, Zielasek J, Toyka KV, Suter U. Hypermyelination and demyelinating peripheral neuropathy in Pmp22-deficient mice. Nat Genet. 1995;11:274–280. doi: 10.1038/ng1195-274. [DOI] [PubMed] [Google Scholar]
- Huxley C, Passage E, Manson A, et al. Construction of a mouse model of Charcot-Marie-Tooth disease type 1A by pronuclear injection of human YAC DNA. Hum Mol Genet. 1996;5:563–569. doi: 10.1093/hmg/5.5.563. [DOI] [PubMed] [Google Scholar]
- Sereda M, Griffiths I, Puhlhofer A, et al. A transgenic rat model of Charcot-Marie-Tooth disease. Neuron. 1996;16:1049–1060. doi: 10.1016/s0896-6273(00)80128-2. [DOI] [PubMed] [Google Scholar]
- Schneider C, King RM, Philipson L. Genes specifically expressed at growth arrest of mammalian cells. Cell. 1988;54:787–793. doi: 10.1016/s0092-8674(88)91065-3. [DOI] [PubMed] [Google Scholar]
- Suter U, Snipes GJ, Schoener-Scott R, et al. Regulation of tissue-specific expression of alternative peripheral myelin protein-22 (PMP22) gene transcripts by two promoters. J Biol Chem. 1994;269:25795–25808. [PubMed] [Google Scholar]
- Hanemann CO, Stoll G, D'Urso D, et al. Peripheral myelin protein-22 expression in Charcot-Marie-Tooth disease type 1a sural nerve biopsies. J Neurosci Res. 1994;37:654–659. doi: 10.1002/jnr.490370513. [DOI] [PubMed] [Google Scholar]
- Yoshikawa H, Nishimura T, Nakatsuji Y, et al. Elevated expression of messenger RNA for peripheral myelin protein 22 in biopsied peripheral nerves of patients with Charcot-Marie-Tooth disease type 1A. Ann Neurol. 1994;35:445–450. doi: 10.1002/ana.410350412. [DOI] [PubMed] [Google Scholar]
- Chapon F, Diraison P, Lechevalier B, et al. Hereditary neuropathy with liability to pressure palsies with a partial deletion of the region often duplicated in Charcot-Marie-Tooth disease, type 1A. J Neurol Neurosurg Psychiatry. 1996;61:535–536. doi: 10.1136/jnnp.61.5.535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ionasescu V, Ionasescu G, Searby C, Barker DF. Is the duplication present in all family members affected with Charcot-Marie-Tooth neuropathy type 1 A. Rom J Neurol Psychiatry. 1993;31:179–187. [PubMed] [Google Scholar]
- Valentijn LJ, Baas F, Zorn I, Hensels GW, de Visser M, Bolhuis PA. Alternatively sized duplication in Charcot-Marie-Tooth disease type 1A. Hum Mol Genet. 1993;2:2143–2146. doi: 10.1093/hmg/2.12.2143. [DOI] [PubMed] [Google Scholar]
- Franssen H, Vermeulen M, Jennekens FG.Chronic inflammatory neuropathiesin Emery AEH (ed): Diagnostic Criteria for Neuromuscular Disorders Royal Society of Medicine Press: London; 199753–59. [Google Scholar]
- Church GM, Gilbert W. Genomic sequencing. Proc Natl Acad Sci USA. 1984;81:1991–1995. doi: 10.1073/pnas.81.7.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hulsebos TJ, Bijlsma EK, Geurts van Kessel AH, Brakenhoff RH, Westerveld A. Direct assignment of the human beta B2 and beta B3 crystallin genes to 22q11.2----q12: markers for neurofibromatosis 2. Cytogenet Cell Genet. 1991;56:171–175. doi: 10.1159/000133080. [DOI] [PubMed] [Google Scholar]
- Riley J, Butler R, Ogilvie D, et al. A novel, rapid method for the isolation of terminal sequences from yeast artificial chromosome (YAC) clones. Nucleic Acids Res. 1990;18:2887–2890. doi: 10.1093/nar/18.10.2887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathijssen IB, Hoovers JM, Mul AN, Man HY, Ket JL, Hennekam RC. Array comparative genomic hybridization analysis of a familial duplication of chromosome 13q: a recognizable syndrome. Am J Med Genet A. 2005;136:76–80. doi: 10.1002/ajmg.a.30758. [DOI] [PubMed] [Google Scholar]
- Verrier JD, Lau P, Hudson L, Murashov AK, Renne R, Notterpek L. Peripheral myelin protein 22 is regulated post-transcriptionally by miRNA-29a. Glia. 2009;57:1265–1279. doi: 10.1002/glia.20846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy A, Yan W, Burns KH, Matzuk MM. Tektin3 encodes an evolutionarily conserved putative testicular microtubules-related protein expressed preferentially in male germ cells. Mol Reprod Dev. 2004;67:295–302. doi: 10.1002/mrd.20025. [DOI] [PubMed] [Google Scholar]
- Sandre-Giovannoli A, Chaouch M, Kozlov S, et al. Homozygous defects in LMNA, encoding lamin A/C nuclear-envelope proteins, cause autosomal recessive axonal neuropathy in human (Charcot-Marie-Tooth disorder type 2) and mouse. Am J Hum Genet. 2002;70:726–736. doi: 10.1086/339274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boerkoel CF, Takashima H, Stankiewicz P, et al. Periaxin mutations cause recessive Dejerine-Sottas neuropathy. Am J Hum Genet. 2001;68:325–333. doi: 10.1086/318208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guilbot A, Williams A, Ravise N, et al. A mutation in periaxin is responsible for CMT4F, an autosomal recessive form of Charcot-Marie-Tooth disease. Hum Mol Genet. 2001;10:415–421. doi: 10.1093/hmg/10.4.415. [DOI] [PubMed] [Google Scholar]
- De Jonghe P, Mersivanova I, Nelis E, et al. Further evidence that neurofilament light chain gene mutations can cause Charcot-Marie-Tooth disease type 2E. Ann Neurol. 2001;49:245–249. doi: 10.1002/1531-8249(20010201)49:2<245::aid-ana45>3.0.co;2-a. [DOI] [PubMed] [Google Scholar]
- Mersiyanova IV, Perepelov AV, Polyakov AV, et al. A new variant of Charcot-Marie-Tooth disease type 2 is probably the result of a mutation in the neurofilament-light gene. Am J Hum Genet. 2000;67:37–46. doi: 10.1086/302962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maier M, Castagner F, Berger P, Suter U. Distinct elements of the peripheral myelin protein 22 (PMP22) promoter regulate expression in Schwann cells and sensory neurons. Mol Cell Neurosci. 2003;24:803–817. doi: 10.1016/s1044-7431(03)00246-x. [DOI] [PubMed] [Google Scholar]
- Maier M, Berger P, Nave KA, Suter U. Identification of the regulatory region of the peripheral myelin protein 22 (PMP22) gene that directs temporal and spatial expression in development and regeneration of peripheral nerves. Mol Cell Neurosci. 2002;20:93–109. doi: 10.1006/mcne.2002.1116. [DOI] [PubMed] [Google Scholar]
- Birney E, Stamatoyannopoulos JA, Dutta A, et al. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447:799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapranov P, Willingham AT, Gingeras TR. Genome-wide transcription and the implications for genomic organization. Nat Rev Genet. 2007;8:413–423. doi: 10.1038/nrg2083. [DOI] [PubMed] [Google Scholar]
- Lee JA, Madrid RE, Sperle K, et al. Spastic paraplegia type 2 associated with axonal neuropathy and apparent PLP1 position effect. Ann Neurol. 2006;59:398–403. doi: 10.1002/ana.20732. [DOI] [PubMed] [Google Scholar]
- Benko S, Fantes JA, Amiel J, et al. Highly conserved non-coding elements on either side of SOX9 associated with Pierre Robin sequence. Nat Genet. 2009;41:359–364. doi: 10.1038/ng.329. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.