DNA sequencing methods have identified a large and growing number of sequence variants that are associated with disease but are of unknown biological significance and remain unclassified with regard to their pathogenic role. A large fraction of these unclassified variants (UVs), estimated between 15 and 50%, could influence RNA splicing. After two decades of basic research on RNA splicing, which have led to a detailed picture of the biochemistry of this process, the impact of splicing defects in human genetics still seems to be underestimated. Moreover, the interpretation of mutations inducing splicing defects remains difficult for the majority of clinical molecular geneticists, mainly because of the complexity of RNA splicing regulation, as illustrated schematically in Figure 1. The best-known splicing mutations are those that affect sequences at the exon/intron boundaries and induce exon skipping, or create new exon boundaries, or activate cryptic splice sites. However, more subtle effects on splicing can be induced by sequence changes that affect exonic or deep intronic splicing-regulatory elements. Thus, intronic as well as exonic changes, including those that are silent at the translation level, can induce splicing mutations. Deep intronic changes can create pseudoexons. The interpretation of splicing mutations often raises questions. For example, the predicted consequences of splicing mutations at the protein level are usually frameshifts or in-frame deletions/insertions and thus are expected to be similar to the consequences of truncating mutations or deletions/insertions caused by other mechanisms, such as insertions/deletions or rearrangements at the gene level. But splicing mutations often show the peculiar features of incomplete expressivity and of tissue specificity.1
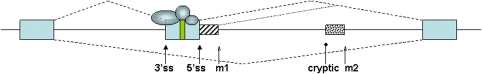
Figure 1.
Schematic overview over disease mechanisms. A hypothetical pre-mRNA is shown. Exons are indicated as boxes, introns as lines. The splicing pattern is depicted by dashed lines. Proteins that aid in exon recognition are indicated as circles. Only three proteins are shown for simplicity. Most of the pre-mRNA will be coated with proteins. An exonic regulatory element is shown by a green rectangle in the alternative exon. Mutations in this exon can alter the usage of the alternative exon and lead to a human disease, even if the mutation does not change the predicted reading frame. The 3′ and 5′ splice sites of the alternative exon (3′ss and 5′ss, respectively) are shown by solid pointed arrows. Mutations of these splice sites generally change exon usage and in most cases studied abolish the use of the alternative exon. In addition, intronic mutations that are indicated by open arrows can generate new exons, if these mutations generate new splice sites. An intronic mutation m1 that is close to the authentic 5′ splice site can lead to the generation of a cryptic exon that is indicated by a striped, smaller box and its splicing pattern is indicated by a dotted line. The deep intronic mutation m2 can lead to the formation of a new exon if it is close enough to a cryptic splice site, indicated by a round arrow. For simplicity, only mutations leading to new 5′ splice sites are shown, but new 3′ splice sites can also be generated by new mutations.
These and other related questions were addressed at the ‘Splicing and genetic diseases' workshop, sponsored by the European Alternative Splicing Network (EURASNET, www.eurasnet.info) in Paris, 1–2 October 2009, which brought together the worlds of clinical molecular geneticists and basic researchers working on alternative splicing.
The conference discussed examples of UVs detected in molecular diagnostic activities on several genes involved in mendelian diseases, such as neurofibromatosis type 1 and tuberous sclerosis (Diana Baralle), spinal muscular atrophy (Alexandra Martins, Rouen), susceptibility to breast/ovarian cancers (Claude Houdayer, Paris; Sylvie Mazoyer, Lyon), hereditary colon cancers (Mario Tosi), Usher syndrome and Duchenne and Becker muscular dystrophy (Sylvie Tuffery-Giraud, Montpellier), and cystic fibrosis (Abdel Aissad and Pascale Fanen, Créteil, France). The examples showed that the effect of mutations within the splice sites could be predicted rather accurately. However, predicting the possible effects on splicing of UVs located at deep intronic positions or at internal positions of exons can be very difficult. Exonic UVs can affect splicing-regulatory elements and induce splicing defects instead of, or in addition to, producing functionally relevant protein changes. It is therefore currently not possible to set general guidelines for predicting the effects of UVs on splicing, and functional assays are necessary to validate bioinformatics predictions or to improve them.
Patient RNA analyses are currently performed only in a minority of molecular diagnostic laboratories. Moreover, the relevant mRNA is sometimes not available, because the gene is expressed only in specialized tissues. Therefore, current efforts are, on one hand, aimed at improving bioinformatics predictions of the effects of UVs on splicing and, on the other hand, at optimizing ex vivo functional splicing assays based on the use of patient DNA and hybrid minigenes.1, 2 To rapidly test mutations in vivo, Stefan Stamm described a high-throughput cloning method for splicing reporters. This method circumvents the need of suitable restriction sites and permits the functional analysis of sequence segments ranging from small exons to large gene portions or entire genes.3
Stefan Stamm also described screening methods and rational design for developing substances that change splice site selection and could be used in therapies based on the somatic correction of splicing defects. The support for current therapeutic strategies based on the modulation of splicing, using drugs4 or antisense oligonucleotides,5 was also provided at the workshop by a report on the natural occurrence of enhanced inclusion of SMN2 exon 7, which correlates with an attenuated form of SMA in several patients (A Martins). This positive modulation of exon 7 splicing is induced by a rare SMN2 variant in a previously unrecognized composite splicing-regulatory element in the center of exon 7.6
During the conference, analysis software for human mutations was presented (André Blavier, Interactive Biosoftware, Rouen; François-Olivier Desmet and Christophe Béroud, Montpellier). Algorithms are quite accurate in predicting the effects of the variants that are located at or near the exon boundaries and of those that generate new splice sites. Therefore, bioinformatics predictions can be used for establishing priorities for analyzing UVs of the BRCA genes at the patient RNA level (Claude Houdayer). However, the effects on splicing of deep intronic variants and of most exonic variants cannot be predicted by the current software, and closer interactions between experts working in basic research on splicing and developers of software are needed to improve the accuracy of bioinformatics predictions.
Clinical geneticists would greatly benefit from the development of criteria for the classification of UVs, including those that may affect splicing.7, 8 Recent achievements toward establishing such criteria were discussed during the workshop by Sean Tavtigian (Lyon, France and Salt Lake City, USA). Statistical methods have been developed to combine data across different data types to obtain an integrated posterior probability that a variant is pathogenic. These methods have been applied to the missense changes of the BRCA1 and BRCA2 genes. Work in progress shows that it should soon be possible to integrate predictions of splice site changes into the calculation of these posterior probabilities. However, it will be difficult to account for exonic variants in general, because they cannot be predicted accurately by current algorithms.
Splicing is not only changed by germline mutations. It can also be altered as a consequence of somatic mutations especially in cancer. Didier Auboeuf (Lyon, France) discussed multiple changes of alternative splicing in tumor progression that can be detected by exon array analyses. This study identified alternative exons associated with metastatic potential in a murine model of mammary tumors. Moreover, a set of alternative splicing events identified in this model system was shown to have prognostic value in breast cancer patients.
The conference showed that tight, cooperative interaction between clinical geneticists and molecular biologists working on splicing has mutual benefits. Both worlds can learn from each other: each human mutation that gives a phenotype is an ‘experiment of nature' that shows how splicing is regulated. Reversely, basic research on splicing can help by predicting whether mutations have an effect on splicing and therefore give insight into the disease pathology. Additional meetings are needed to foster interactions, across the boundaries of particular genetic diseases, between RNA splicing experts and clinical molecular geneticists, and also to discuss practical issues of the interpretation of splicing mutations found in molecular diagnostic laboratories. The authors of this report are organizing another European workshop on splicing and disease, which is expected to take place in France in the fall of 2010.
The authors declare no conflict of interest.
References
- Baralle D, Lucassen A, Buratti E. Missed threads. The impact of pre-mRNA splicing defects on clinical practice. EMBO Rep. 2009;10:810–816. doi: 10.1038/embor.2009.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tournier I, Vezain M, Martins A, et al. A large fraction of unclassified variants of the mismatch repair genes MLH1 and MSH2 is associated with splicing defects. Human Mutat. 2008;29:1412–1424. doi: 10.1002/humu.20796. [DOI] [PubMed] [Google Scholar]
- Kishore S, Khanna A, Stamm S. Rapid generation of splicing reporters with pSpliceExpress. Gene. 2008;427:104–110. doi: 10.1016/j.gene.2008.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumanasekera C, Watt DS, Stamm S. Substances that can change alternative splice-site selection. Biochem Soc Trans. 2008;36:483–490. doi: 10.1042/BST0360483. [DOI] [PubMed] [Google Scholar]
- Aartsma-Rus A, van Ommen GJ. Progress in therapeutic antisense applications for neuromuscular disorders. Eur J Hum Genet. 2010;18:146–153. doi: 10.1038/ejhg.2009.160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vezain M, Saugier-Veber P, Goina E, et al. A rare SMN2 variant in a previously unrecognized composite splicing regulatory element induces exon 7 inclusion and reduces the clinical severity of spinal muscular atrophy. Hum Mutat. 2010;31:E1110–E1125. doi: 10.1002/humu.21173. [DOI] [PubMed] [Google Scholar]
- Tavtigian SV, Byrnes GB, Goldgar DE, Thomas A. Classification of rare missense substitutions, using risk surfaces, with genetic- and molecular-epidemiology applications. Hum Mutat. 2008;29:1342–1354. doi: 10.1002/humu.20896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spurdle AB, Couch FJ, Hogervorst FB, Radice P, Sinilnikova OM. Prediction and assessment of splicing alterations: implications for clinical testing. Hum Mutat. 2008;29:1304–1313. doi: 10.1002/humu.20901. [DOI] [PMC free article] [PubMed] [Google Scholar]