Abstract
The Hox genes encode transcription factors that play a key role in specifying body plans of metazoans. They are organized into clusters that contain up to 13 paralogue group members. The complex morphology of vertebrates has been attributed to the duplication of Hox clusters during vertebrate evolution. In contrast to the single Hox cluster in the amphioxus (Branchiostoma floridae), an invertebrate-chordate, mammals have four clusters containing 39 Hox genes. Ray-finned fishes (Actinopterygii) such as zebrafish and fugu possess more than four Hox clusters. The coelacanth occupies a basal phylogenetic position among lobe-finned fishes (Sarcopterygii), which gave rise to the tetrapod lineage. The lobe fins of sarcopterygians are considered to be the evolutionary precursors of tetrapod limbs. Thus, the characterization of Hox genes in the coelacanth should provide insights into the origin of tetrapod limbs. We have cloned the complete second exon of 33 Hox genes from the Indonesian coelacanth, Latimeria menadoensis, by extensive PCR survey and genome walking. Phylogenetic analysis shows that 32 of these genes have orthologs in the four mammalian HOX clusters, including three genes (HoxA6, D1, and D8) that are absent in ray-finned fishes. The remaining coelacanth gene is an ortholog of hoxc1 found in zebrafish but absent in mammals. Our results suggest that coelacanths have four Hox clusters bearing a gene complement more similar to mammals than to ray-finned fishes, but with an additional gene, HoxC1, which has been lost during the evolution of mammals from lobe-finned fishes.
The Hox genes are a large family of DNA-binding transcription factors that play a crucial role in defining body patterning of metazoans. They are organized into clusters, with each cluster containing up to 13 distinct genes. The order of genes within the cluster is highly conserved throughout evolution, suggesting a selective pressure on the whole cluster. While invertebrate chordates such as ascidians and amphioxus have a single Hox cluster (1, 2), vertebrate chordates have multiple Hox clusters. This finding has led to the suggestion that the complex and diverse morphology of vertebrates has been accomplished through an increase in the number of Hox clusters and Hox genes during vertebrate evolution.
All of the mammals investigated so far have four Hox clusters bearing 39 of the possible 52 genes (refs. 3 and 4; see Fig. 2). Analyses of Hox genes from Xenopus and chicken have suggested that the four-cluster Hox architecture of mammals is conserved in all tetrapods (5–7). In contrast to tetrapods, ray-finned fishes (Actinopterygii) have more than four Hox clusters. Studies in teleost fishes such as zebrafish (Danio rerio), fugu (Fugu rubripes), medaka, and an African cichlid fish have indicated the presence of up to seven clusters (8–11). This finding is postulated to be because of either a large-scale segmental duplication or a whole-genome duplication in the ray-finned fish lineage (8, 11, 12). The duplicated Hox clusters of teleosts, however, contain fewer Hox genes than the ancestral clusters, indicating a large-scale secondary gene loss after cluster duplication. Thus, the seven Hox clusters in zebrafish contain only 47 genes (see Fig. 2).
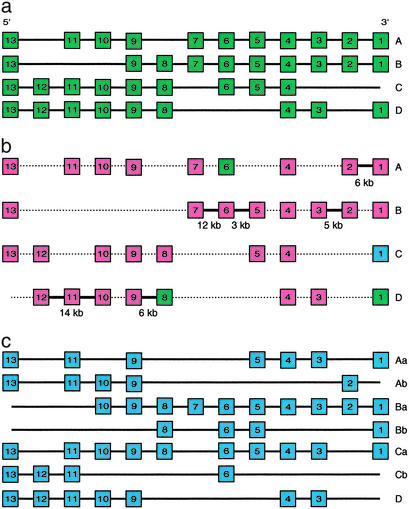
Figure 2.
Hox gene clusters in the mouse (a), Indonesian coelacanth (b), and zebrafish (c). Mouse and zebrafish Hox clusters are shown for comparison. Genes are shown as rectangles. Thick lines between the coelacanth Hox genes represent intergenic regions that we cloned. The sizes of the intergenic regions are shown below the thick line. Coelacanth Hox genes that are unique to the coelacanth and mammals are shown in green, and those unique to the coelacanth and zebrafish are shown in blue.
Multiple Hox clusters have also been identified in primitive jawless vertebrates such as lampreys. Characterization of Hox clusters from two species of lampreys have indicated that they have up to four Hox clusters (13, 14). However, phylogenetic analysis was unable to resolve whether the multiple clusters are a result of independent duplication within the lamprey lineage or in the main branch that gave rise to the tetrapods and the ray-finned fishes (13, 14). Investigations in the horn shark (Heterodontus francisci), a cartilaginous jawed vertebrate, have identified only two Hox clusters so far (15). Further studies are required to clarify whether the sharks have more than two Hox clusters.
The lobe-finned fishes (Sarcopterygii) are the forerunners of tetrapods. They diverged from the ray-finned fishes ≈450 million years ago. The lobe fins of Sarcopterygians are considered to be the intermediary stage in the transition from ray fins to limbs. Thus, the analysis of Hox genes in lobe-finned fishes might shed light on the ancestral state of tetrapod Hox clusters and help to understand the evolutionary origin of tetrapod limbs. Previously, PCR amplification of the homeodomain-encoding region of Hox genes in the African lungfish had identified 14 Hox genes, which were assigned to the four tetrapod clusters (16). However, because of the limited number of variable amino acid positions in the highly conserved homeodomain, the homeodomain sequence alone is not adequate in all cases to unambiguously identify the orthology of Hox genes. Present-day lobe-finned fish are represented by only two groups: the coelacanths (Actinistia) and the lungfishes (Dipnoi). Phylogenetic analysis of mitochondrial sequences and phylogenetic distribution of some molecular markers suggest that coelacanths occupy a basal position among the extant lobe-finned fishes (17, 18). In the present study, we have carried out an extensive PCR survey of Hox genes in the Indonesian coelacanth, Latimeria menadoensis, and determined the complete sequence of the second exon of 33 Hox genes. We established the physical linkage between two clusters of three genes each, and three clusters of two genes each by long-range PCR. Our results suggest that coelacanths have four Hox clusters with a gene complement more similar to mammals than ray-finned fishes.
Materials and Methods
DNA Extraction.
Genomic DNA of the Indonesian coelacanth was extracted from pieces of gills that were preserved in DMSO or ethanol by using the standard protocol. An aliquot of the DNA samples was run on a 0.5% agarose gel to ascertain the quality of DNA.
Cloning and Sequencing of Hox Genes.
We amplified fragments of Hox genes from genomic DNA by PCR using several combinations of degenerate primers flanking the homeobox region (Table 1). PCR was carried out in 20-μl reaction volumes by using AmpliTaq DNA polymerase (Applied Biosystems). A typical PCR cycle consisted of a denaturation step at 95°C for 2 min, 35 cycles of 95°C for 30 sec, 45°C for 1 min, and 72°C for 1 min, and a final elongation step at 72°C for 5 min. PCR products were cloned into a T vector and sequenced on an automated ABI377 or ABI3700 DNA sequencer. The sequences were blast-searched against the nonredundant protein database maintained at the National Center for Biotechnology Information (www.ncbi.nlm.nih.gov) to determine their identity. Unique sequences that showed a similarity to vertebrate Hox genes were selected for determining the complete sequence of the second exon. Since we did not have adequate DNA for generating a good genomic library, we decided to clone the second exon by inverse PCR (19). The circularized libraries of genomic DNA for inverse PCR were prepared by using the following enzymes: AccI, AvaI, BamHI, BclI, BglII, ClaI, EcoRI, HaeIII, HhaI, HindIII, NdeI, PstI, PvuII, SpeI, SspI, TaqI, XbaI, and XmnI. Genomic DNA (≈3.5 μg) was completely digested and the restriction fragments were circularized by ligating their ends. Besides the undiluted aliquot of restriction fragments, two dilutions of restriction fragments (1:50 and 1:2,500) were also incubated in 50 μl of ligation mixture overnight, and 1 μl of the ligation mixture was used as template in inverse PCR. The sequences of primers used in inverse PCR are given in Table 2, which is published as supporting information on the PNAS web site, www.pnas.org. A typical inverse PCR cycle consisted of a denaturation step at 95°C for 2 min, 35 cycles of 95°C for 30 sec, 60°C for 1 min, and 72°C for 2 min, followed by a final elongation step at 72°C for 5 min. The PCR products were cloned into a T vector and sequenced.
Table 1.
PCR primers used to amplify the coelacanth Hox genes
| Target genes | Primers | Sequence (5′ to 3′) |
|---|---|---|
| Hox 1–10 | HoxF1 | GARYTNGARAARGARTT |
| HoxR1 | TGGTTYCARAAYMGNMG | |
| Hox 5 | HoxF5 | GARAARGARTTYCAYTTYAA |
| Hox 6 and 7 | HoxF6 | ACNTAYACNMGNTAYCARAC |
| HoxR6 | TCYTTYTTCCAYTTCAT | |
| Hox 6–8 | HoxF7 | MGRGGNMGRCARACNTA |
| Hox 9–13 | HoxF9 | CGAAAGAAGMGI/CGTI/CCCI/CTAYAC |
| Hox 9–13 | HoxF10 | AAGAARMGNGTNCCITAYAC |
| Hox 11 | HoxF2 | AARAARMGNTGYCCNTAYAC |
| HoxA11 | CA11F1* | AGTGGTCAACGTACAAGGAA |
| Hox 12 | HoxF12 | ACNAARCARCARATHGCNGA |
| Hox 13 | HoxF3 | AARAARMGNGTNCCNTAYAC |
HoxR1 served as the reverse primer for all reactions except for those using HoxF6. R = G or A; Y = T or C; N = G, A, T, or C; M = A or C; and H = A, C, or T.
Based on the first exon sequence of HoxA11 from the African coelacanth Latimeria chalumnae (28).
Cloning of Intergenic Regions.
Physical linkage between coelacanth Hox genes on a particular cluster was demonstrated by amplifying intergenic regions by long-range PCR (Expand 20kbPlus, Roche Diagnostics) by using primers corresponding to the end sequences of the cloned Hox fragments. The sequences of long-range PCR primers that were successful in amplifying intergenic regions are given in Table 3, which is published as supporting information on the PNAS web site. The long-range PCR products were cloned into PCR-TOPO-XL vector (Invitrogen) and their end sequences (600–800 bp) were determined to confirm their identities.
Phylogenetic Analysis.
The amino acid sequences were aligned by using the clustalx Version 1.8 program (20). Regions of sequences that were difficult to align were removed from the data file and the sequences were realigned. These alignments were then used to generate phylogenetic trees by the Neighbor-Joining method by using the suite of programs in phylip Version 3.5 (21). Bootstrap values for the nodes were determined by analyzing 1,000 bootstrap replicate data sets to estimate the strength of the groupings. Orthologous Hox sequences from amphioxus were used as outgroup sequences.
Results and Discussion
The Hox genes in vertebrates consist of two exons, and the highly conserved homeodomain (60 aa) is encoded by the second exon. We used several combinations of degenerate primers, some targeted to several paralogue groups and some to specific groups, to amplify the homeodomain-encoding region of Hox genes from the Indonesian coelacanth (Table 1). We first did several rounds of PCR using “general” degenerate primers, and we then used specific primers designed for paralogue groups that were not represented in our initial survey. A total of 680 PCR fragments (110–158 bp) were cloned and sequenced. By comparing these sequences against each other, we identified a set of unique sequences that were searched against the nonredundant protein database at the National Center for Biotechnology Information, using the blastx algorithm. Our analysis identified 33 different Hox gene fragments, besides the fragments of related genes such as GBX and NKX (data not shown). We then determined the complete sequence of the second exon of all of the Hox fragments by inverse PCR. Some of the inverse PCR fragments included the first intron as well as the first exon. The second exons of the coelacanth Hox genes we cloned code for 71–253 residues, and thus show a wide variation in their lengths. We were able to assign these genes to different paralogue groups based on sequence comparisons and blast analysis.
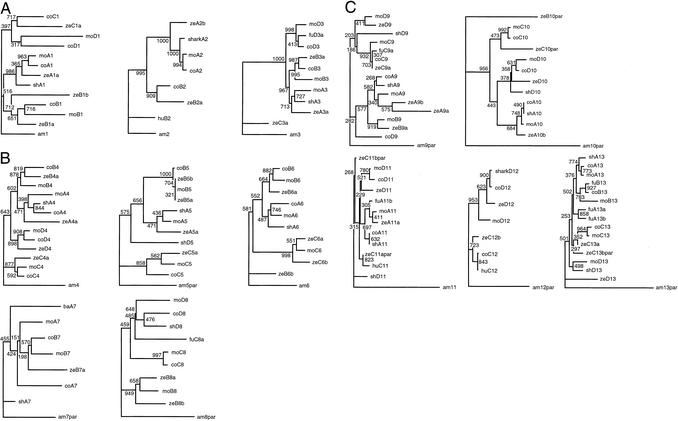
To determine the cluster affiliation and orthology of these Hox genes, we generated phylogenetic trees, using amino acid sequences encoded by the second exons of known chordate Hox genes. Phylogenetic trees were generated separately for individual paralogue groups by using the cognate Hox sequence from amphioxus (Branchiostoma floridae) as an outgroup (Fig. 1). To confirm the groupings of the coelacanth Hox sequences observed in these trees, we generated combined phylogenetic trees for anterior (Hox 1–3), medial (Hox 4–8), or posterior (Hox 9–13) paralogue groups (data not shown). The topologies of coelacanth Hox-bearing branches were very similar in both of the analyses. We did not generate a combined tree for all of the Hox genes because of the difficulty in obtaining a reliable alignment for the divergent sequences.
Figure 1.
Neighbor-joining trees for the anterior (A), medial (B), and posterior (C) Hox groups. The number at each node represents the bootstrap value recovered in 1,000 replicates. The Hox genes from amphioxus, which contains a single Hox gene cluster, was used as an outgroup. Abbreviations for species are as follows: am, amphioxus (B. floridae); ba, striped bass (Morone saxatilis); co, coelacanth (L. menadoensis); fu, fugu (F. rubripes); hu, human (Homo sapiens); mo, mouse (Mus musculus); sh, horn shark (H. francisci); ze, zebrafish (D. rerio). Partial sequences are indicated by the suffix “par.” Hox gene sequences from the amphioxus, striped bass, horn shark, zebrafish, fugu, mouse, and human were retrieved from the GenBank or Ensembl browser (www.ensembl.org). Accession numbers of these sequences are listed in Table 4, which is published as supporting information on the PNAS web site.
To confirm the physical linkage between adjacent coelacanth Hox genes that were cloned, we did long-range PCR using primers complementary to the end sequences of the cloned genes. We tried PCR amplification of intergenic regions between all of the cloned neighboring genes, but we succeeded in obtaining a specific product between only HoxA1 and -A2; HoxB2 and -B3; HoxB5 and -B6; HoxB6 and -B7; HoxD8 and -D9; and HoxD10 and -D12 (Fig. 2). We were able to clone a fragment of coelacanth HoxD11 from the HoxD10-HoxD12 PCR fragment by using the primers HoxF2 and HoxR1 (Table 1), and we then obtained the complete second exon sequence by using walking primers. Thus, we were able to establish the physical linkage between HoxA1 and -A2; HoxB2 and -B3; HoxB5, -B6, and -B7; HoxD8 and -D9; and HoxD10, -D11, and -D12 (Fig. 2). Our attempts to amplify intergenic regions between other Hox genes were unsuccessful, presumably because of the large distances between them, which are not amenable to PCR amplification under the PCR protocol used in this study.
The phylogenetic analyses of Hox sequences were able to clarify the cluster affiliation of all of the coelacanth Hox sequences except two genes, which were putatively identified as HoxA7 and HoxD9 based on their sequence identity (Fig. 1). However, we were able to confirm the identity of HoxD9 based on its physical linkage with HoxD8 (Fig. 2). The remaining gene, HoxA7, belongs to the paralogue group 7, which is known to have only two members (A7 and B7) in all of the tetrapods and ray-finned fishes investigated so far. Our survey of coelacanth Hox clusters identified only two members in this paralogue group. Because we were able to determine the identity of one of them as HoxB7 on the basis of phylogenetic analysis and physical linkage, we conclude that the other gene is likely to be HoxA7. Thirty-two of the coelacanth Hox genes we cloned have orthologs on the four Hox clusters in mammals (Fig. 2). The remaining Hox gene is the ortholog of zebrafish hoxc1. The HoxC cluster of mammals does not contain HoxC1 (Fig. 2). The coelacanth genes we cloned include all members of the six paralogue groups (groups 1, 2, 4, 7, 10, and 12) found on the four Hox clusters in tetrapods. However, we did not identify any additional Hox genes that suggested the presence of more than four Hox clusters, indicating that coelacanths, like tetrapods, have just four Hox clusters. The previous PCR survey in the Australian lungfish had suggested that this lobe-finned fish also has only four Hox clusters (16). Taken together, these results suggest that the ancestral bony vertebrates had four Hox clusters, which have not been duplicated during the evolution of lobe-finned fish and tetrapod lineages. The additional Hox clusters found in the teleosts are, therefore, a result of independent duplication(s) in the ray-finned fish lineage (8–11).
Comparisons of the Hox genes in coelacanth, tetrapods, and ray-finned fishes show that genes for HoxA6, HoxD1, and HoxD8 are present in coelacanth and tetrapods, but are absent in ray-finned fishes such as zebrafish (8) and fugu (9). Further studies on these genes may indicate whether they are related to the unique morphological features that distinguish lobe-finned fishes and tetrapods from ray-finned fishes. The coelacanth HoxA cluster genes we cloned include HoxA7, which is present in the horn shark (15), tetrapods, and ray-finned fishes such as the striped bass (22) and an African cichlid fish (11). However, it is absent on the duplicate hoxa clusters of both zebrafish (8) and fugu (9). Given that classical taxonomic studies group fugu, cichlids, and striped bass under the order Perciformes, and the zebrafish under the order Cypriniformes (23), the absence of HoxA7 ortholog in the fugu and zebrafish suggests that this gene has been lost independently in the fugu and zebrafish lineages. The effect of the loss of this gene on the phenotype of these fishes is unclear.
The HoxC1 found in the coelacanth is unique among the paralogue group 1 genes cloned so far. This gene is absent in tetrapods, and has become a pseudogene in the fugu (9), but it is present in one of the duplicated hoxc clusters in the zebrafish. However, analysis of the structure and function of the zebrafish gene, hoxc1a, has indicated that it may be on the way to becoming a pseudogene (24). Unlike other vertebrate Hox genes that contain an intron, zebrafish hoxc1a is intronless, and, as a result, contains a longer linker region between the homeodomain and the hexapeptide (WMKVKR), which binds to the Pbx cofactors. Furthermore, there are several significant changes in the functionally important domains of zebrafish hoxc1a. In particular, two of the seven diagnostic residues of the paralogue group 1 Hox genes (Fig. 3) and two of the residues in the hexapeptide have been replaced. Functional studies have shown that hoxc1a is less efficient in inducing homeotic transformation (24). Analysis of the coelacanth HoxC1 sequence shows that it has an intron at the right position and encodes all of the seven residues that are diagnostic of the paralogue group 1 homeodomains (Fig. 3), indicating that it is a functional gene. Expression of coelacanth HoxC1 in zebrafish and rodents may provide interesting insights into the function of this gene in lobe-finned fishes.
Figure 3.
clustalx alignment of the amino acid sequences of amphioxus, horn shark, zebrafish, coelacanth, and mouse Hox paralogue group1 genes. The seven residues that are diagnostic of the paralogue group 1 homeodomain are indicated with asterisks. The substitutions in the zebrafish hoxc1a are boxed. The positions of only amphioxus amino acid residues are shown. The abbreviations are the same as those in Fig. 1.
Comparisons of noncoding sequences in the Hox locus between mammals, shark, and ray-finned fishes have identified several conserved putative regulatory elements, suggesting a highly conserved regulatory mechanism of these genes (25, 26). Analysis of the coelacanth HoxB4 intron sequence identified a 90-bp element that is highly conserved in the mouse, fugu, and coelacanth (Fig. 4). This sequence has been identified as an enhancer that mediates the spatial expression pattern of HoxB4 in developing mouse embryos (25, 27). The conservation of this enhancer element in the coelacanth suggests that the regulation, and possibly the function, of HoxB4 is conserved in all three major groups of bony vertebrates.
Figure 4.
clustalx alignment of conserved sequences in the introns of coelacanth, fugu, and mouse HoxB4. Conserved bases are denoted by asterisks. GenBank accession number of the fugu sequence: U92575. The mouse sequence was obtained from the Ensembl browser.
Although the strategy used by us cannot confirm the absence of Hox genes that were not cloned in this study, it can be inferred that coelacanths have four Hox clusters similar to tetrapods, and that their gene complement is more similar to mammalian Hox clusters than to the ray-finned fish Hox clusters, with the exception of HoxC1 gene, which has been lost in the mammalian lineage. The four-cluster architecture of Hox genes is, thus, highly conserved in the lobe-finned fish and tetrapod lineages.
Supplementary Material
Acknowledgments
We thank Ms. Tay Boon Hui and Ms. Diane Tan for technical help. B.V. is an adjunct staff member of the Department of Paediatrics, National University of Singapore. This work was funded by the Agency for Science, Technology, and Research of Singapore.
Footnotes
References
- 1.Garcia-Fernandez J, Holland P W. Nature. 1994;370:563–566. doi: 10.1038/370563a0. [DOI] [PubMed] [Google Scholar]
- 2.Di Gregorio A, Spagnuolo A, Ristoratore F, Pischetola M, Aniello F, Branno M, Cariello L, Di Lauro R. Gene. 1995;156:253–257. doi: 10.1016/0378-1119(95)00035-5. [DOI] [PubMed] [Google Scholar]
- 3.McGinnis W, Krumlauf R. Cell. 1992;68:283–302. doi: 10.1016/0092-8674(92)90471-n. [DOI] [PubMed] [Google Scholar]
- 4.Krumlauf R. Cell. 1994;78:191–201. doi: 10.1016/0092-8674(94)90290-9. [DOI] [PubMed] [Google Scholar]
- 5.Ruddle F H, Bartels J L, Bentley K L, Kappen C, Murtha M T, Pendleton J W. Annu Rev Genet. 1994;28:423–442. doi: 10.1146/annurev.ge.28.120194.002231. [DOI] [PubMed] [Google Scholar]
- 6.Godsave S, Dekker E J, Holling T, Pannese M, Boncinelli E, Durston A. Dev Biol. 1994;166:465–476. doi: 10.1006/dbio.1994.1330. [DOI] [PubMed] [Google Scholar]
- 7.Stein S, Fritsch R, Lemaire L, Kessel M. Mech Dev. 1996;55:91–108. doi: 10.1016/0925-4773(95)00494-7. [DOI] [PubMed] [Google Scholar]
- 8.Amores A, Force A, Yan Y L, Joly L, Amemiya C, Fritz A, Ho R K, Langeland J, Prince V, Wang Y L, et al. Science. 1998;282:1711–1714. doi: 10.1126/science.282.5394.1711. [DOI] [PubMed] [Google Scholar]
- 9.Aparicio S, Chapman J, Stupka E, Putnam N, Chia J M, Dehal P, Christoffels A, Rash S, Hoon S, Smit A, et al. Science. 2002;297:1301–1310. doi: 10.1126/science.1072104. [DOI] [PubMed] [Google Scholar]
- 10.Naruse K, Fukamachi S, Mitani H, Kondo M, Matsuoka T, Kondo S, Hanamura N, Morita Y, Hasegawa K, Nishigaki R, et al. Genetics. 2000;154:1773–1784. doi: 10.1093/genetics/154.4.1773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Malaga-Trillo E, Meyer A. Am Zool. 2001;41:676–686. [Google Scholar]
- 12.Robinson-Rechavi M, Marchand O, Escriva H, Laudet V. Curr Biol. 2001;11:R1007–R1008. doi: 10.1016/s0960-9822(01)00280-9. [DOI] [PubMed] [Google Scholar]
- 13.Force A, Amores A, Postlethwait J H. J Exp Zool. 2002;294:30–46. doi: 10.1002/jez.10091. [DOI] [PubMed] [Google Scholar]
- 14.Irvine S Q, Carr J L, Bailey W J, Kawasaki K, Shimizu N, Amemiya C T, Ruddle F H. J Exp Zool. 2002;294:47–62. doi: 10.1002/jez.10090. [DOI] [PubMed] [Google Scholar]
- 15.Kim C B, Amemiya C, Bailey W, Kawasaki K, Mezey J, Miller W, Minoshima S, Shimizu N, Wagner G, Ruddle F. Proc Natl Acad Sci USA. 2000;97:1655–1660. doi: 10.1073/pnas.030539697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Longhurst T J, Joss J M. J Exp Zool. 1999;285:140–145. doi: 10.1002/(sici)1097-010x(19990815)285:2<140::aid-jez6>3.0.co;2-v. [DOI] [PubMed] [Google Scholar]
- 17.Zardoya R, Cao Y, Hasegawa M, Meyer A. Mol Biol Evol. 1998;15:506–517. doi: 10.1093/oxfordjournals.molbev.a025950. [DOI] [PubMed] [Google Scholar]
- 18.Venkatesh B, Erdmann M V, Brenner S. Proc Natl Acad Sci USA. 2001;98:11382–11387. doi: 10.1073/pnas.201415598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ochman H, Gerber A S, Hartl D L. Genetics. 1988;120:621–623. doi: 10.1093/genetics/120.3.621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Thompson J D, Gibson T J, Plewniak F, Jeanmougin F, Higgins D G. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Felsenstein J. phylip (Phylogeny Inference Package) (Univ. of Washington, Seattle), Version 3.5. 1995. [Google Scholar]
- 22.Snell E A, Scemama J L, Stellwag E J. J Exp Zool. 1999;285:41–49. [PubMed] [Google Scholar]
- 23.Nelson J S. Fishes of the World. New York: Wiley; 1994. [Google Scholar]
- 24.McClintock J M, Carlson R, Mann D M, Prince V E. Development (Cambridge, UK) 2001;128:2471–2484. doi: 10.1242/dev.128.13.2471. [DOI] [PubMed] [Google Scholar]
- 25.Aparicio S, Morrison A, Gould A, Gilthorpe J, Chaudhuri C, Rigby P, Krumlauf R, Brenner S. Proc Natl Acad Sci USA. 1995;92:1684–1688. doi: 10.1073/pnas.92.5.1684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chiu C H, Amemiya C, Dewar K, Kim C B, Ruddle F H, Wagner G P. Proc Natl Acad Sci USA. 2002;99:5492–5497. doi: 10.1073/pnas.052709899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Whiting J, Marshall H, Cook M, Krumlauf R, Rigby P W, Stott D, Allemann R K. Genes Dev. 1991;5:2048–2059. doi: 10.1101/gad.5.11.2048. [DOI] [PubMed] [Google Scholar]
- 28.Chiu C H, Nonaka D, Xue L, Amemiya C T, Wagner G P. Mol Phylogenet Evol. 2001;17:305–316. doi: 10.1006/mpev.2000.0837. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.