Abstract
The cytotoxic T-lymphocyte antigen 4 (CTLA4) is an inhibitory receptor expressed on activated T cells with downregulatory properties. The aim of this study was to analyse whether single-nucleotide polymorphisms (SNPs) within the CTLA4 gene are associated with the diagnosis and disease course of dilated cardiomyopathy (DCM). In two independent cohorts of DCM patients (n=251 and 223) and healthy controls (n=591), the promoter and all four exons of the CTLA4 gene, including their flanking regions, were genotyped, and the resulting allele and genotype distributions of the identified SNPs were compared between the groups. We confirmed two known SNPs in the promoter region (−318C>T) and in exon 1 (+49A>G;Thr17Ala). The allelic frequencies and genotypic distribution of the promoter SNP were similar for DCM patients compared with controls. However, the G/G genotype of the Thr17Ala variant was significantly more frequent in DCM patients compared with controls (37 out of 251 patients (14.7%) versus 44 out of 591 controls (7.4%), P=0.005). The higher frequency of the G/G genotype was confirmed in an additional DCM cohort (29 out of 223 patients (13.0%), P=0.039), indicating that this SNP functions as a risk factor for DCM. At follow-up after 1 year, the ejection fraction and the end-diastolic diameter of the left ventricle did not differ significantly between DCM patients carrying the G/G genotype versus other genotypes (n=199). Our data indicate that the common CTLA4 variant, Thr17Ala, confers susceptibility for DCM, but does not seem to influence the course of the disease 1 year after diagnosis.
Keywords: dilated cardiomyopathy, genotyping, CTLA4, T-cell activation, immune regulation
Introduction
Dilated cardiomyopathy (DCM) is a heterogeneous heart disease with variable aetiological and clinical features characterized by ventricular chamber enlargement and systolic dysfunction that ultimately result in sudden cardiac death or progressive heart failure. The aetiology of the disease is multifactorial and many clinical conditions including genetic factors, viral infections, and toxic agents can lead to DCM.1, 2, 3 There is a growing body of evidence suggesting that numerous single gene mutations have an important function in the pathogenesis of familial forms of DCMs. In particular, genes encoding cytoskeletal, sarcomeric, and nuclear proteins, which are engaged in extracellular anchorage or myocardial contractility, have been linked to DCM.3, 4 Besides base substitutions in the mitochondrial genome, which can also cause a DCM-type disease,5 these mutations critically diminish force generation, transduction, and/or transmission within the contractile apparatus of the myocardium and lead to impaired systolic function. However, these sarcomeric gene defects explain the disease only in some families, leaving other forms of inherited DCM subentities unresolved.
As one of the major factors in DCM pathogenesis involves autoimmune-mediated damage to cardiac tissue resulting from viral infection, candidate genes that are involved in regulating immune reactions have recently come into the focus of genetic research.1, 6 Numerous genetic studies carried out to identify novel DCM susceptibility genes have pointed to genetic variants within the human leukocyte antigen (HLA) locus that confer risk factors for the development or persistence of immune reactions within the myocardium.7 Abnormal expression of HLA antigens and mutations within HLA molecules have been suggested to have a causative function in the pathogenesis of DCM. Circulating auto-antibodies are frequently found in DCM patients that arise from ongoing inflammatory processes against cardiac tissue, and components of the major histocompatibility complex may serve as markers for the propensity to develop immune-mediated myocardial damage.2, 7, 8 From this earlier evidence linking genetic susceptibility for DCM to certain HLA class II alleles, it is conceivable that other proteins with immune regulatory functions may also be involved in the cascade of immune events leading to DCM pathogenesis. One candidate gene involved in DCM encodes the cytotoxic T-lymphocyte antigen 4 (CTLA4), a receptor molecule expressed on activated T lymphocytes, which functions as an important negative regulator of T-cell activation.9 The CTLA4 gene has been associated with a variety of autoimmune disorders including diabetes mellitus, rheumatoid arthritis, and autoimmune endocrinopathies,10, 11, 12 but so far, an association with heart disease has only been investigated once.9 In a small Chinese sample, Liu et al reported on a relationship between CTLA4 polymorphism and genetic susceptibility to idiopathic DCM. In the study presented here, we asked whether CTLA4 genotypes might influence susceptibility to DCM in Caucasians as well and, indeed, we found that a homozygous G allele at position 49 in exon 1 is significantly associated with the disease phenotype, but does not seem to influence the course of the disease within the first year after diagnosis.
Patients and methods
Subjects and clinical evaluation
Patients who were referred to the cardiology department at the University of Marburg with clinical signs of heart failure were consecutively screened to identify individuals with DCM. The diagnostic criteria for DCM were defined as a left ventricular ejection fraction (EF) <45% measured echocardiographically and/or a left ventricular end-diastolic diameter (LVEDD) >56 mm, in the absence of secondary causes of heart failure. Routinely, patients were evaluated by personal and family history, physical examination, 12-lead electrocardiography, chest X-ray, and transthoracic echocardiography including M-mode, two dimensional and Doppler echocardiography. In all DCM patients, clinically relevant coronary artery stenosis was excluded using coronary angiography. In patients from the DCM study cohort, we routinely performed endomyocardial biopsies. Patients who fulfilled the diagnostic criteria for DCM were invited to participate in the study and written informed consent was acquired. As shown in Table 1, the DCM study group consisted of 251 participants, 184 of whom were male (73.3%). The mean age at study recruitment was 50.2 ± 12.9 years. Comorbid immunological diseases were present in 40 patients, with allergy (n=31), psoriasis (n=10), ankylosing spondylitis (n=3), rheumatoid arthritis (n=1), and Hashimoto thyroiditis (n=1) as the main diagnoses. The mean LVEDD at study inclusion was 67.8 ± 8.0 mm and the mean EF was 29.8%, with a standard deviation of 8.3%. The following viruses were routinely tested in endomyocardial biopsies from DCM patients by means of polymerase chain reactions (PCR) (with the percentage of positive results given in brackets): parvovirus B19 (26.4%), Epstein–Barr virus (6.9%), human herpesvirus-6 (1.1%), coxsackievirus B3 (0.7%), herpes simplex virus (0.4%), and cytomegalovirus (0.7%). At 13.2±2.4 months after diagnosis, 199 patients out of the Marburg cohort were reexamined clinically, including non-invasive measurement of EF and LVEDD.
Table 1. Demographic and cardiological parameters of the two independent DCM study cohorts.
| DCM study group (n=251) | DCM additional study group (n=223) | |
|---|---|---|
| Male sex | 184 (73.3%) | 162 (72.6%) |
| Age (years) | 50.2±12.9 | 59.2±13.3 |
| LVEDD (mm) at baseline (n=251) | 67.8±8.0 | 66.7±6.9 |
| LVEDD (mm) at follow-up (n=199) | 64.4±9.4 | n.p. |
| EF (%) at baseline (n=251) | 29.8±8.3 | 31.5±8.6 |
| EF (%) at follow-up (n=199) | 39.2±12.5 | n.p. |
Abbreviations: EF, ejection fraction; LVEDD, left ventricular end-diastolic diameter; n.p., not performed.
Parameters presented as means±SD.
An additional cohort of 223 DCM patients recruited at the University hospital Charité in Berlin was examined for the presence of intragenic polymorphisms at position −318 and +49. The same established criteria for the diagnosis of DCM were used as for the Marburg cohort. A total of 591 clinically healthy subjects without a history of heart failure formed the control group. This independent control sample consisted of blood donors from the local Blood Transfusion Service who came from the same general geographical area as the study population. All control subjects were free of disease as judged by medical examination. Blood samples were obtained from each study participant by venous punction in EDTA anticoagulant-containing tubes and stored at −80°C until further use. The study protocol was approved by the Ethics Committees of the University of Marburg and by the University hospital Charité, respectively.
Laboratory methods
Genomic DNA of each individual was extracted from peripheral blood mononuclear cells by a DNA isolation kit (QIAamp DNA Blood Mini kit from Qiagen, Hilden, Germany) according to the manufacturer's instruction. PCR were performed to amplify DNA fragments from the entire four exons, including their flanking regions. The following oligonucleotide primer pairs with the addition of a GC-clamp to one of each primer were used: ctla4-exon1 5′-GGATCCTGAAAGGTTTTGCTC-3′ and 5′-(GxCy)n-TGCTGAAACAAATGAAACCC-3′, ctla4-exon2 5′-AAGCTAGAAGGCAGAAGGGC-3′ and 5′-(GxCy)n-ACAGTCCTAAAAGTAAACCTCC-3′, ctla4-exon3 5′-GGTATCCCCATCAGACATGG-3′ and 5′-(GxCy)n-TGATGCTCCTGGGGAAGTAG-3′, and ctla4-exon4 5′-ATGGTTAGAAGTGGCTTCCG -3′ and 5′-(GxCy)n-AGAATTGCCTCAGCTCTTGG-3′. PCR reactions were performed in a total volume of 25 μl, including 10x PCR buffer, 1.5 mM MgCl2, 0.15 mM of each dNTP, 0.5 mM of each specific primer pair, 100 ng of genomic DNA, and 1U of Taq DNA polymerase. The PCR conditions were 95°C for 10 min, followed by 35 cycles of denaturation at 95°C for 60 s, annealing at 61°C for 60 s, and extension at 72°C for 60 s, with a final elongation at 72°C for 7 min. Genotyping was carried out by denatured gradient gel electrophoresis using a 20–60% urea/formamide gradient in 12% acrylamide, 1 × Tris/acetic acid/EDTA buffer (300V, 60°C for 6 h). The gels were stained for separated DNA fragments with ethidium bromide. The amplified PCR products were randomly sequenced to validate the genotyping assay.
Single-nucleotide polymorphisms (SNPs) within the promoter region and exon 1 were detected by means of restriction fragment length polymorphism. Briefly, PCR reactions were performed using the following two primer pairs: ctla4-prom 5′-AAATGAATTGGACTGGATGGT-3′ and 5′-TTACGAGAAAGGAAGCCGTG-3′, and ctla4-ex1 5′-CTGAAGACCTGAACACCGCTCC-3′ and 5′-CCAGGTAGGAGAAACACCTCCT-3′. The C>T SNP at position −318 was detected by incubating the amplified PCR product with 3U of the restriction enzyme MseI (New England Biolabs, Frankfurt, Germany) for 4 h, yielding additional 110 and 116 bp fragments in the case of the T allele and leaving the 226 bp product of the C allele intact. The G substitution at position +49 gave rise to a BvbI restriction enzyme recognition site and, therefore, after PCR amplification, the 192 bp PCR product was digested with 3U of BvbI (New England Biolabs) for 4 h at 37°C, which yielded 112 and 80 bp fragments. The resulting DNA fragments were visualized on a 2.4% agarose gel. DNA sequencing using a modified didesoxy method confirmed the presence of the two SNPs.
Statistical analysis
Allele and genotype frequencies were determined in cases and controls by direct counting. Differences in global allele and genotype distributions were tested for significance by means of χ2 tests using 2 × 2 or 2 × 3 contingency tables. The genotype frequencies were tested for Hardy–Weinberg equilibrium by χ2 analyses. Power calculations for α=0.050 were performed using the SigmaStat software for Windows (Systat). P≤0.05 was regarded as statistically significant.
Results
Genotype analysis of all four exons and their flanking non-coding regions revealed SNPs at two known positions within the CTLA4 gene: −318C>T (rs5742909) and +49A>G (rs231775). The distribution of the allelic and genotypic frequencies of the two polymorphisms in both DCM cohorts and the control group is shown in Table 2. Both polymorphisms met the criteria for Hardy–Weinberg equilibrium.
Table 2. The allele and genotype frequency distribution of CTLA4 polymorphisms in two independent samples of DCM patients versus control subjects.
| Polymorphisms (genotypes and alleles) | DCM study group (n=251) | DCM additional study group (n=223) | Controls (n=591) |
|---|---|---|---|
| −318C>T genotype CC:CT:TT | 205:45:1 | 182:30:1 | 494:110:3 |
| Percentage | 81.7%:17.9%:0.4% | 85.4%:14.1%:0.5% | 81.4%:18.1%:0.5% |
| χ2-value | 0.04 | 1.82 | |
| P-value | n.s. | n.s. | |
| −318C>T allele C:T | 455:45 | 394:32 | 1098:116 |
| Percentage | 90.6%:9.4% | 92.5%:7.5% | 90.4%:9.6% |
| χ2-value | 0.07 | 2.49 | |
| P-value | n.s | n.s | |
| +49A>G genotype AA:AG:GG | 90:124:37 | 93:101:29 | 264:283:44 |
| Percentage | 35.9%:49.4%:14.7% | 41.7%:45.3%:13.0% | 44.7%:47.9%:7.4% |
| χ2-value | 13.09 | 6.14 | |
| P-value | 0.001 | 0.047 | |
| +49A>G allele A:G | 304:198 | 287:159 | 811:371 |
| Percentage | 60.6%:39.4% | 64.3%:35.7% | 68.6%:31.4% |
| χ2-value | 9.12 | 2.49 | |
| P-value | 0.003 | n.s. |
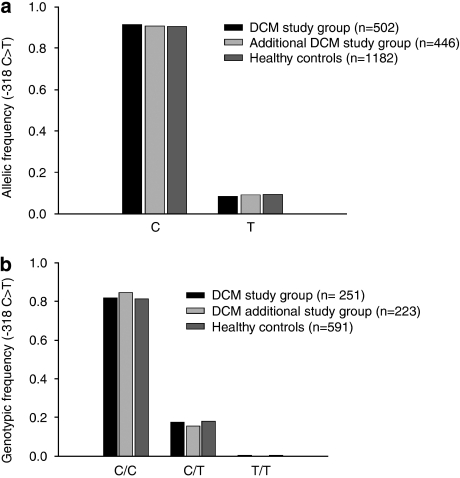
The overall distribution of the −318C>T polymorphism, whether expressed as genotypic or as allelic frequency, found in the three study groups was very similar to that described by others in Caucasian subjects (Figure 1).13 In the majority of DCM patients (81.7%) and healthy controls (81.4%), a C/C genotype at position −318 was identified, whereas only 0.4% of DCM patients and 0.5% of controls were genotyped as T/T (Figure 1b). The high frequency of the C/C genotype was confirmed in the additional DCM study cohort (85.4%) (Figure 1b). There was no significant difference between the allelic and genotypic distribution of the two DCM cohorts compared with normal controls, suggesting that the −318C>T SNP in the promoter region is not associated with DCM.
Figure 1.
The allelic (a) and genotypic (b) frequencies of the −318C>T polymorphism in the promoter of the CTLA4 gene did not significantly differ between two independent DCM study cohorts and healthy controls.
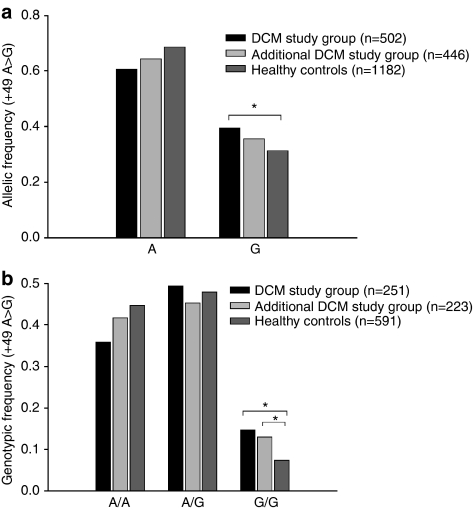
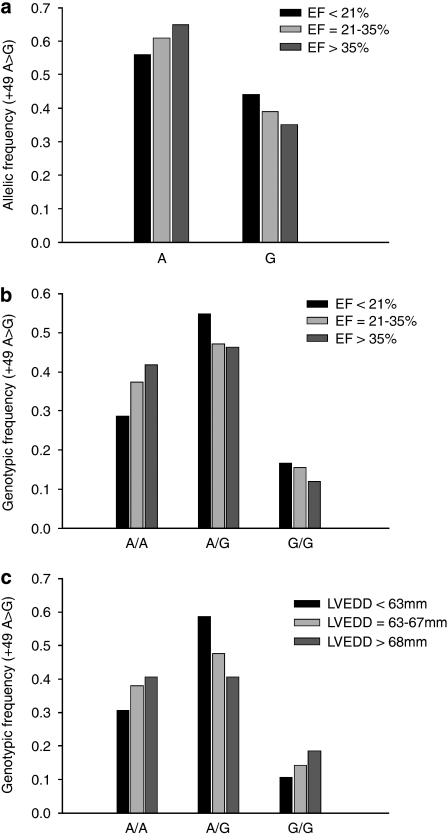
For the G allele in the +49A>G polymorphism, the allelic frequency differed between the two DCM cohorts (39.4% in the Marburg cohort and 35.7% in the Berlin cohort) and controls (31.4%) (Figure 2a). This reached statistical significance in the Marburg cohort, but not in the Berlin cohort (P=0.03 and 0.27, respectively). When we tested for genotype distribution, we found significant differences between both DCM cohorts and healthy control subjects (Figure 2b). The G/G genotype was detected more often in the Marburg cohort than in healthy controls (37 out of 251 patients (14.7%) versus 44 out of 591 controls (7.4%), P=0.005). The higher frequency of the G/G genotype in DCM patients was confirmed in the Berlin cohort (29 out of 223 patients (13.0%), P=0.039), indicating that the homozygous G/G genotype is significantly associated with the diagnosis of DCM. There was no such association between comorbid autoimmune diseases and the genotypic pattern. Even when all DCM patients with comorbid immunological diseases (n=40) were excluded from the DCM study cohort, the association between genotype pattern and DCM diagnosis in this sub-population of ‘pure' DCM patients remained stable (DCM patients n=211; χ2=10.19; P=0.006), suggesting that comorbidity with immunological diseases did not function as a confounder in this relationship. The G/G genotype was more often detected in patients belonging to the lowest EF class (<21%) and the highest LVEDD class (>68 mm). However, these trends did not reach statistical significance (Table 3; Figure 3a–c).
Figure 2.
(a) The allelic distribution of the +49A>G polymorphism in exon 1 of the CTLA4 gene showed significant differences between DCM patients and normal controls. (b) The G/G genotype was more frequently detected in patients from two independent DCM study samples as compared with controls (P<0.05, marked with brackets).
Table 3. Genotype frequencies and allele counts of the CTLA4 +49A>G polymorphism in DCM patients relative to the left ventricular ejection fraction (EF) and end-diastolic diameter (LVEDD) as stratified in three classes.
| Polymorphisms (genotypes and alleles) | EF <21% (n=42) | EF 21–35% (n=142) | EF > 35% (n=67) | Significance |
|---|---|---|---|---|
| Genotypes | ||||
| A/A | 12 (28.57%) | 53 (37.32) | 28 (41.79%) | n.s. |
| A/G | 23 (48.94%) | 67 (47.18%) | 31 (46.27%) | n.s. |
| G/G | 7 (16.67%) | 22 (15.49%) | 8 (11.94%) | n.s. |
| Alleles | ||||
| A | 47 (55.95%) | 173 (60.92%) | 87 (64.93%) | n.s. |
| G | 37 (44.05%) | 111 (39.08%) | 47 (35.07%) | n.s. |
| Polymorphisms (genotypes and alleles) | LVEDD <63 mm (n=75) | LVEDD 63-67 mm (n=63) | LVEDD >67 mm (n=113) | Significance |
| Genotypes | ||||
| A/A | 23 (30.67%) | 24 (38.10%) | 46 (40.71%) | n.s. |
| A/G | 44 (58.67%) | 30 (47.62%) | 46 (40.71%) | n.s. |
| G/G | 8 (10.67%) | 9 (14.29%) | 21 (18.58%) | n.s. |
| Alleles | ||||
| A | 90 (60.00%) | 78 (61.90%) | 138 (61.06%) | n.s. |
| G | 60 (40.00%) | 48 (38.10%) | 88 (38.94%) | n.s. |
Figure 3.
The G allele in the +49A>G polymorphism was more often identified in DCM patients belonging to the lowest EF class (<21%) (a, b) and the highest LVEDD class (>68 mm) (c); however, neither reached statistical significance.
To assess whether our sample size reached the minimum requirements to achieve adequate power, we conducted post hoc power calculations. For the +49A/G SNP, the power of the performed tests for both allele frequency and genotype distribution between the DCM study group and control subjects was above the desired power of 0.8 (0.873 and 0.923, respectively). However, in case of the −318C>T SNP, the power calculations failed to reach the level of significance, suggesting that a larger sample size is needed to exclude false-negative results.
At follow-up, the mean LVEDD was significantly decreased to 64.4 mm, with a standard deviation of 9.4 mm, and the mean EF was increased to 39.2±12.5%. However, parameters did not differ between DCM patients carrying either the G/G, A/A, or G/A genotype, suggesting that in our small sample size, the course of disease was not influenced by this SNP.
Discussion
Given its function as a negative regulator of T-cell activation, CTLA4 has long been implicated in diverse autoimmune conditions, such as diabetes mellitus,14, 15 Graves' disease,16, 17 Hashimoto thyroiditis,18, 19 rheumatoid arthritis,20, 21 systemic lupus erythematosus,22 and others.13, 15, 23 Although autoimmune reactions within the myocardium have been observed in some subentities of DCM, studies on the involvement of CTLA4 in the pathogenesis of the disease are rare. Liu et al9 have performed a genotyping approach in a small sample of DCM patients and identified an SNP at position 49 within the CTLA4 gene that was associated with DCM.
CTLA4 receptor in the pathogenesis of DCM
The CTLA4 receptor is a member of the CD28-B7 immunoglobulin superfamily of immune regulatory molecules with downregulating properties on T-cell activation.24, 25, 26 It is expressed on the plasma membrane of activated T cells and provides an inhibitory signal for T-cell proliferation after binding to B7 receptor molecules on the surface of antigen-presenting cells. Binding of CTLA4 to its B7 ligand interferes with the activation pathway and negatively affects the production of several cytokines, including interleukin-2, and the proliferation of T cells.27, 28 Activation of naive T cells requires two signals: (i) antigen recognition through T-cell receptor binding to the peptide–MHC complex and (ii) an antigen-independent pathway provided by interaction between CD28 expressed on the T-cell surface and B7. The CD28 homologue CTLA4 competes with the costimulatory CD28 receptor for binding to its ligands B7-1 (CD80) and B7-2 (CD86). As a putative mechanism for its inhibitory effect on CD28-mediated T-cell signalling, CTLA4 might exclude CD28 from the immunological synapse, as CTLA4 exhibits significantly higher affinities for both B7 ligands than its CD28 counterpart.28 Under suboptimal activation conditions, CTLA4 seems to attenuate weak signals mediated by the T-cell antigen receptor, thereby contributing to peripheral tolerance. Recently, the crucial function of CTLA4 in maintaining FoxP3+ regulatory T-cell function has been revealed.29, 30
The human CTLA4 gene is known to contain polymorphisms in three distinct regions: a C>T single-base substitution in the promoter at position −318, an A/G dimorphism in exon 1 at position 49, and a multiallelic dinucleotide repeat in the 3′ untranslated region.12, 31, 32 When we screened the four coding exons and the promoter of the CTLA gene for genetic variability in DCM patients and healthy controls, we confirmed the presence of known SNPs in the promoter and in exon 1. We observed that the allelic distribution of the promoter SNP was not associated with the prevalence of DCM. However, our data show a significantly increased frequency of the G/G genotype in exon 1 in two independent populations of DCM patients as compared with healthy controls, indicating the involvement of the Thr17Ala polymorphism in the pathogenesis of DCM.
Comorbid autoimmune diseases did not influence the frequency of the allelic distribution in the DCM study group; thus, the finding of an association between +49 exon 1 variants and DCM supports a functional role of CTLA4 in the development and/or maintenance of particular subentities of DCMs.11 The DCM-associated G/G genotype results in an amino-acid exchange from threonine to alanine in the peptide leader sequence of the CTLA4 receptor. This mutation is identical with the one identified to be engaged in other autoimmune diseases, suggesting that this amino-acid exchange confers susceptibility to autoimmunity in general, but cannot account for determining organ specificity. Ligers et al12 have shown that individuals carrying thymine at position −318 of the CTLA4 promoter or homozygous for adenine at position 49 in exon 1 showed a significantly increased expression of cell-surface CTLA4 receptor molecules on stimulation of T cells.12 Earlier, FACS analysis of CTLA4 expression has revealed a reduced upregulation of CTLA4 from G/G donors after suboptimal stimulation of T cells, compared with cells expressing the wild-type protein.33 Anjos et al34 reported on an incomplete glycosylation and reduced ratio of cell surface/total CTLA4-Ala17 versus CTLA4-Thr17, suggesting that the mutant signal peptide leads to inefficient processing of the autoimmune-predisposing alanine variant in the endoplasmic reticulum.
Our observation of the linking of the CTLA4 gene to DCM suggests that ongoing autoimmune responses against myocardial tissue may be critical factors involved in particular forms of the disease. Unbalanced immune stimulation caused by infiltrating T cells may lead to progressive myocytolysis, apoptotic cell death, and continuous loss of cardiomyocytes, which ultimately damage the heart organ and precipitate DCM. In line with our findings, it had been reported that transgenic mice deficient in CTLA4 showed diffuse and focal lymphocyte infiltration in heart tissue and probably died of myocardial failure as a result of these lesions. Interestingly, in 37 patients from the DCM study cohort (14.7%), endomyocardial biopsy revealed evidence of focal lymphocytic infiltration of the myocardium, closely mimicking the histological features in the hearts of CTLA4-knockout mice.24, 25
Although our study supports independent effects of the +49A>G polymorphism on the pathogenesis of DCM, we do not claim to have defined an aetiological function of this SNP in the disease. The three members of the costimulatory receptor family, CD28, CTLA4, and ICOS, encoded in a short genomic interval, have complementary effects on T-cell activation and their balance has been shown to control the overall outcome of immune responses.35 Butty et al35 have shown that functional variation in the CD28/CTLA4/ICOS region may not be due to polymorphism in a single gene, but to the combination of genetic variants at the three loci. Thus, SNPs in other costimulatory receptors may also be associated with DCM and should be systematically investigated in future studies. Furthermore, studies on DCM-associated polymorphisms in other ethnic populations are warranted, because, owing to diverse environmental and microbial challenges, the association between immune responsiveness and DCM may differ in other geographical regions of the world.35
Study limitations
There are certain limitations in our study design. First, we do not have demographic data from the anonymized control subjects. Although we are confident that all blood donors from our local Blood Transfusion Service were free of disease and particularly did not suffer from symptoms of heart failure, we have no valid information about their ethnic background. Nevertheless, the tested allele and genotype distributions of the CTLA4 SNPs in our control sample closely resemble those reported in other Caucasian populations, but substantially deviate from non-Caucasian samples. The low number of immigrants from non-European countries in the rural geographical area of Marburg suggests that there is no selection bias between cases and controls. In the light of the strong association observed for our single-marker analysis, we believe that our data add weight to the idea that CTLA4 is a candidate gene for the development of DCM. However, the possibility that our observation might derive from an artefact caused by ethnic inhomogeneity resulting from biased stratification cannot be entirely excluded. Second, although the sample size was large enough to detect significant differences for the +49A>G SNP, it might be too small for the −318C>T polymorphism, as indicated from power calculations. Thus, we recommend epidemiological studies in larger, well-defined cohorts based on case–control methods. A third study limitation is related to the follow-up period of 1 year, which might be too short to detect significant differences in disease outcome. Given the aetiological heterogeneity of DCM, longitudinal studies with larger numbers of samples are required to elucidate whether CTLA4 polymorphisms might affect the course of the disease.
Conclusion
Taken together, the epidemiologic data presented in a Caucasian population show evidence for an association of the CTLA4 +49A>G polymorphism with DCM. The genotype carrying the homozygous G/G polymorphism was shown to confer an increased risk of developing the disease. The function of CTLA4 as a susceptibility gene for DCM suggests that, in some patients, upregulated immune reactions in the myocardium contribute to inflammatory responses and favour reparative fibrosis. However, the genotype pattern did not seem to influence the course of the disease within the first year after diagnosis.
The authors declare no conflict of interest.
References
- Maekawa Y, Ouzounian M, Opavsky MA, Liu PP. Connecting the missing link between dilated cardiomyopathy and viral myocarditis. Virus, cytoskeleton, and innate immunity. Circulation. 2007;115:5–8. doi: 10.1161/CIRCULATIONAHA.106.670554. [DOI] [PubMed] [Google Scholar]
- Cihakova D, Rose NR. Pathogenesis of myocarditis and dilated cardiomyopathy. Adv Immunol. 2008;99:95–114. doi: 10.1016/S0065-2776(08)00604-4. [DOI] [PubMed] [Google Scholar]
- Morimoto S. Sarcomeric proteins and inherited cardiomyopathies. Cardiovasc Res. 2008;77:659–666. doi: 10.1093/cvr/cvm084. [DOI] [PubMed] [Google Scholar]
- Richard P, Villard E, Charron P, Isnard R. The genetic bases of cardiomyopathies. J Am Coll Cardiol. 2006;48:A79–A89. [Google Scholar]
- Ruppert V, Nolte D, Aschenbrenner T, Pankuweit S, Funck R, Maisch B. Novel point mutations in the mitochondrial DNA detected in patients with dilated cardiomyopathy by screening the whole mitochondrial genome. Biochem Biophy Res Commun. 2004;318:535–543. doi: 10.1016/j.bbrc.2004.04.061. [DOI] [PubMed] [Google Scholar]
- Mason JW. Myocarditis and dilated cardiomyopathy: an inflammatory link. Cardiovasc Res. 2003;60:5–10. doi: 10.1016/s0008-6363(03)00437-1. [DOI] [PubMed] [Google Scholar]
- Li HS, Ligons DL, Rose NR. Genetic complexity of autoimmune myocarditis. Autoimm Rev. 2008;7:168–173. doi: 10.1016/j.autrev.2007.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maisch B, Richter A, Sandmöller A, Portig I, Pankuweit S, BMBF-Heart Failure Network Inflammatory dilated cardiomyopathy. Herz. 2005;30:535–544. doi: 10.1007/s00059-005-2730-5. [DOI] [PubMed] [Google Scholar]
- Liu W, Li WM, Gao C, Wang XR, Li DM, Sun NL. [Relationship of CTLA-4 exon 1 A49 → G polymorphism with sCTLA-4 and Th1/Th2 bias in idiopathic dilated cardiomyopathy]. (Article in Chinese) Zhonghua Yi Xue Zhi. 2005;85:3221–3224. [PubMed] [Google Scholar]
- Ueda H, Howson JM, Esposito L, et al. Association of the T-cell regulatory gene CTLA4 with susceptibility to autoimmune disease. Nature. 2003;423:506–511. doi: 10.1038/nature01621. [DOI] [PubMed] [Google Scholar]
- Gough SC, Walker LS, Sansom DM. CTLA4 gene polymorphism and autoimmunity. Immunol Rev. 2005;204:102–115. doi: 10.1111/j.0105-2896.2005.00249.x. [DOI] [PubMed] [Google Scholar]
- Ligers A, Teleshova N, Masterman T, Huang WX, Hillert J. CTLA-4 gene expression is influenced by promoter and exon 1 polymorphisms. Genes Immun. 2001;2:145–152. doi: 10.1038/sj.gene.6363752. [DOI] [PubMed] [Google Scholar]
- Kristiansen OP, Larsen ZM, Pociot F. CTLA-4 in autoimmune diseases – a general susceptibility gene to autoimmunity. Genes Immun. 2000;1:170–184. doi: 10.1038/sj.gene.6363655. [DOI] [PubMed] [Google Scholar]
- Marron MP, Raffel LJ, Garchon HJ, et al. Insulin-dependent diabetes mellitus (IDDM) is associated with CTLA4 polymorphisms in multiple ethnic groups. Hum Mol Genet. 1997;6:1275–1282. doi: 10.1093/hmg/6.8.1275. [DOI] [PubMed] [Google Scholar]
- Zhernakova A, Eerligh P, Barrera P, et al. CTLA4 is differentially associated with autoimmune diseases in the Dutch population. Hum Genet. 2005;118:58–66. doi: 10.1007/s00439-005-0006-z. [DOI] [PubMed] [Google Scholar]
- Donner H, Rau H, Walfish PG, et al. CTLA4 alanine-17 confers genetic susceptibility to Graves' disease and to type I diabetes mellitus. J Clin Endocrinol Metab. 1997;82:143–146. doi: 10.1210/jcem.82.1.3699. [DOI] [PubMed] [Google Scholar]
- Kouki T, Sawai Y, Gardine CA, Fisfalen ME, Alegre ML, DeGroot LJ. CTLA-4 gene polymorphism at position 49 in exon 1 reduces the inhibitory function of CTLA-4 and contributes to the pathogenesis of Graves' disease. J Immunol. 2000;165:6606–6611. doi: 10.4049/jimmunol.165.11.6606. [DOI] [PubMed] [Google Scholar]
- Donner H, Braun J, Seidl C, et al. Codon 17 polymorphism of the cytotoxic T lymphcyte antigen 4 gene in Hashimoto's thyroiditis and Addison's disease. J Clin Endocrinol Metab. 1997;82:4130–4132. doi: 10.1210/jcem.82.12.4406. [DOI] [PubMed] [Google Scholar]
- Chistiakov DA, Turakulov RI. CTLA-4 and its role in autoimmune thyroid disease. J Mol Endocrinol. 2003;31:21–36. doi: 10.1677/jme.0.0310021. [DOI] [PubMed] [Google Scholar]
- Seidl C, Donner H, Fischer B, et al. CTLA4 codon 17 dimorphism in patients with rheumatoid arthritis. Tissue Antigens. 1998;51:62–66. doi: 10.1111/j.1399-0039.1998.tb02947.x. [DOI] [PubMed] [Google Scholar]
- Vaidya B, Pearce SH, Charlton S, et al. An association between the CTLA4 exon 1 polymorphism and early rheumatoid arthritis with autoimmune endocrinopathies. Rheumatol. 2002;41:180–183. doi: 10.1093/rheumatology/41.2.180. [DOI] [PubMed] [Google Scholar]
- Barreto M, Santos E, Ferreira R, et al. Evidence for CTLA4 as a susceptibility gene for systemic lupus erythematosus. Eur J Human Genet. 2004;12:620–626. doi: 10.1038/sj.ejhg.5201214. [DOI] [PubMed] [Google Scholar]
- Blomhoff A, Lie BA, Myhre AG, et al. Polymorphisms in the cytotoxic T lymphocyte antigen-4 gene region confer susceptibility to Addison's disease. J Clin Endocrinol Metabo. 2004;89:3474–3476. doi: 10.1210/jc.2003-031854. [DOI] [PubMed] [Google Scholar]
- Tivol EA, Borriello F, Schweitzer AN, Lynch WP, Bluestone JA, Sharpe AH. Loss of CTLA-4 leads to massive lymphoproliferation and fatal multiorgan tissue destruction, revealing a critical negative regulatory role of CTLA-4. Immunity. 1995;3:541–547. doi: 10.1016/1074-7613(95)90125-6. [DOI] [PubMed] [Google Scholar]
- Waterhouse P, Penninger JM, Timms E, et al. Lymphoproliferative disorders with early lethality in mice deficient in Ctla-4. Science. 1995;270:985–988. doi: 10.1126/science.270.5238.985. [DOI] [PubMed] [Google Scholar]
- Chambers CA, Sullivan TJ, Allison JP. Lymphoproliferation in CTLA-4-deficient mice is mediated by costimulation-dependent activation of CD4+ T cells. Immunity. 1997;7:885–895. doi: 10.1016/s1074-7613(00)80406-9. [DOI] [PubMed] [Google Scholar]
- Stamper CC, Zhang Y, Tobin JF, et al. Crystal structure of the B7-1/CTLA-4 complex that inhibits human immune responses. Nature. 2001;410:608–611. doi: 10.1038/35069118. [DOI] [PubMed] [Google Scholar]
- Pentcheva-Hoang T, Egen JG, Wojnoonski K, Allison JP. B7-1 and B7-2 selectively recruit CTLA-4 and CD28 to the immunological synapse. Immunity. 2004;21:401–413. doi: 10.1016/j.immuni.2004.06.017. [DOI] [PubMed] [Google Scholar]
- Tang AL, Teijaro JR, Njau MN, et al. CTLA4 expression is an indicator and regulator of steady-state CD4+FoxP3+ T cell homeostasis. J Immunol. 2008;181:1806–1813. doi: 10.4049/jimmunol.181.3.1806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedline RH, Brown DS, Nguyen H, et al. CD4+ regulatory T cells require CTLA-4 for the maintenance of systemic tolerance. J Exp Med. 2009;206:421–434. doi: 10.1084/jem.20081811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang XB, Zhao X, Giscombe R, Lefvert AK. A CTLA-4 gene polymorphism at position -318 in the promoter region affects the expression of protein. Genes Immun. 2002;3:233–234. doi: 10.1038/sj.gene.6363869. [DOI] [PubMed] [Google Scholar]
- Atabani SF, Thio CL, Divanovic S, et al. Association of CTLA4 polymorphism with regulatory T cell frequency. Eur J Immunol. 2005;35:2157–2162. doi: 10.1002/eji.200526168. [DOI] [PubMed] [Google Scholar]
- Mäurer M, Loserth S, Kolb-Mäurer A, et al. A polymorphism in the human cytotoxic T-lymphocyte antigen 4 (CTLA4) gene (exon 1 +49) alters T-cell activation. Immunogenetics. 2002;54:1–8. doi: 10.1007/s00251-002-0429-9. [DOI] [PubMed] [Google Scholar]
- Anjos S, Nguyen A, Ounissi-Benkalha H, Tessier MC, Polychronakos C. A common autoimmunity predisposing signal peptide variant of the cytotoxic T-lymphocyte antigen 4 results in inefficient glycosylation of the susceptibility allele. J Biol Chem. 2002;277:46478–46486. doi: 10.1074/jbc.M206894200. [DOI] [PubMed] [Google Scholar]
- Butty V, Roy M, Sabeti P, Besse W, Benoist C, Mathis D. Signatures of strong population differentiation shape extended haplotypes across the human CD28, CTLA4, and ICOS costimulatory genes. Proc Natl Acad Sci USA. 2007;104:570–575. doi: 10.1073/pnas.0610124104. [DOI] [PMC free article] [PubMed] [Google Scholar]