Abstract
Objective
To incorporate structural biology, enzyme kinetics, and visualization of protein structures in a medicinal chemistry course to teach fundamental concepts of drug design and principles of drug action.
Design
Pedagogy for active learning was incorporated via hands-on experience with visualization software for drug-receptor interactions and concurrent laboratory sessions. Learning methods included use of clicker technology, in-class assignments, and analogies.
Assessment
Quizzes and tests that included multiple-choice and open-ended items based on Bloom's taxonomy were used to assess learning. Student feedback, classroom exercises, and tests were used to assess teaching methods and effectiveness in meeting learning outcomes.
Conclusion
The addition of active-learning activities increased students' understanding of fundamental medicinal chemistry concepts such as ionization state of molecules, enzyme kinetics, and the significance of protein structure in drug design.
Keywords: drug-receptor interactions, enzyme kinetics, medicinal chemistry, active learning
INTRODUCTION
A molecular level understanding of how a drug works and what it targets are important factors in comprehending the beneficial effects of a drug. Knowledge of how the drug interacts with its target also is important in deciding the therapeutic approach for a patient. The dosage of a drug and its in vivo efficacy are influenced by how strongly the drug binds to a receptor,1 its solubility, and other biopharmaceutical properties. These properties depend on the structure of the drug and its interaction with its receptor. The main molecular targets of drugs are proteins. Traditionally, medicinal chemistry is taught based on the knowledge of the physicochemical properties of the drug itself, and the interaction of drugs with proteins/enzymes is discussed in terms of enzyme kinetics. Many crystal structures of drug/drug-like molecules as their complexes with proteins have been elucidated. These crystal structures give insight into properties of drugs and their interactions with receptors in atomic detail. The affinity of a drug for its receptor and possible modifications of the structures for improved affinity toward the receptor are explained in terms of 3-dimensional (3D) structures. Apart from small molecule drugs, many bio-drugs, such as protein and peptide drugs, are now on the market for therapeutic purposes. At present more than 130 such bio-drugs have been approved by the Food and Drug Administration (FDA) and many more are in development.2
In the mid-1980s and 1990s, the drug discovery approach was changed from a traditional approach to a target-based approach.3 In the traditional approach, possible therapeutic agents are first identified by randomly screening tens of thousands of natural products and synthetic chemicals for biological activity. When a lead compound with potential therapeutic value is found, structural modifications are done, and structure-activity correlations are characterized. However, in rational approach to drug design, the cause of the disease and the enzyme receptor or other macromolecular target responsible for biochemical dysregulation are identified first. Information about the structure of a drug receptor, often along with the structure of its endogenous ligand(s), is studied to identify or design new drug molecules. This new approach requires detailed knowledge of drug targets/receptors and their interactions with drugs. The effectiveness of this approach will be seen in the next 5 to 10 years and seems destined to be compelling, dependent only on the availability of clinical development resources.
With the rapidly increasing emphasis on the discovery and development of bio-drugs and rational drug design, detailed protein structure and function play a major role in drug design and discovery. Protein structures are now considered in the drug discovery process in such detail that the pharmacodynamics properties of a new molecular entity are purposefully designed into its structure. Teaching and understanding of molecular mechanisms in terms of detailed protein structure are largely limited to structural biologists and protein chemists at higher education levels, mainly doctoral programs.
Major changes are occurring in pharmacy curricula in response to these scientific advances and their current and anticipated impact on pharmacotherapy decisions, resulting in a substantial paradigm shift in the way we teach/study drug-target interactions, and the way in which the dynamics of these interactions dictate pharmacotherapeutic choices. Combined knowledge of the drug and the 3-dimensional (3D) structures of the proteins (enzymes, receptors, transporters, biotransforming enzymes), including their chemistry, biochemistry, and structural biology, are needed to fully describe the properties of a drug. Achieving such a significant paradigm shift with respect to content and concepts requires that we begin to adopt new instructional methods in the classroom and teaching laboratory.4,5 According to 2007 Accreditation Council for Pharmacy Education (ACPE) guidelines, biomedical and pharmaceutical sciences should have a clear emphasis in the pharmacy curriculum, and a foundation of this knowledge is important for students to become competent pharmacy practitioners.
This report describes a strategy for teaching in-depth knowledge of structure, intermolecular forces, enzyme function, and kinetics to doctor of pharmacy (PharmD) students. The objective of this particular teaching method is to incorporate structural biology, enzyme kinetics, and visualization of protein structures in a medicinal chemistry course to teach fundamental concepts of drug design and principles of drug action. The topics are taught with the aim of meeting specific outcomes and competencies with regard to the structural and functional characteristics of important molecular targets of drug action as these relate to optimizing pharmacotherapy decisions. On the basis of this knowledge, students should be able to understand the clinical significance of the design and development of drugs. Specifically, the 3 expected pharmacy-related outcomes are the ability to:
Describe the structural and functional characteristics of important molecular targets of drug action: receptors, enzymes, nucleic acids, excitable membranes, transporters, and other biomolecules.
Given the chemical structure of a drug molecule, describe and apply to the solution of therapeutic problems its acid/base properties, water solubility, chemical stability, and stereochemical properties.
Describe the types and bonding strengths of intermolecular forces that occur between a chemical compound and its receptor or enzyme binding site.
DESIGN
The methods described in the article were used in the initial medicinal chemistry course for PharmD students at the University of Louisiana at Monroe from 2006-2009. During 2006-2008, the course was taught as an introductory biochemistry course, with emphasis on the structure of biomolecules and enzyme kinetics. With changes in the curriculum in the fall of 2009, principles of drug action and metabolism were incorporated in an integrated style, along with the structure and function of biomolecules. The methods described herein focus only on teaching the structure of proteins, ligand-receptor interactions, enzyme kinetics, and therapeutically relevant drug design in the course Medicinal Chemistry I (2006–2008) or Principles of Drug Action I (2009). Medicinal Chemistry I was a 3-credit course that met 3 hours per week; Principles of Drug Action I was a 5-credit semester-long course that met 5 hours per week. Students had completed at least 2 years (3 years for the 2009 cohort) of undergraduate courses before entering the PharmD program. All students were required to bring a laptop with access to a wireless network connection to class. Along with this, a module was incorporated into an integrated laboratory course to illustrate, reinforce, and expand the concepts learned in the classroom. For laboratory exercises and in the classroom, PyMol software (DeLano Scientific LLC, San Carlos, CA) enabled the visualization of the 3D structure of molecules.6 The instructional strategies used in the course included lectures, visualization of structures using computer graphics, interactive sessions using clickers, discussion sessions, and laboratory sessions.
The topics for the first part of the course included intermolecular forces; identification of functional groups and structural moieties, including the component amino acids; and ionization states. The topics for the second part of the course included the structure of proteins, function of proteins, allosteric proteins, and details of the structure and function of hemoglobin and myoglobin. Topics for the third part were enzymes, enzyme kinetics, allosteric enzymes, and enzyme-based drug design. In this segment, key concepts of association, dissociation, and kon, koff, Ki, and Kd and their importance in drug action and relationships to intermolecular forces were presented and reinforced. Laboratory sessions were conducted after the topics were covered in the classroom. For laboratory sessions, the students were assigned to groups of 20 to 25 students. In the laboratory sessions, students visualized protein structures in detail with an emphasis on structure-function relationships, and intra/intermolecular forces, and solved graphical problems in enzyme kinetics with the use of data from enzyme kinetics experiments.
Evaluations consisted of 3 or 4 tests, 6 quizzes, and some laboratory exercises, along with class work. After the completion of the lecture/learning of each topic in the classroom, “reflection” or review sessions were conducted. Students were given time to reflect on the topics learned and time to correct their misconceptions through open discussion. Blackboard and Moodle Web-based learning technology tools were used for the course material and communication with students. Practice questions were posted on the Web for students to test their understanding. During the course schedule, some key concepts were reiterated in the class so that students would retain this knowledge and adjust their depth and breadth of understanding as the course topics progressed. A classroom response system (clickers) was used to make the class sessions interactive.7 Tests consisted of questions ranging in format from true/false and multiple choice to open-ended questions, based on Bloom's taxonomy.8
Ligand-Receptor Interactions
Intermolecular forces, ionization state, and 3D structure of proteins.
The nature of the interactions of a drug with a biological macromolecule under physiological conditions ultimately impacts drug dosage regimens.1,9-11 Within the first parts of the course, the concepts of noncovalent interactions in the molecules and their distance-dependences were taught and subsequently refreshed regularly. Most students with 2 years of undergraduate courses have at least superficial knowledge of these fundamental concepts; however, when they are faced with practical problems, many students fail to recognize, or have difficulty in distinguishing, some of the interactions. Different functional groups of the drugs and possible interactions (hydrogen bonding, charge-transfer, hydrophobic, etc) were illustrated in the classroom, and students were asked to identify the interactions involved. Take-home exercises were given to provide in-depth knowledge of these interactions. The principles of these interactions were demonstrated in the structure of proteins.
The ionization states of molecules related to pKa values were taught on the basis of “learn until you get it.”12 Students were given structures of different amino acids and had 5 minutes of class time to write the ionization states of the amino acids at different pH values. When studying the allosteric effect of hemoglobin binding to oxygen, students were given the structures of His146 and Asp94. They were asked the question, “how is the hydrogen bonding affected at high pH?” Students were asked to write down the ionization state of histidine at different pH values and think critically about how a change in pH can lead to loss of a proton and consequent destabilization of the protein. After this exercise, students were asked to view the 3D structure of hemoglobin.
Kd, Ka, kon and koff.
Students quite often struggle to understand the relationship between Kd, kon, koff, and affinity values. On rates and off rates have rapidly gained a fundamental role in the drug creation process, as well as in the better understanding of the pharmacodynamics of already marketed drugs for the purpose of optimization of treatment. For pharmacy students, this means that gaining an understanding of the relationship of these parameters to the dosing regime of a drug is essential. One example that can be used to correlate Kd values, pharmacodynamics, and in vivo efficicacy is provided by a pair of angiotensin II antagonists. Losartan and candesartan are angiotensin II type I receptor (ATR1) antagonists which are used to control blood pressure.1,13,14 In vitro activity suggests that candesartan binds to ATR1 receptor with a Kd value of 7 nM, whereas losartan binds with a Kd value of 350 nM. Candesartan is known to have significantly longer ligand:receptor dissociation half-life compared to losartan. Clinical efficacy of losartan and candesartan in hypertensive patients suggested that candesartan produced significant reduction in blood pressure at lower doses (16 mg per day) compared to losartan (100 mg per day). Thus, the much smaller Kd value with which candesartan binds its receptor and longer residence time (dissociative half-life of greater than 60 minutes for candesartan compared to losartan with 5 minutes) at the receptor can be directly correlated with clinical outcomes of a drug molecule. The development of losartan and candesartan from imidazole-5-acetic acid analogues and S-8308 also illustrates how functional groups were modified in the molecule to fit into the enzyme cavity and to improve the lipophilicity and oral absorption, which lead to improved in vivo activity.13-15
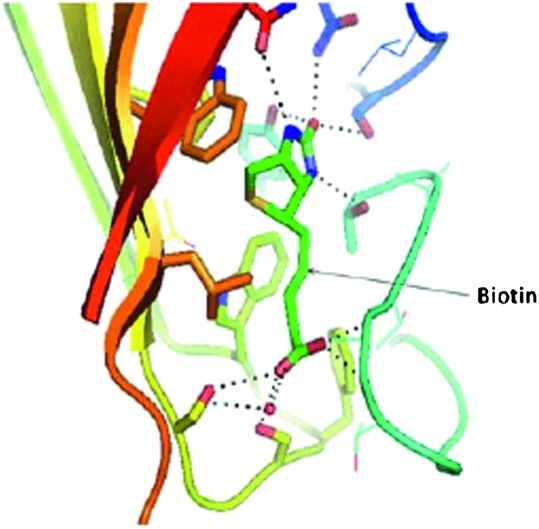
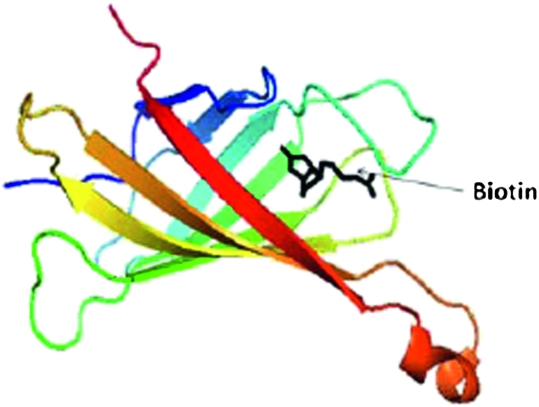
In teaching these concepts, basic chemical kinetics were reviewed, followed by a clear definition of kon and koff, and then illustrations with examples of numerical values. Students were shown a table of mean lifetimes of different protein-ligand complexes with different Kd and kon values from the published literature.9 Using a classroom response system, they were asked questions regarding which one would have stronger binding. To reinforce this concept, students were also shown decay curves of protein-ligand complexes, how the protein-ligand complex breaks down with time, and how that behavior is related to the Kd value. The concept that given similar on-rate values, the longer a ligand stays1 with a protein the smaller is the Kd value (ie, higher affinity) was delivered with a question-answer session using clickers. The concept was also taught with a more tangible analogy of ballroom dancers, where each pair will dance slowly or fast and change partners fast (analogy to off-rates), in which case the mean lifetime of dancing together, and correspondingly the affinity, is less. Finally, a table of Kd values of ligand-receptor complexes from the literature was provided (ranging from 10−2 to 10−10 M), and students were asked to judge which one would be the best ligand for a protein to be a drug candidate. At this stage, students were asked to visualize protein-ligand complexes using Pymol, and carefully scrutinize such noncovalent interactions as hydrogen bonding and hydrophobic interactions, and conclude that the number and strength of such interactions governs the Kd value of the ligand-receptor interaction (along with desolvation and entropy aspects discussed in the course). Relationships between Kd, 3D structure, and intermolecular forces were demonstrated in the classroom with the example of the avidin-biotin complex. Students were provided with a table of different Kd values ranging from 10−3 to 10−11 M from the literature and then shown the Kd value for the biotin-avidin interaction, 10−15 M. The 3D structure of biotin-avidin was shown (protein data bank [PDB] ID, 1AVD). After visualizing the 3D structure of the biotin-avidin complex (Figures 1 and 2), and identifying the hydrogen bonding and hydrophobic interactions between ligand and receptor, the students noted the stability of biotin in the cavity created by the amino acids in avidin. The students were again redirected to the role that affinity contributes in dictating the potency of the drug candidate, and thus, the dosage of the drug necessary for therapy. The noncovalent binding forces between protein-ligand complexes are strong enough to hold the drug for a certain period of time and permit it to have a desired physiological effect, but weak enough to allow the drug to leave the receptor site after it exerts its effect.
Figure 1.
Biotin binding cavity in avidin protein demonstrating the hydrogen bonding and hydrophobic interactions. Dotted lines show hydrogen bonding between biotin and amino acid residues of the protein. Hydrophobic interactions occur between aromatic amino acids and biotin aliphatic groups.
Figure 2.
Biotin in the avidin protein cavity. Non-covalent interactions described in Figure 1 and the cavity shown in this figure ensure the rate of kon for biotin, which leads to high binding affinity. Students use the 3D model in the software and rotate it to identify the interactions.
Michaelis-Menten Kinetics.
After teaching the class on the basic concepts concerning enzymes and enzyme kinetics, the Michaelis-Menten equation was introduced. The following points were clarified with the example of a marble experiment described by Runge et al16: physical meanings of Km and Vmax, turnover number, and obtaining Km and Vmax values using a reciprocal plot. In the marble experiment, 2 plastic containers (1 and 2) are used. A student (acting as an enzyme with eyes closed or blindfolded) transfers the marble (substrate) from plastic container 1 to plastic container 2 (the product). The catalytic event is the transfer of marbles. If there are fewer marbles in container 1 (low substrate concentration, S), the student has to find them first and transfer them to container 2. If there are too many marbles (high S) in container 1, the student can find them easily; however, the rate of transfer is limited by how fast his hands can move to transfer the marbles.
The physical meaning of Km, where it represents 50% of the enzymes occupied by substrate, was clarified. One of the important concepts that was clarified by the marble experiment was why, at higher concentrations of substrate, an enzyme reaches saturation. After demonstrating the marble experiment,16 students identified that if the number of enzymes remains constant and, the substrate concentration is high, the velocity of the reaction must asymptotically approach a maximum. Sets of enzymes with different turnover numbers and Km values were provided to students in the classroom, with opportunities for them to apply their knowledge of Km values and turnover numbers to solidify their understanding.
Allosteric Effect.
In available education materials, the allosteric effect of proteins is usually shown in 2-dimensional (2D) diagrams and plots and described in written text. While most students grasp the concept of the allosteric effect to some extent, illustrating it with 3D structures of proteins added another dimension to students' understanding. Examples provided were hemoglobin and aspartate transcarbamoylase structures. With hemoglobin, the manner in which the α-amino groups at the amino termini and the protonation state of the histidine 146 (His146) residue of the β-chain affect the binding of oxygen to a heme group that is distant from these sites was examined in depth. Details of stabilization by hydrogen bonding and the basis by which protein subunits turn around to cause the allosteric effect were presented.
Laboratory Session
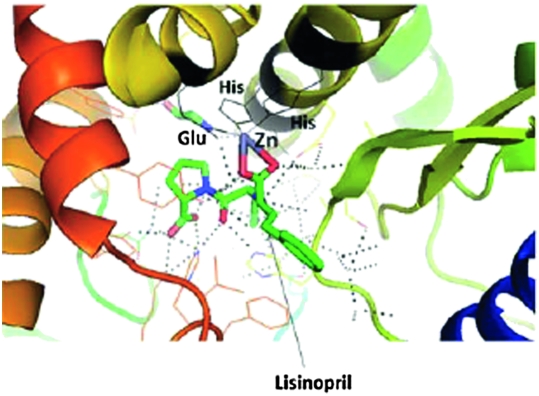
During the years 2006 to 2008, problems based on the topics discussed above were given as assignments, prompting students to use the PyMol software and Microsoft Excel to solve the problems independently. In the fall of 2009, these exercises were incorporated into a session in a parallel laboratory class, one of a longitudinal sequence of courses designed to reinforce students' knowledge, skills, and attitudes necessary for current and future pharmacy practice. In this course, 3 laboratory sessions were conducted on the topics of drug-receptor interactions and enzyme kinetics. In the first session, students practiced recognizing the protein secondary structure, hydrogen bonding, and hydrophobic interactions. In the second session (Appendix 1), students studied 4 examples to understand drug-receptor interactions: (1) HIV protease in complex with an inhibitor; (2) angiotensin-converting enzyme (ACE) bound with an inhibitor; (3) cyclin-dependent kinase with an inhibitor; and (4) human epidermal growth factor receptor-2 (HER-2) with each of the antibody complex drugs trastuzumab and pertuzumab. Students were asked to identify the cofactor zinc ion in the ACE-inhibitor complex and closely examine its interactions with the ligand and protein. After visualizing and studying the structure (Figure 3), students had to answer the questions and complete the assignment. During the first half of the laboratory session, students worked independently. During the second half, discussion among the students was permitted. In the third session (Appendix 2) of the laboratory, students were given 4 problems on competitive and noncompetitive enzyme inhibitors, drawing examples from prostaglandin endoperoxide synthase and hexokinase. The students were asked to plot the data from enzyme kinetics experiments, calculate Km and Vmax values using saturation curves and Hill plots, and state their conclusions with respect to type of inhibition. Excel software was used for plotting the graphs.
Figure 3.
Binding of the drug lisinopril in the catalytic pocket of angiotensin-converting enzyme. Students visualize the enzyme-drug interaction and the importance of the imbedded zinc ion (cofactor in the enzyme). Zinc is held in the enzyme by two histidine residues and a glutamate residue. The Zinc ion also makes contact with the drug lisinopril via its corboxylate moeity. Dotted lines represent the complex hydrogen bonding network between protein and the drug. The figure was created using PyMol software.
ASSESSMENT
Course evaluation was conducted by the university or college and separately by the instructor of the course, providing qualitative and quantitative (Table 1) information. The number of students enrolled in the course varied from 104 to 69 students from fall 2006 to 2009. Based on the qualitative comments of the students, they learned and retained knowledge better when concepts were explained using real-life analogies.
Table 1.
Student Evaluation and Feedback for the Course Medicinal Chemistry I and Principles of Drug Action I
a Based on a scale of 1 to 5 on which 1 =poor and 5 = excellent.
b Response rate = 65%
c Response rate = 82%
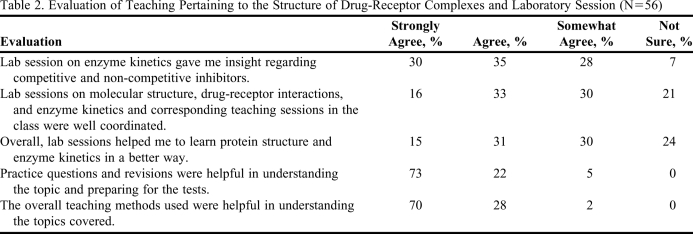
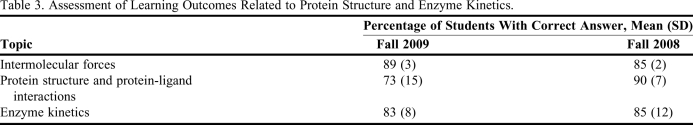
Evaluation of student understanding was accomplished formally (test questions) and informally in class using “clickers.” When students were shown the avidin-biotin structure and then asked to answer clicker questions, 100% of the students participating answered that the high affinity interaction was due to intermolecular forces. When they were asked whether they understood the binding of biotin to avidin better after visualizing the 3D structure, 45% responded yes/they did and only 8% answered that they understood the concept before visualizing the 3D structure. Student feedback on the materials taught is presented in Table 2. Learning outcomes were also assessed using the questions answered in the tests and final examinations (Table 3).17 For the sake of analysis, we divided the questions into 3 categories: questions on (1) intermolecular forces, (2) protein structure and protein-ligand interactions, and (3) enzyme kinetics. From these 3 categories, questions that required a greater depth of thinking, analysis, and synthesis also were analyzed separately. To gain insight into how the laboratory sessions helped the students to understand the concepts, a survey was conducted in fall 2009 (Table 2). Students were asked to evaluate the laboratory sessions and compare their understanding to the classroom sessions. Overall, students understood the concepts taught.
Table 2.
Evaluation of Teaching Pertaining to the Structure of Drug-Receptor Complexes and Laboratory Session (N=56)
Table 3.
Assessment of Learning Outcomes Related to Protein Structure and Enzyme Kinetics.
Score obtained for each year was an average of tests and final examination scores. Results were obtained by analyzing the particular questions answered in the tests and final exam for the year 2008 (79 students) and 2009 (68 students). Students who did not attempt the questions were considered as having given as incorrect answer.
DISCUSSION
One of the challenges facing pharmacy education is to make sure that the medicinal chemistry course taught in the first year of the PharmD curriculum gives a strong foundation on which students can build clinically relevant pharmacy practice experience.18,19 Understanding the inhibition of enzymes or protein-protein interactions benefits greatly from a working knowledge of structure and function. Subtle differences in the conformation of proteins is becoming increasingly important as details of the structural aspects of different subtypes of G-protein coupled receptors and their mechanisms of actions are elucidated. Binding of different small molecules to the same drug target can have different effects (agonist, inverse agonist, partial agonist, antagonist),11 which can be explained only in terms of changes in the 3D structure of proteins.20 Students can gain understanding in this area through instructor- and self-guided visualizations of protein structures complexed with drug molecules, highlighting the importance of functional groups in the drug, their noncovalent interactions, and the relationships to affinity, biopharmaceutical properties, and dosage. Although a student may not be expected to gain confidence in applying this fundamental knowledge without repeated reinforcement via integration of the medicinal chemistry component with clinical pharmacy learning, the methods described in this article considerably enhance and enrich students' foundational education. When students enter their experiential year of the professional program, they may not have retained many of the specifics taught in first-year courses. However, when they encounter a drug that acts on an enzyme or a particular protein, or when they discuss the affinity of the drug and its solubility properties, the students should be able to recognize the importance of the structure of the drugs and its interaction with a receptor. If necessary, they can go back and look at the structural information from the experience gained in this course and comprehend its clinical relevance with medicinal chemistry (examples such as losartan and candesartan provided in this article). The visualization and laboratory experiences gained in this course should remind them that such tools are available to investigate therapeutic problems. Once students start such an investigation, their background knowledge of medicinal chemistry and enzyme kinetics will be refreshed. Evaluation of the laboratory session (Table 2) indicated that a majority of the students benefitted from the teaching methods described in this article. On a scale of 1-5 (1= disagree and 5 = strongly agree), nearly 60% of the students expressed that the laboratory session on enzyme kinetics provided them insight regarding competitive and noncompetitive inhibitors, and that laboratory sessions helped them to learn enzyme kinetics in a better way compared to classroom activities.
Interactive elements incorporated in the course, such as clicker technology and visualization software, discouraged passive learning and required students to become involved with the material. Students' assessment of the teaching methods used in the course (Table 1 and Table 2) suggested that the interactive sessions and laboratory experience helped the students to solidify their understanding. The laboratory sessions conducted helped the students to work independently as well as collaboratively.
Despite the significant enhancements to the course, a fraction of students (Table 3) still encountered difficulties when tested on the material. Most students seemed to understand the concepts; however, when multiple-choice questions with different possibilities were given, some students were unable to judge the best answer. When the questions needed critical thinking or in-depth analysis, many students struggled to answer. To encourage these students and to help them improve their learning process, question-answer sessions were conducted before each test. Although a majority of the students benefited from question-answer sessions, upon evaluation of answers from students for critical-thinking questions on tests, some students exhibited deficiencies. A drop in test performance was noted between 2007 and 2009, possibly due to the change in course curriculum (ie, the percentage of students correctly answering critical-thinking questions dropped from 40% to 30%). With the available data, it is difficult to (quantitatively) compare the old and new curriculum results at this juncture. In the future, to improve the critical-thinking ability of the students, they will be further challenged with critical-thinking questions in the classroom so that they are compelled to solve challenging problems.
A key challenge faced by professional pharmacy students is the diversity of the course content taught in the program: students must shift their thinking process greatly from topics ranging from basic chemistry to microbiology- and pharmacy-related content. Overall, students' performance, course ratings, and comments in informal discussions suggest that active learning using interactive sessions helped them gain understanding of drug-receptor interactions.
CONCLUSION
Obtaining a fundamental understanding of structural basis protein-ligand interactions in their first year is valuable for pharmacy students as they continue their training/education with subsequent courses in medicinal chemistry and principles of drug action. The teaching methods described here enable students to study medicinal chemistry with the help of visualization. The method becomes increasingly valuable for students as standard textbooks such as Foye's Principles of Medicinal Chemistry15 incorporate more information on drug-receptor structures and their interactions in terms of a 3D- rather than a 2D-description of structure.
ACKNOWLEDGMENTS
The author would like to thank Dr. Ronald Hill for his suggestions and Dr. Lesa Lawrence for helping with statistical analysis of teaching evaluations. Drs. Roxie Stewart, Anthony Walker, and Candace Chelette coordinated the laboratory sessions.
Appendix 1. Examples of exercises used in a laboratory session for drug-receptor interaction visualization.
In this exercise you will study the structure of drug receptor interactions using 4 examples.
1) HIV protease with its inhibitor
2) Angiotensin-converting enzyme (ACE) with its inhibitor
3) Cyclin-dependent kinase with its inhibitor
4) Human epidermal growth factor receptor-2 (HER-2) with antibody drugs Trastuzumab and Pertuzumab.
(only 2 exercises are given as examples in the article)
Exercise 1
Open 2UXZhivproteaseinh.pdb using PyMol. The structure of HIV protease with its inhibitor will be shown in stick fashion. Display the ligand site and observe the structure (make the background white and save the image file as .png and insert into a Word file). How many hydrogen bonds are present between the inhibitor and HIV protease with water molecules? Use the “reset”option and look at the entire structure of the complex. Is there any symmetry in the molecule? If so, what is the symmetry in the molecule? Is there any symmetry in the inhibitor structure (drug molecule)? What is the secondary structure of the protein HIV protease? Describe the importance of amino acid residues in the protein and functional groups in the inhibitor in terms of intermolecular forces.
Exercise 2
Open the structure 1O86acelisinorpil.pdb. This is the structure of ACE with its inhibitor (drug) lisinorpil. Display the ligand site. What metal atom is present in the ligand binding site? Identify the amino acids from ACE that interact with the metal atom in the active site. What is the importance of this particular amino acid in binding to the metal ion? Compare the secondary structures of HIV protease and ACE. Describe the importance of amino acid residues in the protein and functional groups in the inhibitor in terms of intermolecular forces.
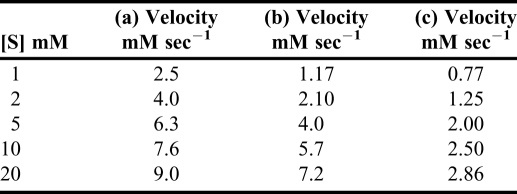
Appendix 2. Example of an exercise related to enzyme kinetics and drug design to understand the enzyme kinetics using numerical data from research papers.
Plot the data from enzyme kinetics experiments and calculate Km and Vmax values using saturation curves and Hill plots. Plot the data obtained in the presence of inhibitor and give a conclusion about the type of inhibitor. Use Excel software.
The following table shows the rate at which a given substrate enters an enzymatic reaction (a) in the absence of any inhibitor, and (b) and (c) in the presence of a constant amount, respectively, of each of 2 inhibitors. First plot the data directly, v against [S]. Also, plot the data using the Lineweaver and Burke method. Label the lines in the plot for competitive and non-competitive inhibitors.
REFERENCES
- 1.Copeland RA, Pompliano DL, Meek TD. Drug-target residence time and its implications for lead optimization. Nat Rev Drug Discov. 2006;5(9):730–739. doi: 10.1038/nrd2082. [DOI] [PubMed] [Google Scholar]
- 2.Leader B, Baca QJ, Golan DE. Protein therapeutics: a summary and pharmacological classification. Nat Rev Drug Discov. 2008;7(1):21–39. doi: 10.1038/nrd2399. [DOI] [PubMed] [Google Scholar]
- 3.Brown D. Unfinished business: target-based drug discovery. Drug Discov Today. 2007;12:1007–1012. doi: 10.1016/j.drudis.2007.10.017. [DOI] [PubMed] [Google Scholar]
- 4.Dee FL. Creating Significant Learning Experiences: An Integrated Approach to Designing College Courses. San Francisco, CA: Jossey-Bass; 2003. [Google Scholar]
- 5.Pan D. Learning to Teach, Teaching to Learn. 4th ed. Singapore: Continental Press; 2001. [Google Scholar]
- 6. DeLano WL. The PyMOL Molecular Graphics System. San Carlos, CA: DeLano Scientific; 2002. http://www.pymol.org. September 16, 2010.
- 7.Duncan D. Clickers in the Classroom: How to Enhance Science Teaching Using Classroom Response Systems. San Francisco, CA: Pearson; 2005. [Google Scholar]
- 8.Bloom BS, editor. Taxonomy of Educational Objectives. The Classification of Educational Goals. Handbook I: Cognitive Domain. New York: McKay; 1956. [Google Scholar]
- 9.Corzo J. Time, the forgotten dimension of ligand binding teaching. Biochem Mol Biol Educ. 2006;34(6):413–416. doi: 10.1002/bmb.2006.494034062678. [DOI] [PubMed] [Google Scholar]
- 10.Cortes A, Cascante M, Cardenas ML, Bowden AC. Relationship between inhibition constants, inhibitor concentration for 50% inhibition and types of inhibition: new ways of analyzing data. Biochem J. 2001;357:263–268. doi: 10.1042/0264-6021:3570263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Patrick GL. An Introduction to Medicinal Chemistry. 4th ed. New York: Oxford University Press; 2008. [Google Scholar]
- 12.Persky AM, Pollack GM. Using answer-until-correct examination to provide immediate feedback to students in a pharmacokinetic course. Am J Pharm Educ. 2008;72(4) doi: 10.5688/aj720483. Article 83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vanderheyden PML, Fierens FLP, Vauquelin G. Angiotensin II type I receptor antagonists: why do some of them produce insurmountable inhibition? Biochem Pharmacol. 2000;60(11):1557–1563. doi: 10.1016/s0006-2952(00)00388-9. [DOI] [PubMed] [Google Scholar]
- 14.Laciurciere Y, Asmar R. A comparison of the efficacy and duration of action of candersartan cilexetil and losartan as assessed by clinic and ambulatory blood pressure after a missed dose in truly hypertensive patients. Am J Hypertens. 1999;12(12):1181–1187. doi: 10.1016/s0895-7061(99)00142-9. [DOI] [PubMed] [Google Scholar]
- 15.Lemke TL, Williams DA, Roche VF, Zito SW. Foye's Principles of Medicinal Chemistry. 6th ed. Philadelphia, PA: Lippincott Williams & Wilkins; 2008. [Google Scholar]
- 16.Runge SW, Hill BJF, Moran WM. A simple classroom teaching technique to help students understand Michaelis-Menten kinetics. Life Sci Educ. 2006;5(4):348–352. doi: 10.1187/cbe.06-04-0160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Angelo TA, Cross KP. Classroom Assessment Techniques: A Handbook for College Teachers. San Francisco CA: Jossey-Bass; 1993. [Google Scholar]
- 18.Alsharif N, Galt KA, Mehana A, Chapman R, Ogunbadeniyi AM. Instructional model to teach clinically relevant medicinal chemistry. Am J Pharm Educ. 2006;70(4) doi: 10.5688/aj700491. Article 91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Roche VF. A receptor-grounded approach to teaching nonsteroidal anti-inflammatory drug chemistry and structure-activity relationship. Am J Pharm Educ. 2009;73(8) doi: 10.5688/aj7308143. Article 143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bokoch MP, Zou Y, Rasmussen SGF, et al. Ligand-specific regulation of the extracellular surface of a G-protein-coupled receptor. Nature. 2010;463(7277):108–112. doi: 10.1038/nature08650. [DOI] [PMC free article] [PubMed] [Google Scholar]