Abstract
Blocking the function of Stat (signal transducer and activator of transcription) proteins, which are critical for antiviral responses, has evolved as a common mechanism for pathogen immune evasion. The poxvirus-encoded phosphatase H1 is critical for viral replication, and may play an additional role in the evasion of host defense by dephosphorylating Stat1 and blocking interferon (IFN)-stimulated innate immune responses. Vaccinia virus (VACV) H1 can inhibit the phosphorylation of the transcription factor Stat1 after IFN-γ stimulation of epithelial cells, greatly attenuating IFN-induced biological functions. In this study, we demonstrate that VACV infection is capable of inhibiting the phosphorylation of Stat1 and Stat2 after stimulation of fibroblasts or bone marrow-derived macrophages with either type I or type II IFNs, but did not inhibit the activation of Stat3 or Stat5 in either cell type. By using recombinant proteins for in vitro assays, we observe that variola virus H1 is more active than VACV H1, although it has similar selectivity for Stat targets. Differential effects of VACV infection were observed on the induction of IFN-stimulated genes, with complete inhibition of some genes by VACV infection, while others were less affected. Despite the IFN-γ-induced expression of some genes in VACV-infected cells, IFN-γ was unable to rescue the VACV-mediated inhibition of MHC class II antigen presentation. Moreover, VACV infection can affect the IFN-induced expression of Stat1-dependent and Stat1-independent genes, suggesting that the virus may target additional IFN-activated pathways. Thus, VACV targets multiple signaling pathways in the evasion of antiviral immune responses.
Introduction
Interferons (IFNs) are a major component of antiviral immunity. Type I IFNs (IFN-α/IFN-β) and type II IFN (IFN-γ) bind to multisubunit receptors, and activate the Jak-Stat signaling pathway. Stat (signal transducer and activator of transcription) protein phosphorylation allows dimerization and translocation to the nucleus where Stat dimers bind DNA and activate transcription. Stimulation with type I IFNs results in Stat1:Stat2 heterodimers, whereas IFN-γ stimulation results in Stat1 homodimers (Schroder and others 2004; Platanias 2005). The critical role of Stat1 in antiviral responses was demonstrated by the exquisite sensitivity of Stat1-deficient and Stat2-deficient mice to several viruses, including mouse hepatitis virus, vesicular stomatitis virus, Sendai virus, influenza virus, and respiratory syncytial virus (Durbin and others 1996; Meraz and others 1996; Garcia-Sastre and others 1998; Goodbourn and others 2000; Durbin and others 2002; Horvath 2004; Kato and others 2007).
The critical role of the Jak-Stat pathway in antiviral responses is also evident in how viruses have evolved to inhibit Stat activation and function at multiple points in the pathway. Mumps virus V protein disrupts the ability of Stat1 to associate with type I and type II IFN receptors (Kubota and others 2002). Measles V protein associates with both Stat1 and Stat2 and blocks function but not activation (Palosaari and others 2003). The V protein of Rubulavirus targets Stat1 and Stat2 for degradation by ubiquitylation (Parisien and others 2002). Rabies virus P protein binds to Stat1 and Stat2, causing them to be retained in the cytoplasm (Brzozka and others 2006). Respiratory syncytial viral proteins, NS1 and NS2, decreased Stat2 protein levels in infected human cells (Lo and others 2005). The NS5 protein of Flavivirus and Japanese encephalitis virus was shown to block phosphorylation of Stat1 possibly by interfering with Jak kinases (Best and others 2005; Lin and others 2006). The C protein of Sendai virus also inhibits the phosphorylation of Stat1 and Stat2 (Komatsu and others 2002; Gotoh and others 2003). Cytomegalovirus M27 decreases levels of Stat2 but does not affect Stat1 function (Zimmerman and others 2005). Therefore, there are multiple mechanisms that can be employed by viruses to block IFN signaling.
Vaccinia virus (VACV), the vaccinating agent used to eradicate smallpox, has a number of distinct mechanisms for evading IFN-stimulated responses. VACV has soluble IFN-γ and IFN-α/IFN-β receptor homologs with broad species specificity that block cytokines from binding to their receptors, although these genes are expressed late in the viral life cycle (Alcami and Smith 1995, 1996; Colamonici and others 1995; Symons and others 1995). VACV also has gene products that interfere with the function of 2′-5′ oligoadenylate synthetase and RNA-dependent protein kinase (Langland and Jacobs 2004). Notably, VACV encodes a dual-specificity phosphatase, VH1, which has been shown to dephosphorylate Stat1 and prevent the induction of IFN-γ-stimulated expression of guanylate-binding protein (GBP) and Stat1 (Najarro and others 2001). The function of VH1 is to dephosphorylate specific tyrosine residues and S/T residues found in VACV proteins including A17 and A14, respectively (Derrien and others 1999; Traktman and others 2000; Mercer and Traktman 2003). VH1 is required for viral replication as mutant VACV lacking VH1 expression has greatly reduced virus production (Najarro and others 2001). Although H1L is a late expressed gene, VH1 was able to have an immediate effect on a new host cell because each infecting virion contains approximately 200 molecules of VH1 (Liu and Traktman 1995). While VACV can inhibit IFN-γ signaling, whether it can affect type I IFN signaling, other cytokine signaling, or a broader array of IFN-induced gene expression has not been examined.
In this study, we demonstrate that VACV inhibits phosphorylation of Stat1 and Stat2 after stimulation with either type I or type II IFNs, but not the activation of other Stat proteins in fibroblasts and primary macrophages. These effects compromise the ability of IFN-γ to increase antigen presentation in VACV-infected cells. Moreover, we demonstrate that VACV infection blocks both Stat1-dependent and Stat1-independent IFN-induced gene expression after a short-term infection, suggesting that VACV also blocks additional IFN-activated pathways.
Materials and Methods
VACV infection, cytokine stimulation, and cell analysis
VACV was sucrose gradient purified and the titers were determined by a standard plaque reduction assay on the human osteosarcoma 143B cell line. NIH 3T3 fibroblasts or BMDM were infected at a multiplicity of infection (MOI) 10 at 37°C for the indicated lengths of time. NIH 3T3 cells were grown in DMEM (Invitrogen, Grand Island, NY) containing 10% fetal bovine serum. Murine bone marrow-derived macrophages (BMDM) from C57BL/6 mice (Harlan Bioproducts, Indianapolis, IN) and 129/Sv or 129/Sv Stat1−/−(Taconic, Germantown, NY) mice were generated, as previously described (Schmidt and others 2006). BMDM cultures were greater than 90% positive for CD11b and F4/80 by flow cytometry. BMDM also expressed CD86 and low levels of I-Ab. For VACV infection (Western Reserve VACV), cells were resuspended in fresh DMEM containing no serum at a concentration of 1.0 × 106/mL. NIH 3T3 cells or BMDM were incubated with or without purified VACV (MOI 10) for 30 min before incubation with or without cytokine [100 ng mIFN-γ (R&D Systems, Inc., Minneapolis, MN) or 100 ng mIFN-β (PBL Biomedical Laboratories, Piscataway, NJ) per 106 cells] for 1 h at 37°C, and cell extracts were used for immunoblotting. BMDM were incubated with or without VACV (MOI 10) for 1 h before incubation with or without cytokine for 4 h at 37°C. Protein extracts were used for immunoblotting, and RNA was used for quantitative PCR. Quantitative PCR was performed as previously described using gene-specific primers (Applied Biosystems, Foster City, CA) with expression levels normalized to β-actin mRNA and presented as expression relative to the unstimulated condition (Mathur and others 2006). For the determination of level of infection by flow cytometry, cells were fixed and permeabilized during staining with PE-conjugated TW2.3 antibody to detect E3L from cell infected with VACV for 5 h. The percentage of E3L positive cells was used as an indication for the efficiency of infection.
Immunoblotting
Total cell extracts were made from NIH 3T3 cells or primary BMDM treated as above. The cells were pelleted and then resuspended in lysis buffer (10% glycerol, 1% IGEPAL, 50 mM Tris pH 7.4, 150 mM NaCl, and 1 mM EDTA). Protein concentrations were determined, and equal amounts of proteins were resolved on a NuPAGE 4–12% Bis-Tris gradient gel (Invitrogen, Carlsbad, CA). The proteins were transferred to a 0.45 μm OPTITRAN BA-S 85 nitrocellulose membrane (PGC Scientifics, Frederick, MD or Schleicher and Schuell, Keene, NH). The blot was probed using anti-phospho-Stat as indicated or stripped and reprobed using anti-total Stat. All antibodies were from Upstate/Millipore (Charlottesville, VA). After the addition of goat anti-rabbit secondary antibodies, the membranes were developed using chemiluminescence, ECL (PerkinElmer LAS, Boston, MA) and exposed to film (Pierce Biotechnology, Rockford, IL).
Cloning and production of recombinant H1 proteins
VACV H1L was cloned by PCR from the cDNA of VACV-infected cells. VACV H1L was mutated to generate a consensus Kozak and translation stop sequences. The cDNA was then cloned into the pRSETB bacterial expression vector. The VACV H1L was mutated into the variola virus (VARV) H1 using site-directed mutagenesis. Active site mutants (C110S) of VACV and VARV H1 were then generated by site-directed mutagenesis. Wild-type and mutant H1L sequences were confirmed. To generate recombinant proteins, pRSETB plasmids expressing VH1 proteins were transformed into BL21LysE (Invitrogen, Carlsbad, CA). Single antibiotic-resistant colonies were used to inoculate 50 mL SOB medium supplemented with 50 μg/mL ampicillin and 35 μg/mL chloramphenicol. These cultures were grown by shaking (200 rev/min) to saturation overnight at 37°C and then diluted 50-fold into several liters of SOB medium supplemented with 50 μg/mL ampicillin. When the cells reached early log phase (OD600 nm = 0.4–0.6), isopropyl-β-d-thiogalactopyranoside (IPTG) was added to a final concentration of 1 mM. One hour later, the cells were recovered by centrifugation at 5000g for 10 min and stored at −80°C. Harvested cell extract was purified using nickel-nitrilotriacetic acid (Ni-NTA) agarose (Qiagen, Valencia, CA), eluted with increasing imidazole concentrations between 50 mM and 150 mM, and active fractions were stored at −20°C in 20% glycerol. Purity was determined by separating proteins on a 4–12% gradient gel after staining with Coomassie blue. Recombinant protein was detected by immunoblotting with anti-Xpress antibody (Invitrogen). Protein concentration was determined using the BCA method. Equal protein concentrations of samples were confirmed using serial dilutions of samples and testing by immunoblotting.
Phosphatase activity assay
The p-nitrophenyl phosphate (pNPP) hydrolysis activity of recombinant H1 was assayed in 5 mL buffer A (100 mM HEPES pH 7.0, 150 mM NaCl, 1 mM EDTA, and 5 mM DTT) containing 5 mg pNPP, incubated at room temperature, and color changes were determined by OD 405 nm at the indicated time points. One unit of phosphatase activity was defined as the amount of enzyme required to hydrolyze 1 μmol pNPP in 1 min. Purified VACV H1 had a specific activity of 0.07 units/mg compared to 0.17 units/mg for VARV H1. To perform Michaelis-Menten kinetics, serial dilutions of pNPP solution were mixed with 5 μg recombinant H1 protein. The mixture was incubated at room temperature for 30 min, and the reaction was stopped with 2 vols of 1 N NaOH. Samples were then assayed for color change at OD 405 nm. Reaction rate is defined as absorbance divided by time. Km and Vmax were determined using a Hanes plot.
Phosphopeptide phosphatase assay
To assay for H1 protein activity on peptide substrates, serial dilutions of recombinant protein were incubated in a 96-well plate with the substrate phosphopeptide in PTPase assay buffer [10 mM Tris pH 8, 2 mM EDTA, 2 mM 2-ME, and 2 mM NaF, 10 μL/mL protease inhibitor cocktail (Roche, Indianapolis, IN)]. The plate was agitated gently (70 rev/min) for 60 min at 31°C, and the reaction was stopped by adding 20 μL of 25 mM sodium vanadate. Samples were then transferred on to a nitrocellulose filter (Hybond ECL nitrocellulose membrane 0.2 μm; Amersham, Piscataway, NJ) using the Bio-Dot microfiltration apparatus (Bio-Rad, Hercules, CA). Phosphopeptides were detected by immunoblotting using antiphosphotyrosine 4G10 monoclonal antibody (mAb) or antiphospho-Stat1 (pY701) antibody (Millipore/Upstate, Charlottesville, VA). Activity was determined using densitometric analysis of the dot-blot assay. Percent dephosphorylation was determined using [control phosphorylation – (phosphorylation following H1 – phosphorylation following H1 C110S)/protein (μg)].
Phosphoprotein phosphatase assay
Total cell extracts (500 μg) from NIH 3T3 cells stimulated with IFN-γ or OSM were incubated with anti-Stat1 or anti-Stat3, respectively, in IP buffer (10% glycerol, 1% IGEPAL, 50 mM Tris pH 7.4, 150 mM NaCl, and 1 mM EDTA) with added protease inhibitors overnight at 4°C. Immune complexes were precipitated with Protein A agarose (Upstate Biotechnology). Precipitates were washed and resuspended in PTPase buffer. Cell extract from NIH 3T3 cells stimulated with IFN-γ and infected with VACV (5 μg) and the indicated recombinant H1 proteins (10 μg) were added to the precipitate and incubated at 31°C for 60 min. Reactions were stopped with SDS-PAGE loading buffer, and immunoblot analysis was performed for the presence of phospho-Stat proteins using specific antibodies.
Antigen presentation
BMDM were stimulated with or without murine IFN-γ for 1 h, then pulsed with increasing concentrations of HEL74–88 peptide in the presence or absence of VACV (MOI 10) for another 5 h. Cells were fixed in 0.5% paraformaldehyde (10 min, on ice) and cocultured at 37°C with HEL-specific T-cell B04 for 24 h. T-cell interleukin-2 (IL-2) production was quantitated using HT-2, an IL-2 dependent T-cell line, as previously described (Guan and others 1991). HT-2 cell proliferation was quantitated via [3H]thymidine incorporation. Data are expressed as counts per minute with all assays performed in triplicate and the mean and SD calculated.
Results
Specificity of VACV inhibition of Stat signaling in vivo
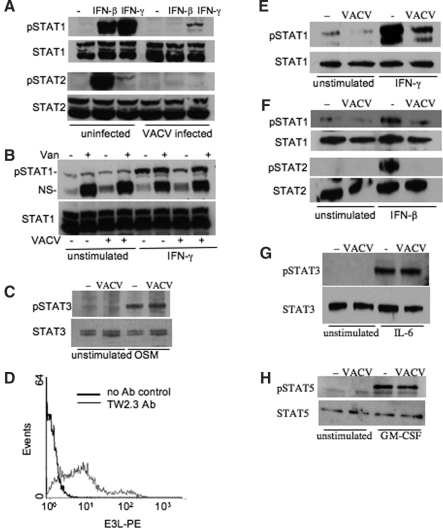
It has been shown previously in HeLa cells that VACV can dephosphorylate Statl (Najarro and others 2001), thus interfering with the establishment of an IFN-γ-induced antiviral state. To further explore the cell type specificity of this function, we tested this in a fibroblast cell line, fibroblasts being a primary source of IFN-β after viral infection. NIH 3T3 cells were cultured in the presence or absence of IFN-γ or IFN-β for the last 60 min of a 90-min VACV infection. Analysis at this early time point avoids the confounding effects of VACV-encoded soluble IFN receptors, which are expressed late in the viral life cycle. VACV typically infected over 95% of the NIH 3T3 cells as assessed by the expression of the viral early gene product E3L using flow cytometry (data not shown). Compared with uninfected IFN-γ-stimulated or IFN-β-stimulated NIH 3T3 cells, the VACV infection greatly diminished the level of phosphorylated Statl without altering the expression of total Stat1 protein in cells (Fig. 1A). Moreover, there was a similar decrease in the phosphorylation of Stat2 caused by VACV infection in IFN-β-stimulated cells (Fig. 1A). To confirm effects we observed were the effects of a phosphatase, we repeated the stimulation of IFN-γ in the presence or absence of sodium orthovanadate (Fig. 1B). Although the orthovanadate had only minor effects on the phosphorylation of Stat1 in unstimulated condition, it prevented decreased Stat1 phosphorylation in VACV-infected samples. A nonspecific band that appeared in the immunoblot was sensitive to orthovanadate in all conditions, confirming the activity of the phosphatase inhibitor (Fig. 1B). In contrast to the effects on Stat1 and Stat2, VACV infection did not inhibit the ability of oncostatin M to induce Stat3 phosphorylation in these cells (Fig. 1C). These results indicate that VACV infection disrupted the activation of Stat1 that originates through signaling from both type I and type II IFNs, and inhibited Stat2 activation following type I IFN signaling.
FIG. 1.
VACV infection inhibits Stat1 and Stat2 phosphorylation. (A) Immunoblots of activated Stat proteins. Uninfected or VACV-infected NIH 3T3 cells were incubated in the presence or absence of IFN-β or IFN-γ for the last 60 min of a 90-min infection. Whole cell extracts were separated by SDS-PAGE and immunoblotted for phospho-Stat1 or phospho-Stat2. Immunoblots were stripped and reprobed for total Stat1 or Stat2. Data are representative of 3–5 experiments. (B) NIH 3T3 cells were cultured as in (A) in the presence or absence of sodium orthovanadate. Extracts were immunoblotted with antibodies to phospho-Stat1 and total Stat1. NS, nonspecific. (C) Immunoblot of pStat3 and total Stat3 from whole cell extracts of NIH 3T3 cells cultured in the presence or absence of oncostatin M (OSM) in VACV-infected or uninfected cells. (D) Histogram of VACV-infected BMDM. BMDM were stained with a-E3L (TW2.3, gray line) or with control antibody (bold line) after 5 h of infection with VACV (MOI 10). Data are representative of three experiments. (E–H) Immunoblot of activated Stat proteins. Uninfected or VACV-infected BMDM were incubated with or without (E) IFN-γ, (F) IFN-β, (G) IL-6, or (H) GM-CSF for the last 60 min of a 90-min infection. Whole cell extracts were separated by SDS-PAGE and immunoblotted for (E and F) phospho-Stat1, (F) phospho-Stat2, (G) phospho-Stat3, or (H) phospho-Stat5. Immunoblots were stripped and reblotted for total Stat proteins. Data are representative of two or three experiments.
We next tested the effect of VACV on Stat protein activation in primary BMDM to assess its ability to alter cytokine signaling in a professional antigen-presenting cell. VACV infected 70–80% of cells in BMDM cultures as assessed by flow cytometric examination of E3L expression (Fig. 1D). To test the effects of VACV infection on IFN-γ signaling, BMDM were cultured in the presence or absence of cytokines for the last 60 min of a 90-min infection with VACV. As was observed in NIH 3T3 cells, VACV infection decreased the level of IFN-γ-induced and IFN-β-induced phospho-Stat1 in BMDM, an effect that was inhibited by phosphatase inhibition (Fig. 1E and 1F and data not shown). Moreover, there was a similar decrease in the phosphorylation of Stat2 caused by VACV infection in the same IFN-β-stimulated cells (Fig. 1F). However, VACV infection did not affect the activation of Stat3 after IL-6 stimulation and had only marginal effects on the activation of Stat5 following GM-CSF stimulation (Fig. 1G and 1H). Thus, VACV inhibited the phosphorylation of Stat1 and Stat2, while it had limited effects on the activation of other Stat proteins.
Activity of VACV and VARV H1 proteins in vitro
As VH1 mediates the dephosphorylation of Stat proteins in VACV-infected cells (Najarro and others 2001), we next wanted to analyze recombinant H1 specificity and determine whether there were differences in activity between VACV and VARV H1 proteins. VACV H1 cDNA was cloned from VACV-infected cells into a bacterial expression vector. The VACV H1 cDNA was mutated by site-directed mutagenesis to generate VARV H1, which differs from VACV H1 by four amino acids (Fig. 2A). The codon of the VACV and VARV H1 active site cysteine was mutated to generate a catalytically inactive enzyme (C110S) (Guan and others 1991). The recombinant H1 and mutant protein products were expressed with a 6× His-affinity tag, which allowed the fusion protein to be purified by using immobilized metal affinity chromatography (IMAC), and an Xpress epitope tag, which allowed immunoblotting using anti-Xpress antibody. The induced cell lysates and purified protein samples were then stained with Coomassie blue following SDS-PAGE that indicated homogeneous purified protein (Fig. 2B). The results showed the presence of one major band that was approximately 17 kDa in mass, the predicted size of H1. The putative H1 proteins were immunoreactive with the anti-Xpress antibody confirming the identity of the fusion proteins (Fig. 2C).
FIG. 2.
Generation and characterization of recombinant H1 protein phosphatase activity. (A) Schematic representation of sequence differences in VACV and VARV H1. The C110 residue that is required for catalytic activity is also indicated. (B) SDS-PAGE (4–12% gradient) and Coomassie blue staining of total lysates (L) or purified proteins (P) from bacteria expressing VACV H1 (1), VARV H1 (2), VACV C110S H1 (3), or VARV C110S H1 (4). (C) Immunoblot analysis of purified recombinant proteins using mouse monoclonal antibody directed against the Xpress tag. P1: VACVH1; P2: VARVH1; P3: VACVC110S H1; P4: VARVC110S H1. (D) Hydrolysis of p-nitrophenyl phosphate by H1 proteins. Hydrolysis is measured as increase in optical density at 405 nm. Data are presented as the mean ± SE of triplicate determinations and are representative of three experiments. (E) Calculation of enzyme activity based on activity at 30 min observed in (D). One unit is the amount of phosphatase required to release 1 nmol phosphate from pNPP in 1 min under the tested assay conditions. Data are presented as mean ± SE. (F) Determination of activity/μg of protein on the indicated peptide substrates (STAT1pY701 peptide, LDDPKRTGpYIKTELISVS; VACV A17pY203 peptide, PYTAGNKVDVDIPTFNSLNTDDpY). Activity was determined as the amount of dephosphorylation per pg protein and was averaged over values at several concentrations of enzyme. A17 phosphopeptides were detected by antiphosphotyrosine 4G10 mAb, and phospho-Stat1 levels were determined with antiphospho-Stat1. Results are representative of three experiments. (G) Stat1 or Stat3 was immunoprecipitated from NIH 3T3 cells stimulated, respectively, with IFN-γ and OSM. Precipitates were incubated with recombinant H1 proteins, and phosphorylation level was determined with immunoblot analysis.
The chromogenic substrate pNPP, an analog of phosphotyrosine, was then used to assay the enzymatic activity of the recombinant VH1 phosphatase proteins. The C110S mutants of either VARV or VACV H1 did not have detectable activity in this assay (Fig. 2D). We used the Lambert-Beer equation to determine the specific activity of the VACV and VARV H1 enzymes. This analysis demonstrated that activity/μg of protein was higher for purified VARV H1 than for VACV H1 (Fig. 2E). We then varied pNPP concentration between 0.01 mM and 5 mM for Michaelis-Menten kinetics. Within 60 min of reaction time, the catalytic efficiency of VARV H1 was about 3-fold higher than VACV H1. Under these conditions, the reaction velocity was linear with respect to enzyme concentration up to at least 10 μg. The enzyme exhibited classical saturation kinetics with respect to pNPP concentration (data not shown). These data suggested that VARV H1 had a higher substrate affinity than VACV H1 (Km of 365 μM for VARV H1 versus Km of 558 μM for VACV H1). Moreover, VARV H1 had a higher maximal reaction rate than VACV H1 (Vmax of 5.85 nmol/min/μg and 2.25 nmol/min/μg, respectively). The Michaelis constants for VACV and VARV H1 phosphatase were 14- and 21-fold lower, respectively, than the corresponding values for the human VACV H1-related phosphatase VHR (Denu and Dixon 1995). The Km for VARV H1 is similar to those described in another report (Tropea and others 2006).
VACV H1 dephosphorylates the C-terminal tyrosine residue of A17 and Ser85 of A14 in vivo (Derrien and others 1999; Mercer and Traktman 2003). VACV H1 also dephosphorylates IFN-γ-induced Stat1 (Najarro and others 2001). To compare the ability of VACV and VARV H1 to dephosphorylate these targets, we used an in vitro phosphatase assay with recombinant H1 and phosphopeptides with sequence derived from Stat1 or A17 as a control. Serially diluted recombinant H1 proteins (from VACV and VARV and C110S mutants of both H1 proteins) were incubated with a fixed amount of each peptide for 1 h, and residual phosphorylation was detected by antiphosphotyrosine or phospho-Stat antibodies in a dot-immunoblot. Although the C110S VACV and VARV mutants showed little or no activity in these assays, both VACV and VARV H1 demonstrated the ability to dephosphorylate about 70% of the pStat1 peptide at the highest enzyme concentration, with the VARV H1 showing slightly higher activity (Fig. 2F). Similar results were observed when a phosphopeptide for the C-terminal amino acids of the VACV A17 protein was used (Fig. 2F). As VARV A17 has an identical sequence to VARV A17, this raises an interesting possibility that VARV H1 also functions on virus targets more efficiently than VACV H1. These results suggest that VARV H1 has higher activity than VACV H1 on relevant targets.
To further examine the specificity of H1 activity we observed in Figure 1, we tested the ability of VACV and VARV H1 proteins to mediate dephosphorylation of native Stat proteins in vitro. Stat1 and Stat3 were immunoprecipitated from NIH 3T3 cells stimulated with IFN-γ and OSM, respectively. Precipitates were then incubated with VARV or VACV H1, or mutant H1 proteins, in phosphatase buffer followed by immunoblotting to assess the level of phospho-Stat protein. Consistent with the data in Figure 1, VACV and VARV H1, but not mutant H1 proteins, were able to dephosphorylate Stat1, but not Stat3 (Fig. 2G). Thus, H1 proteins demonstrate selectivity in substrates targeted for dephosphorylation.
VACV infection blocked IFN-induced Stat1-dependent gene expression in primary macrophages
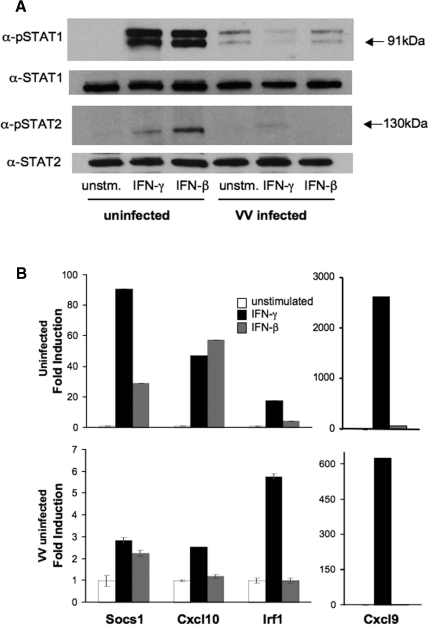
Although VACV infection has been shown to affect the IFN-induced expression of some Stat1 target genes including GBP (Najarro and others 2001), a broad range of IFN targets has not been examined. By using quantitative PCR, we examined the gene expression in BMDM stimulated with IFN for the last 4 h of a 5-h infection with VACV. To confirm that phospho-Stat1 and phospho-Stat2 were still affected by VACV infection at this time point, levels of Stat1 and Stat2 phosphorylation were tested using immunoblot. As demonstrated in Figure 3A, phosphorylation of both Stat proteins was still decreased after 5 h of infection. The effects of VACV infection on the induction of a set of well-described IFN target genes, including Socs1, Cxc19 (MIG), Cxc110 (IP-10), and Irf1, was then tested using quantitative PCR, where gene expression is normalized to the levels of β-actin expression and presented as fold induction over unstimulated cells. Levels of mRNA for β-actin or another housekeeping gene, Hsc70, did not vary greatly over the period of analysis. Each of the target genes was induced by IFN-γ or IFN-β to various degrees (Fig. 3B, upper panel). IFN induction of these four genes was markedly decreased in the VACV-infected cells, with IFN-γ-induced Socs1 expression falling from an 85-fold induction to a 3-fold induction and IFN-γ-induced Cxc110 falling from a 40–50-fold induction to a 2-fold induction. Irf1 and Cxc19 were more modestly affected, falling from a 12-fold induction to a 6-fold induction and a 2500-fold induction to a 600-fold induction, respectively (Fig. 3B). Moreover, while the ability of IFN-β to induce each gene varied within this set, VACV infection eliminated the ability of IFN-β to induce gene expression (Fig. 3B). We then wanted to determine whether stimulating cells with IFNs before VACV infection would result in less susceptibility to inhibition by VACV infection. Cells were stimulated with IFN-γ or IFN-β 1 h before VACV infection and the induction of the same set of genes was examined. VACV infection inhibited gene induction by pretreatment with IFN to similar levels as observed when IFN was added after VACV infection (data not shown). These results demonstrate that IFN-induced gene expression was not completely eliminated by VACV infection and the residual gene induction was dependent on the target gene.
FIG. 3.
IFN-induced gene expression. Cells were stimulated with IFN-β or IFN-γ 1 h after infection or left unstimulated for a total of 5 h, and RNA or protein were isolated. Equal amounts of RNA were used to create cDNA for quantitative PCR. Gene expression was determined in duplicate and reported as an average ± SD. (A) Immunoblots of 5-h infection of BMDM. Uninfected or VACV-infected BMDM were incubated with or without IFN-γ or IFN-β for the last 4 h of a 5-h infection. Whole cell extracts were separated by SDS-PAGE and immunoblotted for phospho-Stat1 or phospho-Stat2. Immunoblots were stripped and reprobed for total Stat1 or total Stat2. Data are representative of two experiments. (B) IFN-induced gene expression. Gene expression in the presence or absence of the indicated IFN is shown in uninfected BMDM (upper panel) or VACV-infected BMDM (lower panel). Gene expression was determined in duplicate and reported as an average ± SD. Data are representative of three experiments.
VACV infection inhibited IFN-induced MHC class II antigen presentation
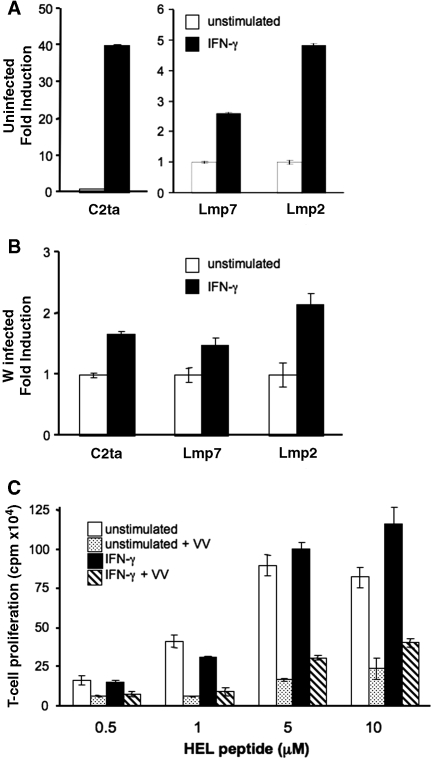
Macrophage stimulation with IFN-γ upregulates antigen processing and presentation pathways. IFNs induce major histocompatibility complex (MHC) I and II, and not surprisingly VACV infection disturbs MHC I and MHC II antigen presentation (Townsend and others 1988; Li and others 2005). Moreover, the induction of CIITA and MHC II by IFNs is absent in Stat1−/− cells (Meraz and others 1996). Thus, the loss of Stat1 activation predicts that VACV infection should have a deleterious effect on IFN-induced enhancement of MHC II in BMDM. We tested the induction of three IFN-γ-inducible antigen-presenting genes, C2ta, Lmp2, and Lmp7, after IFN-γ stimulation of BMDM with or without VACV infection. The robust induction of C2ta because of IFN-γ was eliminated in the VACV-infected cells (Fig. 4A versus 4B). The induction of Lmp2 and Lmp7 was also decreased following VACV infection (Fig. 4A and 4B). This result leads us to predict that VACV infection would interfere with the ability of IFN-γ to enhance antigen presentation or to rescue the effects of VACV-mediated inhibition of MHC class II antigen presentation. Therefore, studies were undertaken to examine class II-restricted antigen presentation in VACV-infected BMDM. To assay the effect of VACV infection on IFN-γ-induced enhancement of antigen presentation, the response of antigen-specific T-cell hybridomas to BMDM incubated with various concentrations of the HEL peptide was tested (Fig. 4C). BMDM were incubated with VACV or mock infected after 1 h of IFN-γ stimulation, and antigen-presenting capability was assessed as previously described (Li and others 2005). At higher concentrations of peptide, IFN-γ was able to slightly enhance antigen presentation. VACV infection of BMDM alone significantly reduced class II antigen presentation, although surface expression of MHC class II assessed by flow cytometry was not significantly decreased (Fig. 4C and data not shown). IFN-γ stimulation only modestly offset this dramatic effect of VACV on antigen presentation. These data show that priming BMDM with IFN-γ for 1 h before VACV infection did not protect BMDM class II antigen presentation function.
FIG. 4.
Effects of VACV infection on MHC class II-restricted antigen presentation. IFN-γ-induced gene expression in BMDM. (A and B) Analysis of the expression of genes involved in MHC-restricted antigen presentation was assessed as in Figure 3B in uninfected (A) and VACV-infected (B) BMDM. Gene expression was determined in duplicate and reported as an average ± SD. Data are representative of two experiments. (C) T-cell proliferation because of BMDM presentation of HEL74–88 peptide. Four concentrations of HEL74–88 peptide ranging from 0.5 μM to 10 μM were incubated with the BMDM. BMDM were incubated under four conditions; unstimulated (open bar), VACV-infected unstimulated (dotted bar), IFN-γ-stimulated (filled bar), and VACV-infected IFN-γ-stimulated (striped bar). Activation of HEL-specific T cells by BMDM was assessed using a bioassay monitoring the proliferation of IL-2-dependent HT-2 cells. Data are representative of three experiments.
VACV infection inhibits IFN-induced Stat1-independent gene expression in primary macrophages
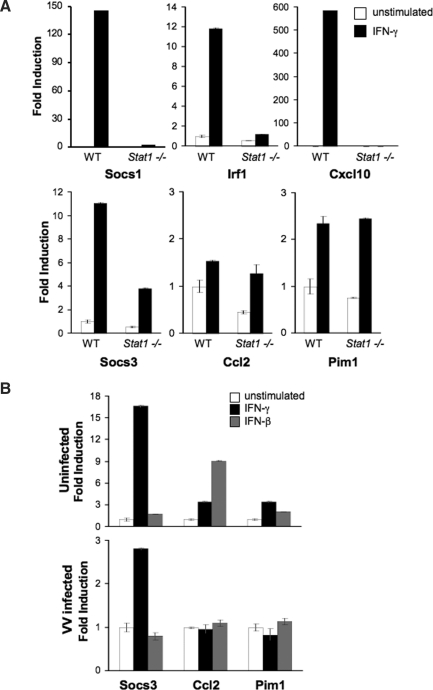
IFN-γ-inducible genes can be characterized as Stat1-dependent or Stat1-independent and established using Stat1-deficient cells. To test the requirement for Stat1 in IFN-γ-induced gene induction in BMDM, wild-type and Stat1−/−BMDM were tested for the induction of genes classified in other cell types as Stat1-dependent and Stat1-independent using quantitative PCR (Schmidt and others 2006). As had been previously shown, the induction of Socs1, Irf1, and Cxc110 was Stat1-dependent. In contrast, IFN-γ-induction of Cc12 and Pim1 were independent of Stat1 (Fig. 5A). As observed in other cell types, IFN-γ-induced Socs3 expression was partially Stat1-dependent (Gil and others 2001; Ramana and others 2002), in that induction was decreased but not absent in Stat1−/− cells (Fig. 5A).
FIG. 5.
Expression of Stat1-independent genes. (A) Wild type or Stat1−/− BMDM were incubated with or without IFN-γ for 5 h. Gene expression was assessed as in Figure 3B. Gene expression was determined in duplicate and reported as an average ± SD. Data are representative of two experiments. (B) BMDM were incubated in the presence or absence of IFN-β or IFN-γ for the last 4 h of a 5-h infection. Gene expression is shown in uninfected BMDM (upper panel) or VACV-infected BMDM (lower panel). Gene expression is determined in duplicate and reported as an average ± SD. Data are representative of three experiments.
To determine whether VACV infection was able to block IFN induction of Stat1-independent genes, BMDM were stimulated with type I or type II IFNs after infection with VACV. The induction of Cc12 and Pim1 by either IFN-γ or IFN-β was essentially eliminated in VACV-infected cells (Fig. 5B). In contrast, the induction of Socs3 was only partially inhibited. The degree of Socs3 induction in the infected BMDM was comparable to the level of induction observed in Stat1−/− BMDM (Fig. 5A and 5B). These results suggest that VACV infection not only blocks Stat1-dependent gene induction, but also affects the IFN-induced Stat1-independent pathways responsible for the induction of Cc12 and Pim1. However, the Stat1-independent pathway responsible for Socs3 induction was only marginally affected by VACV infection, further demonstrating selectivity by VACV, inhibiting some but not all Stat1-independent signaling.
Discussion
During the past century, VACV was used as a vaccine to block the transmission of and eradicate smallpox. Both VARV, the virus that causes smallpox, and VACV are the members of the poxvirus family, with considerable gene homology. Species of Poxviridae share several conserved mechanisms for evading the host immune responses. One protein common among the poxviruses that plays such a role is the dual-specificity phosphatase, VH1. Previous work using HeLa cells demonstrated the ability of VACV to inhibit IFN-γ-induced phosphorylation of Stat1 and that VH1 itself could dephosphorylate Stat1 in vitro (Najarro and others 2001). In this study, we further probe those initial observations showing that VACV infection blocks Stat activation by type I IFNs but not Stat activation by other cytokines, differentially affects IFN-induced gene expression, and that VACV also inhibits some Stat1-independent signaling pathways.
Several cellular protein tyrosine phosphatases have been shown to target Stat protein activation. Of these, there is some selectivity. T-cell-protein tyrosine phosphatase (TC-PTP) targets Stat1 and Stat3 but not Stat5 (ten Hoeve and others 2002). There have been conflicting reports on the ability of TC-PTP to work on Stat6 (ten Hoeve and others 2002; Lu and others 2007). SHP-1 targets Stat6 (Huang and others 2005) and SHP-2 can target Stat1, Stat3, and Stat5 proteins, although some effects may be cell type specific (You and others 1999; Chen and others 2003; Ke and others 2006). LMW-DSP2 and PTPRT have been shown to be Stat3 phosphatases (Sekine and others 2006; Zhang and others 2007). PTP-BL has also recently been shown to be a physiological phosphatase for multiple Stat proteins (Nakahira and others 2007). However, none of these phosphatases have a similar specificity in two distinct cell types to what we have shown for H1, which targets Stat1 and Stat2, but not other Stat proteins. While being able to target Stat1 is clearly advantageous for the virus in being able to evade IFN responses, the reason for the inability of H1 to target other Stat proteins is less clear. Dual-specificity phosphatases are also well characterized as inhibiting mitogen-activated protein kinase (MAPK) kinase pathways, although we did not observe any inhibition of MAPK pathway activation in VACV-infected cells. VACV infection of antigen-presenting cells actually appears to increase MAPK activation (Renukaradhya and others 2005). These data suggest that in addition to the requirement for H1 in viral replication, this phosphatase plays a specific role in immune evasion by blocking Stat activation in response to both type I and type II IFNs.
Although the VARV H1 protein has been previously described (Tropea and others 2006), this study represents the first comparison between VACV and VARV H1 proteins. While the specificity of both phosphatases appears similar, VARV H1 has greater activity in both chromogenic substrate and peptide assays. VARV H1 only has four amino acids different from the VACV H1, although one change is directly adjacent to the active site cysteine. This is similar to other VARV proteins, such as complement inhibitor and TNF-binding proteins, which have been shown to be more active than homologs from less pathogenic poxviruses (Rosengard and others 2002; Gileva and others 2006). It is intriguing to speculate that the increased VARV H1 activity contributes to the pathogenesis of VARV, either by increased activity on viral substrates or by increased interference with Stat function. In this respect, H1 remains an attractive target for pox-virus antiviral compounds.
Previous results indicated that VACV infection efficiently blocked IFN-γ-induced gene expression in epithelial cells. Our results, using a broader range of IFN-induced targets, demonstrate that the inhibition of gene induction varies with the target. Induction of Socs1, Cxc110, and C2ta are almost completely blocked by VACV infection (Figs. 3 and 4). In contrast, there was still considerable induction of Irf1 and Cxc19 by IFN-γ following VACV infection, although the fold induction was decreased about 70%. These results suggest that some, but not all, IFN-stimulated functions are compromised and the differences may depend on the sensitivity of distinct gene promoters to Stat1 levels. To further test biologic functions of IFN-γ in antigen-presenting cells, we also tested whether IFN-γ could rescue the inhibition of MHC class II antigen presentation observed after VACV infection (Li and others 2005). However, IFN-γ only modestly affected antigen presentation during VACV infection of BMDM. These data suggest that VACV inhibits the IFN-stimulated immune response, although some target genes are more efficiently inhibited than others.
IFN-induced genes can be classified as Stat1-dependent or Stat1-independent. We tested the ability of VACV infection to block the induction of Stat1-independent genes, and observed that the induction of two of the three genes examined was blocked by VACV infection. This suggests that VACV has mechanisms to block additional IFN-stimulated pathways. Although Stat1-independent pathways are not well characterized, a number of transcription factors have been suggested to mediate gene activation in the absence of Stat1. A small number of Stat1-independent genes can be regulated by Stat3 (Qing and Stark 2004; Ramana and others 2005). However, as shown in Figure 1, VACV infection did not block the activation of Stat3 in the cells examined in these studies. The activity of IRF9, AP-1, and nonphosphorylated Stat2 have also been suggested to mediate this process (Hahm and others 2005; Ousman and others 2005; Gough and others 2007). However, we observed no VACV-induced changes in the nuclear or cytoplasmic localization of IRF9, c-fos, c-jun, or Stat2, or in the DNA-binding activity of AP-1 complexes in extracts from BMDM (data not shown). Thus, VACV infection inhibits a novel IFN-activated Stat1-independent pathway.
Poxviruses have evolved multiple mechanisms to evade or prevent the IFN response. During early stages of VACV infection, viral gene products block host antiviral enzymes along with the activation of Stat1-dependent IFN signaling pathways, which would inhibit viral replication within an infected cell (Smith and others 1997). Late in infection, pox-viruses produce soluble IFN receptors that would further block IFN responses to facilitate viral spread. Together, these mechanisms target specific antiviral pathways and promote reproductive success during poxvirus replication.
Acknowledgments
We thank B. Champ, J. Eltz, and K. Gillett-Heacock in the VACV core, K. Toomey and V.T. Thieu for technical assistance. We thank P. Traktman for providing a protocol for purifying H1 protein, and B. Chan for reviewing this manuscript. This work was supported by National Institutes of Health grant POl AI056097.
References
- Alcami A. Smith GL. Vaccinia, cowpox, and camelpox viruses encode soluble gamma interferon receptors with novel broad species specificity. J Virol. 1995;69:4633–4639. doi: 10.1128/jvi.69.8.4633-4639.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alcami A. Smith GL. Soluble interferon-gamma receptors encoded by poxviruses. Comp Immunol Microbiol Infect Dis. 1996;19:305–317. doi: 10.1016/0147-9571(96)00013-6. [DOI] [PubMed] [Google Scholar]
- Best SM. Morris KL. Shannon JG. Robertson SJ. Mitzel DN. Park GS. Boer E. Wolfinbarger JB. Bloom ME. Inhibition of interferon-stimulated JAK-STAT signaling by a tick-borne flavivirus and identification of NS5 as an interferon antagonist. J Virol. 2005;79:12828–12839. doi: 10.1128/JVI.79.20.12828-12839.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brzozka K. Finke S. Conzelmann KK. Inhibition of interferon signaling by rabies virus phosphoprotein P: activation-dependent binding of STAT1 and STAT2. J Virol. 2006;80:2675–2683. doi: 10.1128/JVI.80.6.2675-2683.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y.Wen R.Yang S.Schuman J.Zhang EE.Yi T.Feng GS.Wang D.2003Identification of Shp-2 as a Stat5A phosphatase J Biol Chem 27816520–16527.12615921 [Google Scholar]
- Colamonici OR. Domanski P. Sweitzer SM. Larner A. Buller RM. Vaccinia virus B18R gene encodes a type I interferon-binding protein that blocks interferon alpha transmembrane signaling. J Biol Chem. 1995;270:15974–15978. doi: 10.1074/jbc.270.27.15974. [DOI] [PubMed] [Google Scholar]
- Denu JM. Dixon JE. A catalytic mechanism for the dual-specific phosphatases. Proc Nat1 Acad Sci USA. 1995;92:5910–5914. doi: 10.1073/pnas.92.13.5910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derrien M. Punjabi A. Khanna M. Grubisha O. Traktman P. Tyrosine phosphorylation of A17 during vaccinia virus infection: involvement of the H1 phosphatase and the F10 kinase. J Virol. 1999;73:7287–7296. doi: 10.1128/jvi.73.9.7287-7296.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durbin JE. Hackenmiller R. Simon MC. Levy DE. Targeted disruption of the mouse Stat1 gene results in compromised innate immunity to viral disease. Cell. 1996;84:443–450. doi: 10.1016/s0092-8674(00)81289-1. [DOI] [PubMed] [Google Scholar]
- Durbin JE. Johnson TR. Durbin RK. Mertz SE. Morotti RA. Peebles RS. Graham BS. The role of IFN in respiratory syncytial virus pathogenesis. J Immunol. 2002;168:2944–2952. doi: 10.4049/jimmunol.168.6.2944. [DOI] [PubMed] [Google Scholar]
- Garcia-Sastre A. Durbin RK. Zheng H. Palese P. Gertner R. Levy DE. Durbin JE. The role of interferon in influenza virus tissue tropism. J Virol. 1998;72:8550–8558. doi: 10.1128/jvi.72.11.8550-8558.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gil MP. Bohn E. O'Guin AK. Ramana CV. Levine B. Stark GR. Virgin HW. Schreiber RD. Biologic consequences of Stat1-independent IFN signaling. Proc Nat1 Acad Sci USA. 2001;98:6680–6685. doi: 10.1073/pnas.111163898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gileva IP. Nepomnyashchikh TS. Antonets DV. Lebedev LR. Kochneva GV. Grazhdantseva AV. Shchelkunov SN. Properties of the recombinant TNF-binding proteins from variola, monkeypox, and cowpox viruses are different. Biochim Biophys Acta. 2006;1764:1710–1718. doi: 10.1016/j.bbapap.2006.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodbourn S. Didcock L. Randall RE. Interferons: cell signalling, immune modulation, antiviral response and virus counter-measures. J Gen Virol. 2000;81:2341–2364. doi: 10.1099/0022-1317-81-10-2341. [DOI] [PubMed] [Google Scholar]
- Gotoh B. Takeuchi K. Komatsu T. Yokoo J. The STAT2 activation process is a crucial target of Sendai virus C protein for the blockade of alpha interferon signaling. J Virol. 2003;77:3360–3370. doi: 10.1128/JVI.77.6.3360-3370.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gough DJ. Sabapathy K. Ko EY. Arthur HA. Schreiber RD. Trapani JA. Clarke CJ. Johnstone RW. A novel c-Jun-dependent signal transduction pathway necessary for the transcriptional activation of interferon gamma response genes. J Biol Chem. 2007;282:938–946. doi: 10.1074/jbc.M607674200. [DOI] [PubMed] [Google Scholar]
- Guan KL. Broyles SS. Dixon JE. A Tyr/Ser protein phosphatase encoded by vaccinia virus. Nature. 1991;350:359–362. doi: 10.1038/350359a0. [DOI] [PubMed] [Google Scholar]
- Hahm B. Trifilo MJ. Zuniga EI. Oldstone MB. Viruses evade the immune system through type I interferon-mediated STAT2-dependent, but STAT1-independent, signaling. Immunity. 2005;22:247–257. doi: 10.1016/j.immuni.2005.01.005. [DOI] [PubMed] [Google Scholar]
- Horvath CM. Weapons of STAT destruction. Interferon evasion by paramyxovirus V protein. Eur J Biochem. 2004;271:4621–4628. doi: 10.1111/j.1432-1033.2004.04425.x. [DOI] [PubMed] [Google Scholar]
- Huang Z. Coleman JM. Su Y. Mann M. Ryan J. Shultz LD. Huang H. SHP-1 regulates STAT6 phosphorylation and IL-4-mediated function in a cell type-specific manner. Cytokine. 2005;29:118–124. doi: 10.1016/j.cyto.2004.10.004. [DOI] [PubMed] [Google Scholar]
- Kato A. Kiyotani K. Kubota T. Yoshida T. Tashiro M. Nagai Y. Importance of the anti-interferon capacity of Sendai virus C protein for pathogenicity in mice. J Virol. 2007;81:3264–3271. doi: 10.1128/JVI.02590-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ke Y. Lesperance J. Zhang EE. Bard-Chapeau EA. Oshima RG. Muller WJ. Feng GS. Conditional deletion of Shp2 in the mammary gland leads to impaired lobulo-alveolar outgrowth and attenuated Stat5 activation. J Biol Chem. 2006;2281:34374–34380. doi: 10.1074/jbc.M607325200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komatsu T. Takeuchi K. Yokoo J. Gotoh B. Sendai virus C protein impairs both phosphorylation and dephosphorylation processes of Stat1. FEBS Lett. 2002;511:139–144. doi: 10.1016/s0014-5793(01)03301-4. [DOI] [PubMed] [Google Scholar]
- Kubota T. Yokosawa N. Yokota S. Fujii N. Association of mumps virus V protein with RACK1 results in dissociation of STAT-1 from the alpha interferon receptor complex. J Virol. 2002;76:12676–12682. doi: 10.1128/JVI.76.24.12676-12682.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langland JO. Jacobs BL. Inhibition of PKR by vaccinia virus: role of the N- and C-terminal domains of E3L. Virology. 2004;324:419–429. doi: 10.1016/j.virol.2004.03.012. [DOI] [PubMed] [Google Scholar]
- Li P. Wang N. Zhou D. Yee CS. Chang CH. Brutkiewicz RR. Blum JS. Disruption of MHC class II-restricted antigen presentation by vaccinia virus. J Immunol. 2005;175:6481–6488. doi: 10.4049/jimmunol.175.10.6481. [DOI] [PubMed] [Google Scholar]
- Lin RJ. Chang BL. Yu HP. Liao CL. Lin YL. Blocking of interferon-induced Jak-Stat signaling by Japanese encephalitis virus NS5 through a protein tyrosine phosphatase-mediated mechanism. J Virol. 2006;80:5908–5918. doi: 10.1128/JVI.02714-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu KL. Traktman P. The dual-specificity phosphatase encoded by vaccinia virus, VH1, is essential for viral transcription in vivo and in vitro. J Virol. 1995;69:7823–7834. doi: 10.1128/jvi.69.12.7823-7834.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo MS. Brazas RM. Holtzman MJ. Respiratory syncytial virus nonstructural proteins NS1 and NS2 mediate inhibition of Stat2 expression and alpha/beta interferon responsiveness. J Virol. 2005;79:9315–9319. doi: 10.1128/JVI.79.14.9315-9319.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu X. Chen J. Sasmono RT. Hsi ED. Sarosiek KA. Tiganis T. Lossos IS. T-cell protein tyrosine phosphatase, distinctively expressed in activated-B-cell-like diffuse large B-cell lymphomas, is the nuclear phosphatase of STAT6. Mol Cell Biol. 2007;27:2166–2179. doi: 10.1128/MCB.01234-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathur AN. Chang HC. Zisoulis DG. Kapur R. Belladonna ML. Kansas GS. Kaplan MH. T-bet is a critical determinant in the instability of the IL-17-secreting T-helper phenotype. Blood. 2006;108:1595–1601. doi: 10.1182/blood-2006-04-015016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meraz MA. White JM. Sheehan KC. Bach EA. Rodig SJ. Dighe AS. Kaplan DH. Riley JK. Greenlund AC. Campbell D. Carver-Moore K. DuBois RN. Clark R. Aguet M. Schreiber RD. Targeted disruption of the Stat1 gene in mice reveals unexpected physiologic specificity in the JAK-STAT signaling pathway. Cell. 1996;84:431–442. doi: 10.1016/s0092-8674(00)81288-x. [DOI] [PubMed] [Google Scholar]
- Mercer J. Traktman P. Investigation of structural and functional motifs within the vaccinia virus A14 phosphoprotein, an essential component of the virion membrane. J Virol. 2003;77:8857–8871. doi: 10.1128/JVI.77.16.8857-8871.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Najarro P. Traktman P. Lewis JA. Vaccinia virus blocks gamma interferon signal transduction: viral VH1 phosphatase reverses Stat1 activation. J Virol. 2001;75:3185–3196. doi: 10.1128/JVI.75.7.3185-3196.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakahira M. Tanaka T. Robson BE. Mizgerd JP. Grusby MJ. Regulation of signal transducer and activator of transcription signaling by the tyrosine phosphatase PTP-BL. Immunity. 2007;26:163–176. doi: 10.1016/j.immuni.2007.01.010. [DOI] [PubMed] [Google Scholar]
- Ousman SS. Wang J. Campbell IL. Differential regulation of interferon regulatory factor (IRF)-7 and IRF-9 gene expression in the central nervous system during viral infection. J Virol. 2005;79:7514–7527. doi: 10.1128/JVI.79.12.7514-7527.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palosaari H. Parisien JP. Rodriguez JJ. Ulane CM. Horvath CM. STAT protein interference and suppression of cytokine signal transduction by measles virus V protein. J Virol. 2003;77:7635–7644. doi: 10.1128/JVI.77.13.7635-7644.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parisien JP. Lau JF. Rodriguez JJ. Ulane CM. Horvath CM. Selective STAT protein degradation induced by paramyxoviruses requires both STAT1 and STAT2 but is independent of alpha/beta interferon signal transduction. J Virol. 2002;76:4190–4198. doi: 10.1128/JVI.76.9.4190-4198.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Platanias LC. Mechanisms of type-I- and type-II-interferon-mediated signalling. Nat Rev Immunol. 2005;5:375–386. doi: 10.1038/nri1604. [DOI] [PubMed] [Google Scholar]
- Qing Y. Stark GR. Alternative activation of STAT1 and STAT3 in response to interferon-gamma. J Biol Chem. 2004;279:41679–41685. doi: 10.1074/jbc.M406413200. [DOI] [PubMed] [Google Scholar]
- Ramana CV. Gil MP. Schreiber RD. Stark GR. Stat1-dependent and -independent pathways in IFN-gamma-dependent signaling. Trends Immunol. 2002;23:96–101. doi: 10.1016/s1471-4906(01)02118-4. [DOI] [PubMed] [Google Scholar]
- Ramana CV. Kumar A. Enelow R. Stat1-independent induction of SOCS-3 by interferon-gamma is mediated by sustained activation of Stat3 in mouse embryonic fibroblasts. Biochem Biophys Res Commun. 2005;327:727–733. doi: 10.1016/j.bbrc.2004.12.074. [DOI] [PubMed] [Google Scholar]
- Renukaradhya GJ. Webb TJ. Khan MA. Lin YL. Du W. Gervay-Hague J. Brutkiewicz RR. Virus-induced inhibition of CD1d1-mediated antigen presentation: reciprocal regulation by p38 and ERK. J Immunol. 2005;175:4301–4308. doi: 10.4049/jimmunol.175.7.4301. [DOI] [PubMed] [Google Scholar]
- Rosengard AM. Liu Y. Nie Z. Jimenez R. Variola virus immune evasion design: expression of a highly efficient inhibitor of human complement. Proc Nat1 Acad Sci USA. 2002;99:8808–8813. doi: 10.1073/pnas.112220499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt NW. Thieu VT. Mann BA. Ahyi AN. Kaplan MH. Bruton's tyrosine kinase is required for TLR-induced IL-10 production. J Immunol. 2006;177:7203–7210. doi: 10.4049/jimmunol.177.10.7203. [DOI] [PubMed] [Google Scholar]
- Schroder K. Hertzog PJ. Ravasi T. Hume DA. Interferon-gamma: an overview of signals, mechanisms and functions. J Leukoc Biol. 2004;75:163–189. doi: 10.1189/jlb.0603252. [DOI] [PubMed] [Google Scholar]
- Sekine Y. Tsuji S. Ikeda O. Sato N. Aoki N. Aoyama K. Sugiyama K. Matsuda T. Regulation of STAT3-mediated signaling by LMW-DSP2. Oncogene. 2006;25:5801–5806. doi: 10.1038/sj.onc.1209578. [DOI] [PubMed] [Google Scholar]
- Smith GL. Symons JA. Khanna A. Vanderplasschen A. Alcami A. Vaccinia virus immune evasion. Immunol Rev. 1997;159:137–154. doi: 10.1111/j.1600-065x.1997.tb01012.x. [DOI] [PubMed] [Google Scholar]
- Symons JA. Alcami A. Smith GL. Vaccinia virus encodes a soluble type I interferon receptor of novel structure and broad species specificity. Cell. 1995;81:551–560. doi: 10.1016/0092-8674(95)90076-4. [DOI] [PubMed] [Google Scholar]
- ten Hoeve J. de Jesus Ibarra-Sanchez M. Fu Y. Zhu W. Tremblay M. David M. Shuai K. Identification of a nuclear Stat1 protein tyrosine phosphatase. Mol Cell Biol. 2002;22:5662–5668. doi: 10.1128/MCB.22.16.5662-5668.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Townsend A. Bastin J. Gould K. Brownlee G. Andrew M. Coupar B. Boyle D. Chan S. Smith G. Defective presentation to class I-restricted cytotoxic T lymphocytes in vaccinia-infected cells is overcome by enhanced degradation of antigen. J Exp Med. 1988;168:1211–1224. doi: 10.1084/jem.168.4.1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traktman P. Liu K. DeMasi J. Rollins R. Jesty S. Unger B. Elucidating the essential role of the A14 phosphoprotein in vaccinia virus morphogenesis: construction and characterization of a tetracycline-inducible recombinant. J Virol. 2000;74:3682–3695. doi: 10.1128/jvi.74.8.3682-3695.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tropea JE. Phan J. Waugh DS. Overproduction, purification, and biochemical characterization of the dual specificity H1 protein phosphatase encoded by variola major virus. Protein Expr Purif. 2006;50:31–36. doi: 10.1016/j.pep.2006.05.007. [DOI] [PubMed] [Google Scholar]
- You M. Yu DH. Feng GS. Shp-2 tyrosine phosphatase functions as a negative regulator of the interferon-stimulated Jak/STAT pathway. Mol Cell Biol. 1999;19:2416–2424. doi: 10.1128/mcb.19.3.2416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X. Guo A. Yu J. Possemato A. Chen Y. Zheng W. Polakiewicz RD. Kinzler KW. Vogelstein B. Velculescu VE. Wang ZJ. Identification of STAT3 as a substrate of receptor protein tyrosine phosphatase T. Proc Natl Acad Sci USA. 2007;104:4060–4064. doi: 10.1073/pnas.0611665104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann A. Trilling M. Wagner M. Wilborn M. Bubic I. Jonjic S. Koszinowski U. Hengel H. A cytomegaloviral protein reveals a dual role for STAT2 in IFN-γ signaling and antiviral responses. J Exp Med. 2005;201:1543–1553. doi: 10.1084/jem.20041401. [DOI] [PMC free article] [PubMed] [Google Scholar]