Abstract
In communities with high rates of consanguinity and consequently high prevalence of recessive phenotypes, homozygosity mapping with SNP arrays is an effective approach for gene discovery. In 20 Palestinian kindreds with prelingual nonsyndromic hearing loss, we generated homozygosity profiles reflecting linkage to the phenotype. Family sizes ranged from small nuclear families with two affected children, one unaffected sibling, and parents to multigenerational kindreds with 12 affected relatives. By including unaffected parents and siblings and screening 250 K SNP arrays, even small nuclear families yielded informative profiles. In 14 families, we identified the allele responsible for hearing loss by screening a single candidate gene in the longest homozygous region. Novel alleles included missense, nonsense, and splice site mutations of CDH23, MYO7A, MYO15A, OTOF, PJVK, Pendrin/SLC26A4, TECTA, TMHS, and TMPRSS3, and a large genomic deletion of Otoancorin (OTOA). All point mutations were rare in the Palestinian population (zero carriers in 288 unrelated controls); the carrier frequency of the OTOA genomic deletion was 1%. In six families, we identified five genomic regions likely to harbor novel genes for human hearing loss on chromosomes 1p13.3 (DFNB82), 9p23–p21.2/p13.3–q21.13 (DFNB83), 12q14.3–q21.2 (DFNB84; two families), 14q23.1–q31.1, and 17p12–q11.2 (DFNB85).
Keywords: hearing, deafness, DFNB, homozygosity mapping, mutation, protein modeling
Introduction
Recessively inherited phenotypes are common in the Palestinian population as the result of a high frequency of consanguineous marriages.1, 2 Palestinian families expressing such traits have cooperated with research studies to identify the critical genes for many phenotypes. In particular, Palestinian families with nonsyndromic hearing loss (NSHL) have been a valuable genetic resource for the identification of genes essential for normal hearing.3, 4, 5, 6, 7, 8 Homozygosity mapping with SNPs has proven an effective approach to the identification of these genes, because persons with a recessive phenotype and born of a consanguineous union are likely to be homozygous both for the causative mutation and for polymorphisms in the genomic region surrounding the mutation. In this study, we used high-density SNP arrays to genotype the DNA of 155 relatives from 20 consanguineous families with autosomal recessive NSHL. Deafness-associated homozygosity profiles revealed loci likely to harbor novel genes for NSHL in 6 families and homozygous mutations in known genes for NSHL in 14 families. In all families, the deafness-causing mutation was present in the longest homozygous segment of the deaf individuals' genomes.
Materials and methods
Subjects
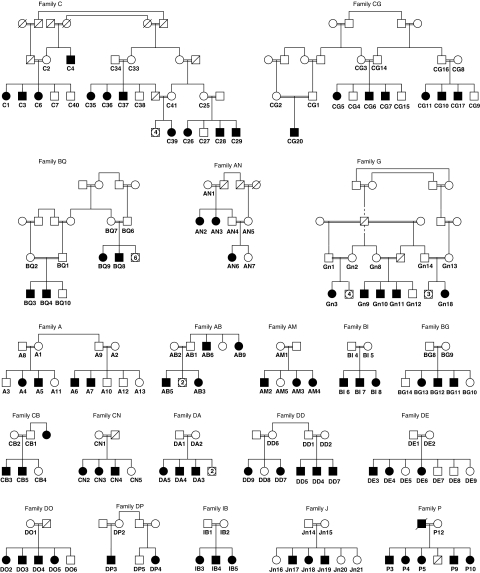
Subjects with prelingual bilateral hearing loss (n=238) were ascertained through schools for deaf children in the West Bank: Eftah School for the Deaf in Bethlehem, Al Amad School for the Deaf in Hebron, and the Princess Basma Rehabilitation Center for Deaf Children in Jerusalem. In addition, families were referred to the Hereditary Research Laboratory at Bethlehem University from the clinic of Dr Wael Salhab and from the Dar Al-Kalima Audiology Clinic in Bethlehem by Mr Michael Rahil. Families were contacted and informed consent was obtained from parents with assent from older children. The project was approved by the Human Subjects Committee of Bethlehem University and by the Human Subjects Division of the University of Washington. Pedigrees were constructed with 80% of families reporting consanguineous marriages. Audiological exams were performed to exclude hearing loss attributed to infection or trauma and to evaluate the severity and laterality of the hearing loss. Additional ascertainment was carried out for the extended families of 20 probands reporting affected relatives (Figure 1). For these 20 kindreds, all family members willing to participate were enrolled (Supplementary Table 1). Complete physical, visual, and vestibular exams were carried out to exclude syndromic deafness. Population controls comprised 288 Palestinian adults with normal hearing and came from two sources: (i) persons attending West Bank clinics for preventive health services (n=201) and (ii) Palestinian individuals living in Israel who contributed DNA to the National Laboratory for the Genetics of Israeli Populations at Tel Aviv University (n=87).
Figure 1.
Inherited prelingual hearing loss in Palestinian families. Hearing loss in affected individuals (filled symbols) is prelingual with thresholds >90 dB. Double bars between parents indicate marriages within the same hamula, or clan. Only enrolled individuals and their parents are shown. Based on complete pedigree information, segregation of hearing loss is consistent with recessive inheritance in each family.
Excluding known Palestinian deafness alleles
Genomic DNA was prepared by salting out.9 Genomic DNA from 238 probands and 58 additional deaf relatives from 20 extended families were fully sequenced for both exons of connexin 26, as described.10 In addition, all individuals were tested for deafness-associated alleles previously identified in families of Palestinian origin: Otoancorin.1067A>T, SLC26A4.716T>A, SLC26A4.1001G>T, TMPRSS3.988delA, TRIOBP.1039C>T, TRIOBP.1741C>T,8, 11 Otoferlin.2416T>A,3 TECTA.IVS9(+1)G>A,4 Otoancorin.IVS12(+2)G>T,5 the β-satellite insertion of TMPRSS3,6 and Whirlin.2332C>T.7
Homozygosity mapping
Genomic DNA samples (250 ng) were hybridized on Affymetrix GeneChip (Santa Clara, CA, USA) Human Mapping 250K NspI arrays according to the manufacturer's protocol (http://www.affymetrix.com). Arrays were scanned using the Affymetrix Gene Chip Scanner 3000 with the Gene Chip Operating System 1.4 (GCOS). Data were analyzed using the Gene Chip DNA Analysis Software 2.0 (GDAS) and classified by chromosome and sorted on the basis of the physical position of the SNPs. Homozygous regions were defined as segments where SNPs were homozygous for one allele among affected relatives but discordant (heterozygous or homozygous of the complement allele) among unaffected relatives in the pedigree. Enhanced stringency requirements for SNP quality were applied, with only SNPs with a quality score better than 0.06 from GDAS and GCOS being retained, thus using about 67% of the SNPs in homozygosity analyses. A segment was considered continuously homozygous if there was no more than one heterozygous SNP in any 10-SNP window. Homozygous segments of 2 Mb or longer were retained for further analysis. LOD scores12 were calculated for these homozygous segments for each family, exploiting genotypes of all informative SNPs. The LOD score for a family depends on the family's structure and parameters of the recessive model and is the same for all informative homozygous regions in the genome.
Sequencing known genes
Previously described deafness genes13 mapping within homozygous regions greater than 2 Mb were evaluated by Sanger DNA sequencing of PCR amplicons. Primers were designed to encompass all annotated exons and 50–100 bp of flanking intronic sequence.8 Primers for Cadherin23, ESRRB, Otoferlin, Pejvakin, Myosin 7A, Myosin 15A, SLC26A4, TECTA, TMC1, TMHS, and TMPRSS3 are available on request. For families C, AN, and CB, both genomic DNA and RNA from white cells were available; for the other families, only genomic DNA was available for analysis. As each new mutation was identified, the additional 218 deaf probands and 288 hearing controls were screened by PCR-RFLP and direct sequencing.
Analysis of copy number variation
DNA samples from affected individuals were PCR amplified with unique STSs from the nonduplicated portion of the Otoancorin gene, exons 1–19 (chr16: 21 597 336–21 647 927). Primers are available on request. Genomic boundaries of the deletion in family DO and in Palestinian controls were determined by high-resolution arrayCGH on NimbleGen 2.1 million probe arrays. Genomic samples were analyzed14 using NA15724 as the reference genome for hybridizations. In controls, genomic deletion was evaluated by SYBR Green real-time PCR. Briefly, 10 ng of genomic DNA was amplified with primer sets located in exon 6 of Otoancorin and in intron 12 of Albumin (which served as an endogenous control) on an Applied Biosystems (Foster City, CA, USA) 7300 Real-Time PCR Instrument. Samples were run in quadruplicate alongside a known 2-copy control and a known 1-copy carrier from family DO. Raw Ct values were collected and the relative fold difference between test samples and controls was determined using the 2−ΔΔCt method.15 Samples with a 0.5-fold decrease in signal at Otoancorin relative to the known two-copy sample were analyzed by arrayCGH to determine break points.
Results
In this cohort of 286 families, hearing loss is attributed to connexin 26 mutations in 8% of families (23/286) and to other previously described alleles of Palestinian origin in 9% of families (25/286). From the families of the unresolved 238 probands, we selected 20 informative kindreds in which parents reported consanguineous marriages and patterns of hearing loss were consistent with recessive inheritance (Figure 1). From these 20 kindreds, 78 individuals with NSHL, 28 of their hearing siblings, and 49 parents enrolled in the project (Supplementary Table 1).
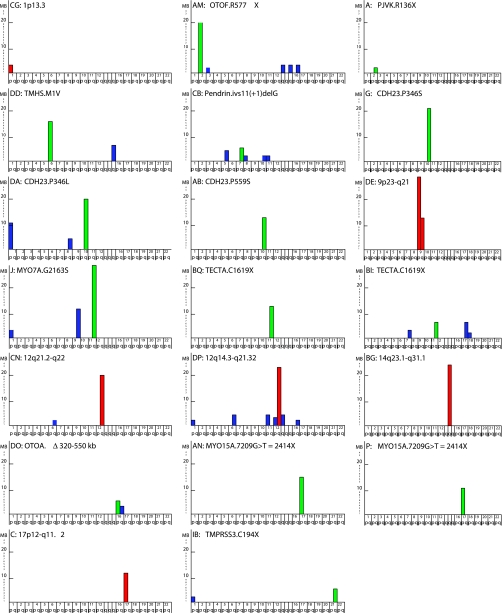
Each participant provided a sample of whole blood from which DNA was extracted and genotyped on Affymetrix 250K SNP arrays. We generated homozygosity profiles specific to the deaf individuals in each family, defining a deafness-associated homozygous peak as a contiguous genomic region >2 Mb across which SNPs were homozygous for one allele among affected relatives but discordant (heterozygous or homozygous for the alternate allele) among unaffected parents or sibs. For nine families, a single homozygous peak was observed, in five families two peaks were observed, and in six families multiple peaks were observed (Figure 2).
Figure 2.
Genomic regions of homozygosity associated with hearing loss. Each colored bar represents a genomic region homozygous for the same haplotype in all deaf individuals in a family, and heterozygous or homozygous for the alternate allele among unaffected parents or siblings in the same family. Chromosome arms are indicated on X-axes; lengths in Mb of the homozygous regions are indicated on Y-axes. Green bars represent homozygous regions with a deleterious mutation in a known deafness gene, as indicated in Table 1. Red bars represent homozygous regions likely to harbor novel deafness genes as indicated in Table 2. Blue bars indicate other regions of homozygosity.
In 16 of the 20 families, the longest homozygous region included a gene known to be associated with hearing loss (Table 1). For each family, the relevant gene was fully sequenced from genomic DNA of the proband, revealing in 14 families alleles that were very likely to be deleterious. These included premature truncating alleles of Pejvakin, TECTA, TMPRSS3, and Otoferlin, a splice site mutation leading to a large in-frame deletion of the Pendrin/SLC26A4 message, and nonsynonymous mutations in MYO15A, MYO7A, CDH23, and TMHS, which were further evaluated by splicing analysis and molecular modeling. Detailed analyses of these point mutations are presented in Supplementary Information. One family (DO) harbored a large genomic deletion of Otoancorin. For three families (DE, BG, C), homozygous regions harbored known deafness genes, but full sequence analysis of these candidate genes did not reveal any rare alleles segregating with hearing loss. These linkage regions likely harbor as yet undiscovered genes for hearing loss. Finally, in three other families (CG, DP, CN), hearing loss mapped to homozygous regions that did not include known genes for human hearing loss, thereby also likely reflecting new recessive deafness loci in the Palestinian population. In summary, six of the 20 families are likely to represent five novel genes for NSHL.
Table 1. Genomic regions homozygous in deaf individuals revealing mutations in known genes for hearing loss.
| Kindred | Peaks >2 Mb | Longest homozygous region | Homozygous mutation in family | Frequency of mutation in the Palestinian population | ||||
|---|---|---|---|---|---|---|---|---|
| Length (Mb) | Location NCBI build 36.1 (hg18) | DFN gene in region | Nucleotide change | Amino-acid change | Heterozygotes in 288 controls | Homozygotes in 218 unrelated deaf | ||
| AM | 5 | 19.4 | chr2: 14 733 739–34 121 329 | OTOF | c4157 C>T | Arg 577 STOP | 0 | 0 |
| A | 1 | 1.9 | chr2: 177 555 791–179 471 954 | PJVK | c762 C>T | Arg 136 STOP | 0 | 1 |
| DD | 2 | 14.6 | chr6: 23 791 422–38 418 869 | TMHS | c1 A>G | Met 1 Val | 0 | 0 |
| CB | 5 | 5.2 | chr7: 105 307 599–110 520 953 | SLC26A4 | c1341 ΔG | Δ aa 422–447 | 0 | 0 |
| G | 1 | 19.8 | chr10: 53 479 033–73 231 404 | CDH23 | c1427 C>T | Pro 346 Ser | 0 | 0 |
| DA | 3 | 19.2 | chr10: 71 683 886–90 894 091 | CDH23 | c1428 C>T | Pro 346 Leu | 0 | 0 |
| AB | 1 | 11.7 | chr10: 72 774 642–84 459 990 | CDH23 | c2065 C>T | Pro 559 Ser | 0 | 1 |
| J | 3 | 26.4 | chr11: 69 115 451–95 560 647 | MYO7A | c6487 G>A | Gly 2163 Ser | 0 | 2 |
| BQ | 1 | 11.4 | chr11: 110 396 930–121 744 055 | TECTA | c4857 C>A | Cys 1619 STOP | 0 | See BI |
| BI | 4 | 5.9 | chr11: 117 111 049–123 047 230 | TECTA | c4857 C>A | Cys 1619 STOP | 0 | 2 |
| DO | 2 | 5.4 | chr16: 21 035 675–26 477 060 | OTOA | Complete Δ | Complete Δ | 3 | 0 |
| P | 1 | 10.1 | chr17: 16 806 979–26 862 502 | MYO15A | c7545 G>T | Asp 2403 fsX2414 | 0 | See AN |
| AN | 1 | 13.6 | chr17: 17 292 606–30 871 427 | MYO15A | c7545 G>T | Asp 2403 fsX2414 | 0 | 5 |
| IB | 2 | 5.4 | chr21: 40 357 695–45 755 537 | TMPRSS3 | c783T>A | Cys 194 STOP | 0 | 0 |
Homozygosity for a genomic deletion of Otoancorin at 16p12
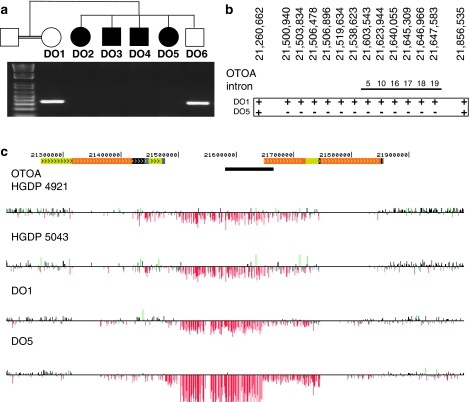
The largest homozygous peak in family DO was a 5.4 Mb region of chromosome 16p12.1–p12.2, including the gene Otoancorin. The genomic sequence flanking Otoancorin has multiple clusters of high-identity segmental duplications, which likely gave rise to a deletion spanning the Otoancorin gene through the nonallelic homologous recombination (Figure 3).16 The largest Otoancorin isoform comprises 28 exons. Exons 1–19 map to a single location in the human genome, but exons 20–28 are located in a 68 kb region that is segmentally duplicated, at greater than 99% identity, at chr16: 22 464 054–22 532 760, approximately 820 kb centromeric of the original locus.
Figure 3.
Genomic deletion of Otoancorin (OTOA). (a) Family DO pedigree and PCR amplification of a unique STS in OTOA intron 17 at chr16: 21 645 309. The STS is present in the unaffected mother and sibling and absent from all affected siblings. (b) Presence (+) or absence (−) of multiple STSs within and flanking OTOA in genomic DNA from unaffected mother DO1 and deaf child DO5. (c) High-resolution arrayCGH of 16p12.2 (chr16: 21 200 000–22 000 000) in genomic DNA from Palestinian control individuals HGDP4921 and HGDP5043 and from family DO unaffected mother DO1 and deaf child DO5. Deviations from zero of log2 ratios of probe intensities are depicted by vertical lines, with ratios >1.5 SDs from the mean ratio colored red and green to represent relative losses and gains, respectively. Segmental duplications of increasing similarity (90–98, 98–99, and >99%) are represented by gray, yellow, and orange bars, respectively. The size of the OTOA locus (black bar) is 82 kb; the size of the deletion is 320–550 kb. In the Palestinian hearing population, the carrier frequency of the deletion is 1% (3/288 controls).
From the genomic DNA of DO2, DO3, DO4, and DO5 (all of whom are deaf), PCR amplification of Otoancorin exons 1–19 failed to produce a product (Figures 3a and b). In contrast, amplification of these exons from genomic DNA of DO1 and DO6 yielded products of normal size. This pattern suggests that the deaf individuals are homozygous for a genomic deletion spanning these exons. DNA from the same individuals was further analyzed by high-density genome-wide arrayCGH, revealing a deletion of approximately 500 kb that completely deletes the Otoancorin gene (Figure 3c). Deaf individuals in family DO were homozygous for the deletion and their parents heterozygous for the deletion. The deletion is flanked by segmental duplications located at chr16: 21 314 758–21 419 562 and chr16: 21 746 863–21 851 057. The identity between these two loci is approximately 99.7%, suggesting that the nonallelic homologous recombination is the likely mechanism of deletion, as has been described for other genomic regions.16 The deletion was observed as a heterozygote in 1.0% of unrelated Palestinian controls (3/288). A comparison of the arrayCGH intensity values suggests that the mutations in family DO and in the controls have the same genomic breakpoints (Figure 3c).
Novel deafness loci
In six families, homozygosity mapping suggests that hearing loss is due to mutations in as-yet-unidentified genes. The six families represent five new loci (ie, two families map to the same region). We identified the critical regions of linkage, calculated LOD scores for each of the six families, and reserved DFNB loci (HUGO Gene Nomenclature Committee) for families with LOD scores greater than 3.0 (Table 2).
Table 2. Chromosomal regions of new loci for nonsyndromic hearing loss.
| DFNB | Kindred | Chromosome | Size (Mb) | Flanking markers | LOD score | |||
|---|---|---|---|---|---|---|---|---|
| Start (hg18) | End (hg 18) | |||||||
| 82 | CG | 1 | 108 108 849 | 111 251 931 | 3.1 | rs17542571 | rs1936942 | 5.16 |
| 83 | DE | 9 | 9 959 023 | 26 430 090 | 16.5 | rs4742645 | rs1571364 | 3.07 |
| 9 | 34 401 026 | 77 904 900 | 28.5 | rs10738927 | rs12001902 | |||
| 84 | CN/DP | 12 | 74 498 486 | 86 977 739 | 12.5 | rs10879987 | rs2471512 | 3.45 |
| 85 | C | 17 | 15 212 798 | 26 490 848 | 11.1 | rs230884 | rs12603885 | 7.25 |
| - | BG | 14 | 57 474 255 | 80 664 133 | 23.2 | rs41514351 | rs3873938 | |
Sizes of linked regions exclude gaps at centromeres.
DFNB82
Hearing loss in family CG maps to a 3.1 Mb region of chromosome 1p13.3 with an LOD score of 5.16 (Supplementary Figure 3a). The DFNB82 region overlaps the DFNB32 locus by 833 kb.17 DFNB32 was described in a Tunisian family with profound congenital hearing loss; the critical gene is not yet known. In DNA from family CG, we sequenced the two genes in the region of overlap (VAV3 and SLC25A24), but found no novel variants segregating with NSHL. It is therefore likely that DFNB82 and DFNB32 reflect different genes for NSHL.
DFNB83
Hearing loss in family DE maps to two regions of chromosome 9: a 9p region of 16.5 Mb and a pericentromeric region of 28.5 Mb (excluding the unannotated 15 Mb gap at the centromere; supplementary Figure 3b). The LOD score is 3.07. The chromosome 9 pericentromeric region includes the NSHL gene TMC1.18 The 9p region includes the MTAP gene, which is disrupted by a de novo balanced translocation in a child with syndromic deafness.19 We sequenced all exons and conserved intronic regions of TMC1 and MTAP in family DE, but did not identify novel variants segregating with deafness in either gene. The DFNA47 locus maps within the DFNB83 region at chr9:13 046 167–21 980 675.20 The gene responsible for DFNA47 is not known. It is possible that mutations of the same gene will be responsible for recessive and dominant hearing loss.
DFNB84
Deaf individuals in families CN and DP have overlapping regions of homozygosity and identical haplotypes over a 12.5 Mb region of chromosome 12q (chr12: 74 498 486–86 977 739) with an LOD score of 3.45 (supplementary Figure 3c). No genes for inherited hearing loss have been described in this genomic interval and it therefore represents a novel locus for recessive NSHL.
DFNB85
Family C is a large kindred with 42 individuals enrolled. Hearing loss in family C maps to an 11.1 Mb region of chromosome 17p12–q11.2 with an LOD score of 7.25 (supplementary Figure 3d). MYO15A lies in this region and was therefore an excellent candidate for the NSHL gene in this family. However, full sequencing of MYO15A from genomic DNA and lymphoblast cDNA from homozygous affected persons and heterozygous obligate carrier relatives revealed no functional mutations. Furthermore, the two alleles of MYO15A were expressed at the same levels in lymphoblast cDNA from obligate carriers, as determined by the evaluation of cSNP rs854772 in exon 40. Although it is possible that MYO15A may harbor a cryptic mutation in a regulatory element driving cochlear-specific expression, clustering of functionally related genes suggests that it is likely that a second gene for NSHL is among the 165 other genes in this region.
Chromosome 14q locus
Homozygosity mapping of family BG revealed a single 23 Mb region of homozygosity on chromosome 14a (Supplementary Figure 3e). This region includes the gene ESRRB responsible for DFNB35.21 Full sequencing of ESRRB in genomic DNA from affected relatives and carriers did not reveal any novel variants in the coding region or flanking regulatory regions. We hope to recruit additional informative relatives from family BG in an effort to reduce the size of the homozygous interval.
Discussion
For the analysis of recessively inherited hearing loss in these consanguineous families, homozygosity mapping with high-density SNP arrays yielded highly informative homozygosity profiles. In 14 families, the critical mutation was identified by sequencing a single gene mapping within the largest homozygous region. Even in small families with few deaf individuals (eg, families AM and CB), each with multiple homozygous peaks, the critical allele was still identifiable by searching exclusively in the largest region.
None of the ten newly described alleles explain a high proportion of hearing loss among Palestinian families. In the Palestinian hearing population, heterozygous carriers were observed for only one allele, the large genomic deletion of OTOA. This region of chromosome 16p12.2 represents a ‘hot spot' for genomic deletion because of its location between two identical segmental duplications. This specific deletion and the reciprocal duplication have been observed in 0.5–1.0% of European controls.16 It has recently been shown that Stereocilin (DFNB15) is also subject to genomic rearrangements mediated by segmental duplications at chromosome 15q15.3. A homozygous deletion of 90 kb, including STRC and four other genes, is responsible for a syndromic form of deafness with male infertility.22, 23 The carrier frequency of the STRC deletion has been estimated at 1.6%. It is possible that homozygous deletions of OTOA and STRC will be enriched in deaf individuals from other consanguineous communities.
The six families in which NSHL maps to five new loci represent an excellent opportunity to identify novel genes essential for normal hearing. The genomic regions harboring these genes are large (generally >10 Mb), but the critical gene can be identified readily with next generation sequencing approaches. Consanguineous kindreds, even small families, prove extremely valuable for the identification of novel alleles and novel loci. Of the families evaluated in this project, 30% are likely to reveal additional genes for hearing loss. This proportion suggests that a large number of genes critical to hearing remain to be discovered.
Acknowledgments
We thank the families for their generous participation in this study. We thank Dr Wael Salhab for a clinical evaluation of the patients, and Michael Rahil of the Dar Al-Kalima Audiology Clinic for audiological analyses. We thank the technical staff of the Hereditary Research Laboratory (Bethlehem University), Rob Hall at the Center for Array Technology (University of Washington), Gregory Cooper, and Heather Mefford for helpful comments and suggestions. This study was supported by the NIH/NIDCD Grant R01DC005641.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies the paper on European Journal of Human Genetics website (http://www.nature.com/ejhg)
Supplementary Material
References
- Jaber L, Halpern GJ, Shohat T. Trends in the frequencies of consanguineous marriages in the Israeli Arab community. Clin Genet. 2000;58:106–110. doi: 10.1034/j.1399-0004.2000.580203.x. [DOI] [PubMed] [Google Scholar]
- Zlotogora J, Barges S, Bisharat B, Shalev SA. Genetic disorders among Palestinian Arabs. 4: Genetic clinics in the community. Am J Med Genet A. 2006;140:1644–1646. doi: 10.1002/ajmg.a.31342. [DOI] [PubMed] [Google Scholar]
- Yasunaga S, Grati M, Cohen-Salmon M, et al. A mutation in OTOF, encoding otoferlin, a FER-1-like protein, causes DFNB9, a nonsyndromic form of deafness. Nat Genet. 1999;21:363–369. doi: 10.1038/7693. [DOI] [PubMed] [Google Scholar]
- Mustapha M, Weil D, Chardenoux S, et al. An alpha-tectorin gene defect causes a newly identified autosomal recessive form of sensorineural pre-lingual non-syndromic deafness, DFNB21. Hum Mol Genet. 1999;8:409–412. doi: 10.1093/hmg/8.3.409. [DOI] [PubMed] [Google Scholar]
- Zwaenepoel I, Mustapha M, Leibovici M, et al. Otoancorin, an inner ear protein restricted to the interface between the apical surface of sensory epithelia and their overlying acellular gels, is defective in autosomal recessive deafness DFNB22. Proc Natl Acad Sci USA. 2002;99:6240–6245. doi: 10.1073/pnas.082515999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott HS, Kudoh J, Wattenhofer M, et al. Insertion of beta-satellite repeats identifies a transmembrane protease causing both congenital and childhood onset autosomal recessive deafness. Nat Genet. 2001;27:59–63. doi: 10.1038/83768. [DOI] [PubMed] [Google Scholar]
- Mburu P, Mustapha M, Varela A, et al. Defects in whirlin, a PDZ domain molecule involved in stereocilia elongation, cause deafness in the whirler mouse and families with DFNB31. Nat Genet. 2003;34:421–428. doi: 10.1038/ng1208. [DOI] [PubMed] [Google Scholar]
- Shahin H, Walsh T, Sobe T, et al. Mutations in a novel isoform of TRIOBP that encodes a filamentous-actin binding protein are responsible for DFNB28 recessive non-syndromic hearing loss. Am J Hum Genet. 2006;78:144–152. doi: 10.1086/499495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller SA, Dykes DD, Polesky HF. A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res. 1988;16:1215. doi: 10.1093/nar/16.3.1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shahin H, Walsh T, Sobe T, et al. Genetics of congenital deafness in the Palestinian population: multiple connexin 26 alleles with shared origins in the Middle East. Hum Genet. 2002;110:284–289. doi: 10.1007/s00439-001-0674-2. [DOI] [PubMed] [Google Scholar]
- Walsh T, Abu Rayan A, Abu Sa'ed J, et al. Genomic analysis of a heterogeneous Mendelian phenotype: multiple novel alleles for inherited hearing loss in the Palestinian population. Hum Genomics. 2006;2:203–211. doi: 10.1186/1479-7364-2-4-203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ott J.Analysis of Human Genetic Linkage3rd edn.,Johns Hopkins Univ Press: Baltimore [Google Scholar]
- Hereditary Hearing Loss Homepage ( http://webh01.ua.ac.be/hhh/ ).
- Selzer RR, Richmond TA, Pofahl NJ, et al. Analysis of chromosome breakpoints in neuroblastoma at sub-kilobase resolution using fine-tiling oligonucleotide array CGH. Genes Chromosomes Cancer. 2005;44:305–319. doi: 10.1002/gcc.20243. [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT Methods. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Itsara A, Cooper GM, Baker C, et al. Population analysis of large copy number variants and hotspots of human genetic disease. Am J Hum Genet. 2009;84:148–161. doi: 10.1016/j.ajhg.2008.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masmoudi S, Tlili A, Majava M, et al. Mapping of a new autosomal recessive nonsyndromic hearing loss locus (DFNB32) to chromosome 1p13.3–22 1. Eur J Hum Genet. 2003;11:185–188. doi: 10.1038/sj.ejhg.5200934. [DOI] [PubMed] [Google Scholar]
- Kurima K, Peters LM, Yang Y, et al. Dominant and recessive deafness caused by mutations of a novel gene, TMC1, required for cochlear hair-cell function. Nat Genet. 2002;30:277–284. doi: 10.1038/ng842. [DOI] [PubMed] [Google Scholar]
- Williamson RE, Darrow KN, Michaud S, et al. Methylthioadenosine phosphorylase (MTAP) in hearing: Gene disruption by chromosomal rearrangement in a hearing impaired individual and model organism analysis. Am J Med Genet A. 2007;143A:1630–1639. doi: 10.1002/ajmg.a.31724. [DOI] [PubMed] [Google Scholar]
- D'Adamo P, Donaudy F, D'Eustacchio A, Di Iorio E, Melchionda S, Gasparini P. A new locus (DFNA47) for autosomal dominant non-syndromic inherited hearing loss maps to 9p21–22 in a large Italian family. Eur J Hum Genet. 2003;11:121–124. doi: 10.1038/sj.ejhg.5200929. [DOI] [PubMed] [Google Scholar]
- Collin RW, Kalay E, Tariq M, et al. Mutations of ESRRB encoding estrogen-related receptor beta cause autosomal-recessive nonsyndromic hearing impairment DFNB35. Am J Hum Genet. 2008;82:125–138. doi: 10.1016/j.ajhg.2007.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Malekpour M, Al-Madani N, et al. Sensorineural deafness and male infertility: a contiguous gene deletion syndrome. J Med Genet. 2007;44:233–240. doi: 10.1136/jmg.2006.045765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knijnenburg J, Oberstein SA, Frei K, et al. A homozygous deletion of a normal variation locus in a patient with hearing loss from non-consanguineous parents. J Med Genet. 2009;46:412–417. doi: 10.1136/jmg.2008.063685. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.