Abstract
Association study (especially the genome-wide association study) now has a key function in identification and characterization of disease-predisposing genetic variant(s), which customarily involve multiple single nucleotide polymorphisms (SNPs) in a candidate region or across the genome. Case–control association design remains the most popular and a challenging issue in the statistical analysis is the optimal use of all information contained in these SNPs. Previous approaches often treated gene–gene interaction as deviation from additive genetic effects or replaced it with SNP–SNP interaction. However, these approaches are limited for their failure of consideration of gene–gene interaction or gene–gene co-association at gene level. Although the co-association of the SNPs within a candidate gene can be detected by principal component analysis-based logistic regression model, the detection of co-association between genes in genome remains uncertain. Here, we proposed a canonical correlation-based U statistic (CCU) for detecting gene-based gene–gene co-association in the case–control design. We explored its type I error rates and power through simulation and analyzed two real data sets. By treating gene as a functional unit in analysis, we found that CCU was a strong alternative to previous approaches. We discussed the performance of CCU as a gene-based gene–gene co-association statistic and the prospect of further improvement.
Keywords: gene-based, gene–gene co-association, canonical correlation
Introduction
Association study (especially the genome-wide association study, GWAS) now has a key function in identification and characterization of disease-predisposing genetic variant(s), which customarily involves multiple single nucleotide polymorphisms (SNPs) in a candidate region or across the genome. Case–control association design, which remains the most popular and a challenging issue in the statistical analysis, makes optimal use of all information contained in these SNPs. In human complex diseases, correlations exist not only between the SNPs in the candidate genes but also between the genes in the genome because of linkage disequilibrium (LD) (or SNP–SNP interactions) and co-association (or interaction) between genes. Both gene–gene co-association and interaction could imply that the two genes share their role in causing diseases (or trait), or a de facto high dependency or correlation between two genes in disease predisposition.1 The role in the etiology of complex diseases is the basis for constructing gene networks. Co-association between genes can be seen as joint effect of genes contributing to the disease or trait, and can be measured based on the correlation between genes. This is in contrast to the definition or measure of interaction (epistasis), which is somewhat confusing. Moore delineated among genetic, biological and statistical epistases; differences in genetic and biological epistases among individuals in a population give rise to statistical epistasis.2 However, the practical difficulty in interpreting biological epistasis through statistical epistasis can lead to controversy regarding the relationship between them.
The classic statistical interaction as defined by Fisher3 and developed further by Cockerham4 and Kempthorne5 treats gene–gene interaction as deviation from additive genetic effects.6 Modeling a trait as an additive combination of its single-locus main effects and interaction terms is likely to limit the power to detect interaction.7
Several methods for detection of gene–gene interaction are worthy of note. In particular, multifactor dimensionality reduction (MDR)8 is a data-mining method8, 9, 10, 11, 12, 13, 14 However, the heavy dependence on data structure, complicated procedure and lack of clear biological interpretation of the detected gene–gene interactions had limited applications of data-mining methods in complex disease association study.7 Parametric methods are more powerful than nonparametric methods provided valid assumptions are made.7 In this regard, LD1, 7 and entropy-based15, 16 methods have clearer biological interpretation and are powerful. However, all these methods share a limitation in common. Although developed for detecting gene–gene interactions, they are practically testing for SNP–SNP interactions, which are insufficient for interpretation of gene–gene interaction. Multiple variants in a gene have made it difficult to be tagged by a single SNP, whereas SNP–SNP interaction may not truly reflect many potential factors such as LD between SNPs. Furthermore, all SNP-based methods have to tackle the multiple-testing problem.
Recently, several groups have proposed to combine principal component analysis (PCA) with logistic regression17, 18, 19 to explore contribution of set of SNPs within a candidate gene on the disease (trait), namely the co-association of the SNPs to disease (trait). However, the detection of co-association between genes in genome remains uncertain. Gene–gene co-association is not equivalent to gene–gene interaction but could imply gene–gene interaction or joint effect of two genes. In the study of etiological gene networks of disease pathogenesis, gene–gene co-association is much more meaningful as it could render a priori topological structure (or model) for establishing biological pathways and gene networks of the disease.
In this study, we develop a gene-based statistic for detecting gene–gene co-association between cases and controls. We use canonical correlation analysis (CCA)20 to obtain systematic correlations between two genes through a linear transformation of all SNPs in each gene. We also develop a statistic for detecting gene–gene co-association and investigate its performance under different disease models and for a range of sample sizes and various degrees of correlations between two genes in cases and controls through simulation study. Finally, we analyze two real data sets and make comparisons with the results of MDR, LD-based statistic and logistic regression analysis.
Materials and Methods
Data simulation
Computer program MS21 was used to generate haplotypes in two associated genes, which were paired at random to generate individuals' genotypes. Under the null hypothesis, a population with 20 000 individuals was generated. Cases and controls were selected randomly from the population, according to sample sizes 100(100)1000, 1000(1000)5000; 10 000 simulations were repeated at each sample size to study the characteristics of distribution and type I error rates of canonical correlation-based U statistic (CCU). Under the alternative hypothesis, two populations were generated by specifying different parameters, to call MS. One is taken as case population and the other as control population. Cases and controls were randomly sampled separately from each population, with sample size similar to those for the null hypothesis and 10 000 simulations were repeated for each sample size to explore power of CCU.
Quantification
Assume a case–control study (n individuals in each group), and gene A with p SNPs and gene with B q SNPs. We code the genotypes according to specific genetic model.22, 23 For instance, under joint additive–additive model and cases, SNP genotypes in gene A and gene B are quantified as xDik=2,1,0 and yDik=2,1,0, i=1,2,…,p, j=1,2,…,q, k=1,2,…,n (2 for mutant homozygote, 1 for heterozygote, and 0 wild-type homozygote), respectively, In controls, xCik=2,1,0, yCik=2,1,0, i=1,2,…,p, j=1,2,…,q, k=1,2,…,n are similarly obtained. This quantification of genotype data avoids complicated haplotype deduction.
Test statistic
As noted earlier, we focus on the difference of correlation between two genes in cases and controls as a measure of co-association of the two genes contributing to the disease. We use canonical correlation20 for this measure. Let the afore-mentioned genotyped data of case–control study be coded as (X1D, X2D,…,XpD) and (Y1D, Y2D,…,YqD) for gene A and gene B for cases, and (X1C, X2C,…,XpC) and (Y1C, Y2C,…,YqC) for controls. The maximum canonical correlation coefficient rD (1≥rD≥0) between (X1D, X2D,…,XpD) and (Y1D, Y2D,…,YqD) obtained by CCA could be taken as a measure of gene-based gene–gene co-association in cases, and rC (1≥rC≥0) from (X1C, X2C,…,XpC) and (Y1C, Y2C,…,YqC) be a measure of gene–gene co-association in controls (Appendix A in Supplementary information). Our test of gene–gene co-association contributing to disease is then turned to a test of the difference between rD and rC. The transformation  24, 25 in analogy to Fisher's simple correlation coefficient(s) transformation
24, 25 in analogy to Fisher's simple correlation coefficient(s) transformation 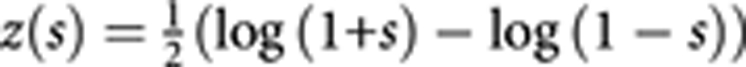 26 are used to canonical correlation coefficients to approximate normal distribution27, 28 (Appendix B in supplementary information), that is,
26 are used to canonical correlation coefficients to approximate normal distribution27, 28 (Appendix B in supplementary information), that is,  and
and  . A CCU for detecting statistical significance of the difference of gene-based gene–gene co-association between cases and controls is then as follows,
. A CCU for detecting statistical significance of the difference of gene-based gene–gene co-association between cases and controls is then as follows,
 |
which is asymptotically normal distributed as N(0,1) (Appendix C in Supplementary information).
Applications
We conducted two real data analyses. The first concerned heroin addiction of self-reported positive response on first use of heroin among 91 individuals in positive group and 245 individuals in negative group, who were all of Han Chinese origin recruited in Shanghai Voluntary Drug Dependence Treatment Center.29 Twenty SNPs in regions of the three genes, μ-opioid receptor gene (OPRM1), κ-opioid receptor gene (OPRK1) and δ-opioid receptor gene (OPRD1), were genotyped. The second was a GWAS of North American Rheumatoid Arthritis (RA) Consortium involving 868 cases and 1194 controls.30 On the basis of the previous result from GAW16, four genes (C5, VEGFA, PADI4, PTPN22) were selected to detect gene–gene co-association with RA susceptibility. There were eight, four, six and nine SNPs genotyped in each gene, respectively.
Results
Type I error rates
Results for the joint additive–additive model are shown in Table 1, noting that when sample size of case–control study is equal to or larger than 200, the normal distribution of CCU under the null hypothesis was confirmed by normal tests, but not so for sample size being less than 200, and type I error rates of CCU are not appreciably different from the nominal levels at 0.01, 0.05, 0.1 and 0.2. Results for joint dominant–dominant model are shown in Table 2 and similar to that from joint additive–additive model. For both joint additive–additive model and joint dominant–dominant model, the type I error rates are close to given nominal levels when sample size of case–control study is larger than 300 (Tables 1 and 2). CCU is normally distributed and the results showed that it is insensitive to model misspecification under null hypothesis.
Table 1. Performance of CCU under the null hypothesis (joint additive–additive model).
| Normality test | Type I error rates (%) | ||||||
|---|---|---|---|---|---|---|---|
| Sample size | D | W2 | A2 | 1 | 5 | 10 | 20 |
| 100 | <0.01 | <0.005 | <0.005 | 0.06 | 0.06 | 0.13 | 0.97 |
| 200 | 0.14 | >0.25 | >0.25 | 1.12 | 5.36 | 10.33 | 20.65 |
| 300 | >0.15 | >0.25 | >0.25 | 1.04 | 5.08 | 9.93 | 19.87 |
| 400 | >0.15 | >0.25 | >0.25 | 0.96 | 5.20 | 10.06 | 20.07 |
| 500 | >0.15 | >0.25 | >0.25 | 1.10 | 5.48 | 10.05 | 19.95 |
| 600 | >0.15 | >0.25 | >0.25 | 1.03 | 5.09 | 9.93 | 19.51 |
| 700 | >0.15 | >0.25 | >0.25 | 1.12 | 5.40 | 10.35 | 20.55 |
| 800 | >0.15 | >0.25 | >0.25 | 1.09 | 5.18 | 9.90 | 19.93 |
| 900 | >0.15 | >0.25 | >0.25 | 0.89 | 5.23 | 9.92 | 19.60 |
| 1000 | >0.15 | >0.25 | >0.25 | 0.85 | 4.80 | 9.63 | 19.10 |
| 2000 | >0.15 | >0.25 | >0.25 | 1.00 | 4.77 | 9.95 | 20.06 |
| 3000 | >0.15 | 0.25 | >0.25 | 1.02 | 4.95 | 10.29 | 20.23 |
| 4000 | >0.15 | >0.25 | >0.25 | 1.21 | 5.21 | 9.87 | 19.24 |
| 5000 | >0.15 | 0.15 | 0.14 | 0.86 | 4.83 | 9.91 | 20.17 |
D, Kolmogorov–Smirnov D test.
W2, Cramer-von Mises W2 test.
A2, Anderson–Darling A2 test.
Table 2. Performance of CCU under the null hypothesis (joint dominant–dominant model).
| Normality test | Type I error rates (%) | ||||||
|---|---|---|---|---|---|---|---|
| Sample size | D | W2 | A2 | 1 | 5 | 10 | 20 |
| 100 | 0.01 | <0.005 | <0.005 | 0.13 | 0.13 | 0.14 | 0.20 |
| 200 | 0.06 | 0.07 | 0.08 | 1.13 | 4.96 | 9.89 | 19.85 |
| 300 | >0.15 | >0.25 | 0.25 | 1.08 | 5.23 | 9.85 | 19.30 |
| 400 | >0.15 | >0.25 | >0.25 | 0.99 | 4.85 | 9.64 | 19.77 |
| 500 | 0.08 | 0.16 | 0.19 | 0.99 | 5.02 | 9.85 | 19.24 |
| 600 | >0.15 | 0.17 | 0.12 | 1.14 | 5.27 | 9.86 | 19.92 |
| 700 | >0.15 | >0.25 | >0.25 | 0.95 | 4.99 | 10.13 | 20.27 |
| 800 | >0.15 | >0.25 | >0.25 | 0.93 | 4.97 | 9.97 | 19.85 |
| 900 | >0.15 | >0.25 | >0.25 | 1.04 | 5.02 | 10.00 | 20.04 |
| 1000 | >0.15 | >0.25 | >0.25 | 1.18 | 5.32 | 10.31 | 20.18 |
| 2000 | >0.15 | >0.25 | >0.25 | 0.92 | 4.86 | 10.04 | 20.52 |
| 3000 | >0.15 | >0.25 | >0.25 | 0.98 | 5.07 | 10.17 | 19.77 |
| 4000 | >0.15 | >0.25 | >0.25 | 1.17 | 5.13 | 10.05 | 20.14 |
| 5000 | >0.15 | >0.25 | >0.25 | 1.00 | 5.20 | 10.10 | 19.99 |
D, Kolmogorov–Smirnov D test.
W2, Cramer-von Mises W2 test.
A2, Anderson–Darling A2 test.
Power
Under joint additive–additive model, the power of CCU is not only a monotonically increasing function of sample size (Figure 1a and b) but also relates to correlations between two genes in cases and controls. Power calculations are performed for control groups sampled from the same population, whereas case groups from several populations, and the larger the deviation of canonical correlations between genes in cases and controls, the larger the power (Figure 1a). More interestingly, power varies when both cases and controls are from several different populations, whereas the deviation of correlations between cases and controls is similar, where the correlations in cases and controls are both larger, and the power is much higher (Figure 1b). Power performance of CCU under joint dominant–dominant model shows similar to those under joint additive–additive model (Figures 1c and d). The results imply that CCU proposed in this study is insensitive to model misspecification.
Figure 1.
Power performance of CCU under joint additive–additive model and joint dominant–dominant model (α=0.05). Under joint additive–additive model: (a) the solid squares represent power evaluation of which cases and controls are from populations with gene–gene correlation of 0.324 and 0.434, respectively, and solid triangles represent that of 0.670 and 0.434, whereas hollow diamonds represent that of 0.129 and 0.434. (b) The solid squares represent power evaluation of which cases and controls are from populations with gene–gene correlation of 0.708 and 0.625, respectively, the solid triangles represent that of 0.345 and 0.259, the hollow diamonds represent that of 0.896 and 0.806, and inverse triangles represent that of 0.434 and 0.324. Under joint dominant–dominant model: (c) the solid squares represent power evaluation of which cases and controls are from populations with gene–gene correlation of 0.127 and 0.192, respectively, and the solid triangles represent that of 0.394 and 0.192. (d) The solid squares represent power evaluation of which cases and controls are from populations with gene–gene correlation of 0.534 and 0.394, respectively, the solid triangles represent that of 0.830 and 0.699, and hollow diamonds represent that of 0.699 and 0.534.
Applications
For the heroin addiction data, the result of MDR method (Appendix D in Supplementary) showed that rs678849, rs797397 and rs12404612 in OPRD1, rs6985606 in OPRK1 and rs510769 in OPRM1 were likely to interact with each other (Table 3). The results of CCU and LD-based statistic under joint additive–additive genetic model are summarized in Table 4. CCU suggested that gene–gene co-associations between OPRD1 and OPRM1 and that between OPRD1 and OPRK1 were significant with heroin-induced positive response on first use (Table 4), whereas LD-based statistic suggested SNP–SNP interaction in OPRD1 and OPRM1 and that in OPRK1 and OPRM1 were significant. For the RA data, the results of CCU, LD-based statistic and logistic regression analysis (Appendix D in Supplementary) under joint additive–additive genetic model are shown in Table 5. CCU suggested that co-associations of C5-PADI4, C5-PTPN22 and VEGFA-PADI4 were significant with RA susceptibility, whereas LD-based statistic suggested SNP–SNP interactions in C5-PADI4, C5-PTPN22 and VEGFA-PADI4; logistic regression analysis could only detect SNP–SNP interaction in VEGFA-PADI4.
Table 3. Results of detected interaction among OPRD1, OPRK1 and OPRM1 by MDR.
| Model | Train Bal. Acc. | Test Bal. Acc. | CV consist |
|---|---|---|---|
| rs482387(OPRD1), rs696522(OPRM1) | 0.6223 | 0.4690 | 10(3) |
| rs482387(OPRD1), rs1799971(OPRM1), rs696522(OPRM1) | 0.6705 | 0.4806 | 10(3) |
| rs797397(OPRD1), rs12404612(OPRD1), rs6985606(OPRK1), rs510769(OPRM1) | 0.7332 | 0.4786 | 10(5) |
| rs678849(OPRD1), rs797397(OPRD1), rs12404612(OPRD1), rs6985606(OPRK1), rs510769(OPRM1) | 0.8111 | 0.4929 | 10(6) |
Train Bal. Acc., training balanced accuracy.
Test Bal. Acc., testing balanced accuracy.
CV consist, cross-validation consistency.
Table 4. Results of detected gene–gene co-association among OPRD1, OPRK1 and OPRM1 by CCU and their SNP–SNP interaction by LD-based statistic.
| CCU | LD-based statistic | ||||
|---|---|---|---|---|---|
| Interaction | Measure | U-value | P-value | SNP–SNP | P-value |
| OPRD1-OPRK1 | 0.52 | 3.7071 | 0.0002 | NULL | NULL |
| 0.25 | |||||
| OPRD1-OPRM1 | 0.63 | 2.6100 | 0.0091 | rs482387–rs3778151 | 0.0001a |
| 0.32 | |||||
| OPRK1-OPRM1 | 0.44 | 1.4376 | 0.1505 | rs16918941–rs3778151 | 0.0003a |
| 0.29 | |||||
Significant after multiple testing.
NULL none pair of SNP–SNP interaction was significant after multiple testing.
Table 5. Results of detected gene–gene co-association among C5, VEGFA, PADI4 and PTPN22 by CCU and their SNP–SNP interaction by LD-based statistic and logistic regression analysis.
| CCU | LD-based statistic | Logistic regression analysis | |||||
|---|---|---|---|---|---|---|---|
| Interaction | Measure | U-value | P-value | SNP–SNP | P-value | SNP–SNP | P-value |
| C5-PADI4 | 0.16 | 2.0608 | 0.0393 | rs3824535–rs11203405 | <0.0001 | N | N |
| 0.10 | rs10818768–rs4387213 | <0.0001 | |||||
| C5-PTPN22 | 0.16 | 1.9815 | 0.0475 | rs3739836–rs11485101 | <0.0001 | N | N |
| 0.11 | rs3824535–rs11811771 | 0.0003 | |||||
| VEGFA-PADI4 | 0.18 | 2.7999 | 0.0051 | rs3025033–rs1120340 | <0.0001 | rs3025010–rs6659366 | 0.0005 |
| 0.10 | |||||||
N, none pair of SNP–SNP interaction was significant after multiple testing.
Discussion
Gene-based association study
Currently, the level of association most commonly seen in the literature is SNP. These SNP-based methods, such as logistic regression analysis, MDR,8 LD-based1, 7 and entropy15, 16 statistics, have practical limitations. First, as there are multiple variants in a gene, one single SNP (or tagging SNP) is inadequate to represent the effect of the gene in the whole genome as a functional unit. Second, replication at SNP level runs a high risk of false negative results because of different functional variants within the replication sample or subpopulation. Third, the problem of multiple testing can greatly reduce the power of SNP-based methods. Neale and Sham31 suggested a move toward gene-based approach, for the reasons that genes are the functional unit of the human genome and the positions, sequence and function of genes are highly consistent across diverse human populations. This scope is considerably greater than that of either an SNP or a haplotype. Gene-based association study explicitly accounts for biological function of a gene, so it takes the problem of nonreplication up to the gene level. Effectively, gene-based association study alleviates the burden of multiple testing in into two stages: handling of multiple variants within a gene and multiple genes across the genome.31
In an earlier attempt to detect association at gene level, several groups have proposed to combine PCA with logistic regression tests (LRT).17, 18, 19 Such a PCA–LRT approach involved two basic steps. First, PCA was used to compute combination of correlated SNPs that capture the underlying correlation structure of a candidate region. Then, logistic regression model test was used to assess the association between principal components scores and disease. This approach captures the co-association between SNPs within a candidate gene and is less computationally demanding compared to haplotype-based analysis. It takes advantages of principal components to avoid multicolinearity between SNPs. Studies showed that PCA–LRT was typically as or more powerful than both genotype- and haplotype-based methods. For a candidate gene, however, PCA–LRT could only detect co-association between SNPs. The CCU statistic in this paper detects gene–gene co-association, which could suggest true joint effect between two genes. Furthermore, gene–gene co-associations can render a priori topological structure (or model) for establishing etiological gene network of the disease pathogenesis.
Relationship between gene–gene interaction and co-association
The definition of gene–gene interaction or epistasis is somewhat inconsistent. Gene–gene interaction is typically defined as statistical deviation from additive genetic effects,6 whereas Zhao et al7 defined interaction between two unlinked loci (or genes) for a qualitative trait as the deviation of the penetrance for a haplotype at two loci from the product of the marginal penetrance of the individual alleles that span the haplotype. In epidemiology,32, 33 gene–gene interaction refers to the extent to which the joint effect of two genes on disease (or trait) differs from the independent effect of each gene. In terms of their causal effects on disease incidence, two genes may act independently or interact to augment (in case of synergism) or deduct (in case of antagonism) the effect of one another. To determine the presence of interaction between two genes in a case–control association study, a product term is customarily added to the logistic regression model: Logit(P/1−P))=β0+β1Agene+β2Bgene+γAgene × Bgene, where γ is the measurement of the interaction. This model implies that the interaction (γ) between gene A and gene B assumes independence between them. However, two genes in the genome are often correlated with each other in specific pathways or networks to cause a disease. Co-association could be more appropriate to measure the joint effect of two genes contributing to a disease such that it is based on the correlation between genes (such as CCU statistic in this paper), implying that the genes share their role for causing a disease, and the shared feature is the de facto dependency or correlation between two genes. Gene–gene co-association thus extends the concept of gene–gene interaction and/or gene–gene correlation. Gene–gene co-association is much more meaningful as it could render a priori topological structure (or model) for establishing pathways or networks between genes to the disease.
Characteristics of CCU statistic
In this article, we have developed a statistic for detecting gene-based gene–gene co-association using cases and controls. The proposed statistic (CCU) has advantages for the following reasons. First and foremost, it is gene based and integrates the effect between two functional units (two genes) in the human genome. By considering co-association between two genes, the result of CCU is expected to be closer to a biological interpretation. Second, it reduces the problem of nonreplication from SNP to a gene level.,31 Last but not the least, the dimensions of the genotyped data are substantially reduced, as is the problem of multiple testing.
Our simulation showed that CCU has good performance under null hypothesis. Under alternative, power is shown to be a monotonically increasing function of sample size and gene–gene co-association between cases and controls. In the analysis of the heroin addiction data, CCU suggested that gene–gene co-association between OPRD1 and OPRM1 and that between OPRD1 and OPRK1 were significant (Table 4), as with LD-based statistic suggesting that SNP–SNP interaction in OPRD1 and OPRM1 was significant but not in OPRD1 and OPRK1. Gene–gene interaction between OPRD1 and OPRM1 had been detected in many studies34, 35, 36, 37, 38, 39 and for that between OPRD1 and OPRK1, Jordan and Devi40 had provided biochemical and pharmacological evidence for the heterodimerization of the two fully functional opioid receptors, which suggests the result of CCU is credible. Analysis of the RA data showed that CCU is much more efficient than traditional logistic regression analysis (Table 5). Most interestingly, an association study between single SNP and RA susceptibility (Table 6) showed that none of the SNPs in the four genes' regions was significant, implying that a single SNP was unable to represent the gene, so SNP–SNP interactions detected by LD-based statistic or logistic regression analysis were doubtful.
Table 6. Single SNP association with RA susceptibility in C5, VEGFA, PADI4 and PTPN22.
| Gene | SNP | Control | Case | χ2 | P-value |
|---|---|---|---|---|---|
| C5 | rs2239540 | 1194 | 868 | 2.889 | 0.236 |
| rs3739836 | 1193 | 868 | 2.366 | 0.306 | |
| rs3824535 | 1194 | 868 | 3.432 | 0.180 | |
| rs10818768 | 1193 | 868 | 8.728 | 0.013 | |
| rs10760260 | 1194 | 868 | 1.137 | 0.566 | |
| rs10985840 | 1193 | 867 | 1.140 | 0.565 | |
| rs618746 | 1194 | 868 | 1.368 | 0.505 | |
| rs12337223 | 1192 | 867 | 1.111 | 0.574 | |
| VEGFA | rs833069 | 1183 | 861 | 0.277 | 0.871 |
| rs3025010 | 1122 | 757 | 3.568 | 0.168 | |
| rs3025033 | 1193 | 866 | 2.446 | 0.294 | |
| rs3025035 | 1194 | 868 | 0.730 | 0.694 | |
| PADI4 | rs2800687 | 1193 | 867 | 5.841 | 0.054 |
| rs2883272 | 1194 | 868 | 1.550 | 0.461 | |
| rs2526822 | 1193 | 868 | 5.352 | 0.069 | |
| rs6659366 | 1191 | 865 | 3.748 | 0.153 | |
| rs11203405 | 1193 | 868 | 1.818 | 0.403 | |
| rs4387213 | 1193 | 867 | 1.568 | 0.457 | |
| PTPN22 | rs971173 | 1192 | 868 | 6.592 | 0.037 |
| rs1217390 | 1191 | 864 | 8.929 | 0.012 | |
| rs878129 | 1194 | 868 | 3.462 | 0.177 | |
| rs11811771 | 1193 | 866 | 1.809 | 0.405 | |
| rs11102703 | 1194 | 868 | 3.248 | 0.197 | |
| rs7545038 | 1190 | 865 | 3.683 | 0.159 | |
| rs1503832 | 1194 | 868 | 3.534 | 0.171 | |
| rs12127377 | 1193 | 868 | 3.273 | 0.195 | |
| rs11485101 | 1194 | 868 | 1.876 | 0.391 |
Limitation and future development
The CCU statistic could only catch linear correlation between two genes, which may be insufficient to represent gene–gene co-association. For example, in the heroin addiction data, the co-associations measured between C5 and PADI4 in cases and controls were 0.16 and 0.125, the power could not be very high as shown in power calculation (Figure 1). To improve power, further work should focus on searching for approaches that could catch nonlinear co-association between genes. CCU could only deal with pairwise gene–gene co-association. Future investigation on multigene co-association is needed. In general, adoption of gene-based approach to association analysis and replication is becoming feasible with many advantages.17 CCU is likely to be a preferred option for the genetic dissection of complex diseases.
Acknowledgments
This work was supported by the grant from National Natural Science Foundation of China (30871392). We thank Dr Zhang Dandan (Fudan University) and NARAC for supplying us with the data. We also thank the reviewer of this paper for the suggestion of using the term ‘association' to fit our CCU statistic.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies the paper on European Journal of Human Genetics website (http://www.nature.com/ejhg)
Supplementary Material
References
- Wu XS, Jin L, Xiong MM. Composite measure of linkage disequilibrium for testing interaction between unlinked loci. Eur J Hum Genet. 2008;16:1160. doi: 10.1038/sj.ejhg.5202004. [DOI] [PubMed] [Google Scholar]
- Moore JH. A global view of epistasis. Nat Genet. 2005;37:13–14. doi: 10.1038/ng0105-13. [DOI] [PubMed] [Google Scholar]
- Fisher RA. The correlation between relatives on the supposition of Mendelian inheritance. Philos Trans R Soc. 1918;52:399–433. [Google Scholar]
- Cockerham CC. An extension of the concept of partitioning hereditary variance for analysis of covariances among relatives when epistasis is present. Genetics. 1954;39:859–882. doi: 10.1093/genetics/39.6.859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kempthorne O. The correlation between relatives in a random mating population. Proc R Soc Lond B Biol Sci. 1954;143:103–113. [PubMed] [Google Scholar]
- Cordell HJ. Epistasis: what it means, what it doesn't mean, and statistical methods to detect it in humans. Hum Mol Genet. 2002;11:2463–2468. doi: 10.1093/hmg/11.20.2463. [DOI] [PubMed] [Google Scholar]
- Zhao J, Jin L, Xiong M. Test for interaction between two unlinked loci. Am J Hum Genet. 2006;79:831–845. doi: 10.1086/508571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brassat D, Motsinger AA, Caillier SJ, et al. Multifactor dimensionality reduction reveals gene-gene interactions associated with multiple sclerosis susceptibility in African Americans. Genes Immun. 2006;7:310–315. doi: 10.1038/sj.gene.6364299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore JH, Gilbert JC, Tsai CT, et al. A flexible computational framework for detecting, characterizing, and interpreting statistical patterns of epistasis in genetic studies of human disease susceptibility. J Theor Biol. 2006;241:252–261. doi: 10.1016/j.jtbi.2005.11.036. [DOI] [PubMed] [Google Scholar]
- Nelson MR, Kardia SLR, Ferrell RE, Sing CF. A combinatorial partitioning method to identify multilocus genotypic partitions that predict quantitative trait variation. Genome Res. 2001;11:458–470. doi: 10.1101/gr.172901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore JH, Hahn LW. A cellular automata approach to detecting interactions among single-nucleotide polymorphisms in complex multifactorial diseases. Pac Symp Biocomput. 2002;7:53–64. [PubMed] [Google Scholar]
- Chen CH, Chang CJ, Yang WS, Chen CL, Fann CS. A genome-wide scan using tree-based association analysis for candidate loci related to fasting plasma glucose levels. BMC Genet. 2003;4 (Suppl 1:S65. doi: 10.1186/1471-2156-4-S1-S65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ccok NR, Zee RYL, Ridker PM. Tree and spline based association analysis of gene-gene interaction models for ischemic stroke. Stat Med. 2004;23:1439–1453. doi: 10.1002/sim.1749. [DOI] [PubMed] [Google Scholar]
- Bureau A, Dupuis J, Falls K, et al. Identifying SNPs predictive of phenotype using random forests. Genet Epidemiol. 2005;28:171–182. doi: 10.1002/gepi.20041. [DOI] [PubMed] [Google Scholar]
- Dong CZ, Chu X, Wang Y, et al. Exploration of gene-gene interaction effects using entropy-based methods. Eur J Hum Genet. 2008;16:229–235. doi: 10.1038/sj.ejhg.5201921. [DOI] [PubMed] [Google Scholar]
- Kang G, Yue W, Zhang J, Cui Y, Zuo Y, Zhang D. An entropy-based approach for testing genetic epistasis underlying complex diseases. J Theor Biol. 2008;250:362–374. doi: 10.1016/j.jtbi.2007.10.001. [DOI] [PubMed] [Google Scholar]
- Zhang FY, Wagener D. An approach to incorporate linkage disequilibrium structure into genomic association analysis. J Genet Genomics. 2008;35:381–385. doi: 10.1016/S1673-8527(08)60055-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gauderman WJ, Murcray C, Gilliland F, Conti DV. Testing association between disease and multiple SNPs in a candidate gene. Genet Epidemiol. 2007;31:383–395. doi: 10.1002/gepi.20219. [DOI] [PubMed] [Google Scholar]
- Oh S, Park T. Association tests based on the principal-component analysis. BMC Proc. 2007;1 (Suppl 1:S130. doi: 10.1186/1753-6561-1-s1-s130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hotelling H. Relations between two sets of variants. Biometrika. 1936;28:321–377. [Google Scholar]
- Hudson RR. Generating samples under a Wright-Fisher neutral model of genetic variation. Bioinformatics. 2002;18:337–338. doi: 10.1093/bioinformatics/18.2.337. [DOI] [PubMed] [Google Scholar]
- Marchini J, Donnelly P, Cardon LR. Genome-wide strategies for detecting multiple loci that influence complex diseases. Nat Genet. 2005;37:413–417. doi: 10.1038/ng1537. [DOI] [PubMed] [Google Scholar]
- Li WT, Reich J. A complete enumeration and classification of two-locus disease models. Hum Hered. 2000;50:334–349. doi: 10.1159/000022939. [DOI] [PubMed] [Google Scholar]
- Lawley DN. Tests of significance in canonical analysis. Biometrika. 1959;46:59–66. [Google Scholar]
- Konishi S. Normalizing transformations of some statistics in multivariate analysis. Biometrika. 1981;68:647–651. [Google Scholar]
- Fisher RA. On the ‘probable error' of a coefficient of correlation deduced from a small sample. Metron. 1921;1:3–32. [Google Scholar]
- Konishi S. An approximation to the distribution of the sample correlation coefficient. Biometrika. 1978;65:654–656. [Google Scholar]
- Konishi S. Normalizing and variance stabilizing transformations for intraclass correlations. Ann Inst Stat Math. 1985;37:87–94. [Google Scholar]
- Zhang D, Shao C, Shao M, et al. Effect of M-opioid receptor gene polymorphisms on heroin-induced subjective responses in a Chinese population. Biol Psychiatry. 2007;61:1244–1251. doi: 10.1016/j.biopsych.2006.07.012. [DOI] [PubMed] [Google Scholar]
- Plenge RM, Seielstad M, Padyukov L, et al. TRAF1-C5 as a risk locus for rheumatoid arthritis—a genomewide study. N Engl J Med. 2007;357:1199–1209. doi: 10.1056/NEJMoa073491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neale B, Sham P. The future of association studies: gene-based analysis and replication. Am J Hum Genet. 2004;75:353–362. doi: 10.1086/423901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miettinen O. Confounding and effect-modification. Am J Epidemiol. 1974;100:350–353. doi: 10.1093/oxfordjournals.aje.a112044. [DOI] [PubMed] [Google Scholar]
- Ahlbom A, Alfredsson L. Interaction: A word with two meanings creates confusion. Eur J Epidemiol. 2005;20:563–564. doi: 10.1007/s10654-005-4410-4. [DOI] [PubMed] [Google Scholar]
- Sora I, Funada M, Uhl GR. The mu-opioid receptor is necessary for [D-Pen2,D-Pen5]enkephalin-induced analgesia. Eur J Pharmacol. 1997;324:R1–R2. doi: 10.1016/s0014-2999(97)10016-4. [DOI] [PubMed] [Google Scholar]
- Becker A, Grecksch G, Brodemann R, et al. Morphine self-administration in mu-opioid receptor-deficient mice. Naunyn-Schmiedebergs Arch Pharmacol. 2000;361:584–589. doi: 10.1007/s002100000244. [DOI] [PubMed] [Google Scholar]
- Gomes I, Jordan BA, Gupta A, Trapaidze N, Nagy V, Devi LA. Heterodimerization of mu and delta opioid receptors: A role in opiate synergy. J Neurosci. 2000;20:RC110. doi: 10.1523/JNEUROSCI.20-22-j0007.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kieffer BL, Gaveriaux-Ruff C. Exploring the opioid system by gene knockout. Prog Neurobiol. 2002;66:285–306. doi: 10.1016/s0301-0082(02)00008-4. [DOI] [PubMed] [Google Scholar]
- Gomes I, Gupta A, Filipovska J, Szeto H, Pintar J, Devi L. A role for heterodimerization of mu and delta opiate receptors in enhancing morphine analgesia. Proc Natl Acad Sci USA. 2004;101:5135–5139. doi: 10.1073/pnas.0307601101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snook L, Milligan G, Kieffer B, Massotte D. mu-delta opioid receptor functional interaction: Insight using receptor-G protein fusions. J Pharmacol Exp Ther. 2006;318:683–690. doi: 10.1124/jpet.106.101220. [DOI] [PubMed] [Google Scholar]
- Jordan BA, Devi LA. G-protein-coupled receptor heterodimerization modulates receptor function. Nature. 1999;399:697–700. doi: 10.1038/21441. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.



