Abstract
A recent genome-wide association study conducted by the International Multiple Sclerosis Genetic Consortium (IMSGC) identified, among others, a number of putative multiple sclerosis (MS) susceptibility variants at position 1p22. Twenty-one SNPs positively associated with MS were located at the GFI-EVI5-RPL5-FAM69A locus. In this study, we performed an analysis and fine mapping of this locus, genotyping eight Tag-SNPs in 732 MS patients and 974 controls from Spain. We observed an association with MS in three of eight Tag-SNPs: rs11804321 (P=0.008, OR=1.29; 95% CI=1.08–1.54), rs11808092 (P=0.048, OR=1.19; 95% CI=1.03–1.39) and rs6680578 (P=0.0082, OR=1.23; 95% CI=1.07–1.41). After correcting for multiple comparisons and using logistic regression analysis to test the addition of each SNP to the most associated SNPs, we observed that rs11804321 alone was sufficient to model the association. This Tag-SNP captures two SNPs in complete linkage disequilibrium (r2=1), both located within the 17th intron of the EVI5 gene. Our findings agree with the corresponding data of the recent IMSGC study and present new genetic evidence that points to EVI5 as a factor of susceptibility to MS.
Keywords: Multiple sclerosis, Tag-SNP analysis, polymorphisms, GFI1-EVI5-RPL5-FAM69A genes, association, GWAS
INTRODUCTION
Multiple sclerosis (MS) is a complex disease presumed to be autoimmune and characterized by inflammation and demyelination with axonal and neuronal degeneration. MS is the most common central nervous system disease in young adults.1 The prevalence and incidence rates in Spain are around 77/1 00 000 habitants and 5.3/1 00 000 habitants per year, respectively, similar to that found in Britain.2, 3 Susceptibility to MS is thought to be conferred by the combination of genetic and environmental factors.1, 4
The best-established region implicated in predisposition to MS is the major histocompatibility complex on chromosome 6p21, specifically the HLA-DRB1* 1501 class II allele, but this accounts for less than 50% of MS genetics.1, 5 Recently, other regions have been implicated in MS susceptibility and replicated in different independent populations such as the interleukin 7 receptor alpha (IL7RA),6, 7, 8 the interferon regulatory factor 5 (IRF5) gene9 and the interleukin-2 receptor alpha (IL2RA).8, 10 However, except for IL7RA, the causal SNP of the new determined risk loci and the functional effect in the encoded proteins are unknown.
A recent genome-wide association study conducted by the International Multiple Sclerosis Genetics Consortium (IMSGC) has identified a number of putative MS susceptibility genes,8 among them, 21 SNPs that are located in the locus containing the growth factor-independent 1 (GFI1), ecotropic viral integration site 5 (EVI5), ribosomal protein L5 (RPL5) and family with sequence similarity 69 (FAM69). The association of two of these SNPs has been replicated in a posterior study with 1318 MS Canadian patients.11 In this study, our aim was to validate the association of the GFI1-EVI5-RPL5-FAM69A locus in a Caucasian Spanish population and to try to identify the gene responsible for the association with MS by a Tag-SNP strategy that captures 279 common variants at a pairwise r2≥0.6.
MATERIALS AND METHODS
Study subjects
Case samples comprised 732 patients with clinically defined MS according to Poser's criteria.12 They were obtained from four public hospitals: Hospital Clínico of Granada (n=126), Hospital Virgen de las Nieves Of Granada (n=165), Hospital Carlos Haya of Málaga (n=365) and Hospital Virgen de la Macarena of Seville (n=76); all three cities located within a 200 km radius in the South of Spain. The mean age of cases at the moment of sample collection was 36 years and the mean age of controls at interview was 38 years. The percentage of females was 68% for cases and 68% for controls. All of them were classified as relapsing remitting (RR) or secondary progressive (SP) MS cases. Controls were 974 blood donors with no history of inflammatory disease visiting the blood banks of Granada (n=823), Seville (n=71) and Málaga (n=80). The study was approved by the ethics committees of each of the hospitals participating in the study and written informed consent was obtained from all participants.
Genotyping
High-molecular-weight DNA was isolated from whole blood using the Flexigene Kit (Qiagen, Hilden, Gemany) according to the manufacturer's protocol. The eight SNPs were genotyped by TaqMan technology under conditions recommended by the manufacturer (Applied Biosystems, Foster City, CA, USA).
Sample power calculation
First, we computed power calculations using the Genetic Power Calculator (http://pngu.mgh.harvard.edu/~purcell/gpc/). Thus, we determined that the case–control sample had 84% power for detecting a risk allele with 20% frequency and a dominant genotype relative risk of 1.3. To assess the power for detecting association due to linkage disequilibrium (LD) with a causal loci, we also carried out power calculations for an indirect association study that uses Tag-SNPs. We estimated that our study was able to detect a susceptibility locus (80% power) with an odds ratio (OR) of 1.4 if the MAF is 0.2 and an OR of 1.3 if the MAF is 0.3, assuming a dominant effect at an unobserved locus, an alpha (α) value of 0.05 and r2=0.85 for the ability of haplotypes to predict the allele count at the causal locus.
Statistical analysis
Departure from Hardy–Weinberg equilibrium for all the biallelic SNP markers was tested using an exact test.13 For individual SNP association analyses, genotype frequencies were assessed by means of the MAX statistic. This test fits dominant, recessive and additive models and considers the most significant among them. To avoid multiple comparisons by fitting three different models, P-values are then computed using the exact distribution of the MAX statistic.14 Logistic regression models were used to estimate crude ORs and 95% confidence intervals (95% CI). To determine the SNP that is more associated with MS, multiple logistic regression models were computed. By using the likelihood ratio test, we compared the effect of each SNP adjusted by the most promising SNP found in the crude analysis. These analyses were performed using the SNPassoc R package.15 To avoid false-positive results due to multiple testing and to ensure that the SNPs analyzed are not in complete disequilibrium, we applied the Benjamini–Hochberg method,16 which is robust against positive dependence and controls the false discovery rate.17
RESULTS
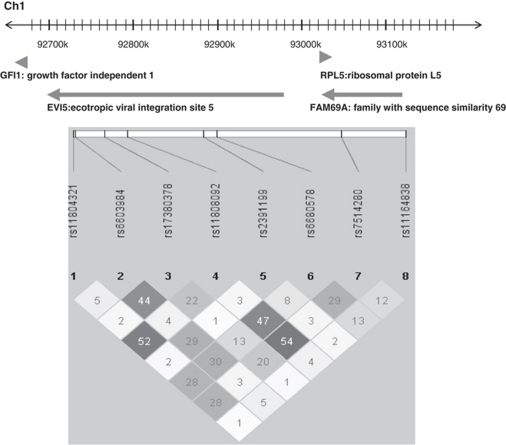
We selected the region between SNPs rs11164607 and rs17380908 that covers 455413 base pairs containing the GFI1-EVI5-RPL5-FAM69A risk locus, which is associated with MS in the IMSGC study.8 To analyze this region, we chose eight SNPs by pairwise tagging from the HapMap B35 CEU population, which captured 279 markers with r2≥0.6 (mean r2=0.911) and a minor allele frequency (MAF) ≥0.1.
We proceeded to genotype the Tag-SNPs in a Caucasian population of 732 MS patients and 974 controls from the South of Spain. The results of genotypic and carrier distribution between cases and controls are shown in Table 1. The genotype frequencies assessed by the MAX statistic and the best-fitted model are also shown in Table 1. Three polymorphisms were associated with MS: rs11804321 (P=0.008, OR=1.29; 95% CI=1.08–1.54), rs11808092 (P=0.048, OR=1.19; 95% CI=1.03–1.39) and rs6680578 (P=0.0082, OR=1.23; 95% CI=1.07–1.41); from these, two survived Benjamini–Hochberg correction, rs11804321 and rs6680578. We performed a logistic regression analysis to test the addition of each SNP to both SNPs (independently) to determine the primarily associated marker (Table 2). We found that rs11804321 alone was sufficient to model the association with MS because, as observed in Table 2, the model that adds the locus rs6680578 to rs11804321 was not statistically significant (P from likelihood ratio test equal to 0.3074). This means that the effect found in the single analysis of rs6680578 was confounded by the effect of the polymorphism rs11804321. In addition, we also observed that the variant rs11804321 remained statistically significant after being adjusted for the other SNPs (Table 2, second analysis). Therefore, the rs11804321 polymorphism, among the ones analyzed with minor allele frequency ≥0.1 and r2≥0.6 with rs11804321, showed the strongest association in the GFI1-EVI5-RPL5-FAM69 locus and had an independent effect on MS susceptibility.
Table 1. Tag-SNP mapping of the GFI1-EVI5-RPL5-FAM69 locus.
| Genotypes | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Alleles | Multiple sclerosis | Healthy controls | P-value | Additive | |||||||||
| SNP | 1 | 2 | N | 11 | 12 | 22 | N | 11 | 12 | 22 | MAX-statistic (best genetic model) | Benjamini– Hochberg | OR (95% CI) |
| 1. rs11804321 | C | T | 681 | 0.05 | 0.36 | 0.59 | 871 | 0.03 | 0.3 | 0.66 | 0.008 (add) | 0.034 | 1.29 (1.08,1.54) |
| 2. rs6603984 | A | G | 724 | 0.04 | 0.32 | 0.64 | 911 | 0.04 | 0.31 | 0.65 | 0.751 (dom) | ns | 1.00 (0.84,1.19) |
| 3. rs17380378 | C | T | 698 | 0.01 | 0.18 | 0.8 | 881 | 0.01 | 0.18 | 0.81 | 0.952 (dom) | ns | 1.03 (0.82,1.29) |
| 4. rs11808092 | A | C | 726 | 0.1 | 0.44 | 0.45 | 889 | 0.08 | 0.41 | 0.51 | 0.048 (add) | ns | 1.19 (1.03,1.39) |
| 5. rs2391199 | T | C | 700 | 0.01 | 0.15 | 0.83 | 884 | 0.01 | 0.16 | 0.83 | 0.909 (rec) | ns | 0.96 (0.76,1.22) |
| 6. rs6680578 | T | A | 732 | 0.22 | 0.49 | 0.29 | 907 | 0.17 | 0.49 | 0.35 | 0.008 (add) | 0.034 | 1.23 (1.07,1.41) |
| 7. rs7514280 | T | C | 728 | 0.09 | 0.45 | 0.46 | 908 | 0.09 | 0.42 | 0.49 | 0.346 (rec) | ns | 1.07 (0.92,1.25) |
| 8. rs11164838 | C | T | 730 | 0.17 | 0.46 | 0.37 | 978 | 0.19 | 0.47 | 0.34 | 0.335 (add) | ns | 0.91 (0.80,1.04) |
Abbreviations: add, additive; CI, confidence interval; dom, dominant; N, number of samples; NS, Nonsignificant; OR, odds ratio; rec, recessive.
Risk allele is bold and underlined.
Table 2. Regression analyses of eight Tag-SNPs in the GFI1-EVI5-RPL5-FAM69 region genotyped in up to 732 affected individuals and 974 control simples.
| Add locus to rs11804321a | Add rs11804321 to locusb | ||||
|---|---|---|---|---|---|
| SNP | A1 | P | OR (95% CI) | P | OR (95% CI) |
| rs11804321 | C | NA | NA | NA | NA |
| rs6603984 | A | 0.7164 | 1.03 (0.85,1.23) | 0.0092 | 1.28 (1.06,1.54) |
| rs17380378 | C | 0.4577 | 1.09 (0.86,1.38) | 0.0048 | 1.30 (1.08,1.55) |
| rs11808092 | A | 0.8394 | 0.98 (0.78,1.22) | 0.0208 | 1.35 (1.05,1.75) |
| rs2391199 | T | 0.9669 | 1.01 (0.79,1.28) | 0.0044 | 1.30 (1.08,1.56) |
| rs6680578 | T | 0.3074 | 1.09 (0.92,1.29) | 0.0306 | 1.26 (1.02,1.55) |
| rs7514280 | T | 0.6027 | 0.95 (0.79,1.14) | 0.0064 | 1.34 (1.09,1.65) |
| rs11164838 | C | 0.2561 | 0.92 (0.80,1.06) | 0.0055 | 1.29 (1.08,1.55) |
Abbreviations: CI, confidence interval; NA, not available; OR, odds ratio.
Results for models assuming additive effects.
Results adjusted for rs11804321.
Results adjusted for locus tested.
The IMSGC8 analyzed 34 SNPs in this region, 32 of them were represented in HapMap B35. On the basis of the LD for these 32 SNPs from the HapMap, we grouped them into six Tag-SNPs (SNP2, 4, 5, 6, 7, 8), as shown in Table 3. The other two Tag-SNPs were not represented by any IMSGC marker. Thirteen out of 21 IMSGC-associated SNPs were tagged by SNP6 and four by SNP4. SNP7 and SNP8 were not associated in our cohort. Twelve SNPs not associated in the IMSGC work were tagged by SNP2 and SNP5, which also tested negative for association with MS in this study. The 87 HapMap SNPs tagged by SNP6 were localized in the four genes and intergenic regions of the 1p22 locus, whereas the 23 tagged by SNP4 were located in the EVI5 gene. The two SNPs tagged by SNP1 are in the 17th intron of the EVI5 gene. The frequency and LD map of the eight Tag-SNPs in our population were similar to the ones obtained from the HapMap CEU genotypes (Figure 1).
Table 3. Tagged SNPs from the GWAS and location.
| IMSGC-GWAS | ||||||
|---|---|---|---|---|---|---|
| SNP | SNPs r2≥0.6 | Risk allele | MAF | Allelic P-value | OR 95% CI | Gene localization |
| 1. rs11804321 | EVI5 | |||||
| 2. rs6603984 | rs6692187* | 0.2 | 0.9 | EVI5-GFI1 | ||
| rs4970712* | 0.2 | 0.7 | ||||
| rs4970705* | 0.2 | 0.9 | ||||
| rs1556562* | 0.2 | 0.8 | ||||
| 3. rs17380378 | EVI5-FAM69 | |||||
| 4. rs11808092 | EVI5 | |||||
| rs12743520 | A | 0.28 | 0.000047 | 1.14 (1.07–1.21) | ||
| rs11800848 | C | 0.28 | 0.00021 | 1.12 (1.06–1.2) | ||
| rs1415296 | C | 0.29 | 0.00037 | 1.12 (1.05–1.19) | ||
| rs11578004* | C | 0.28 | 0.0005 | 1.24 (1.09–1.4) | ||
| 5. rs2391199 | EVI5-FAM69 | |||||
| rs2107521* | 0.09 | 0.1 | ||||
| rs4847280* | 0.08 | 0.2 | ||||
| rs6604015* | 0.09 | 0.1 | ||||
| rs2391164* | 0.09 | 0.1 | ||||
| rs10747448* | 0.09 | 0.1 | ||||
| rs2027060* | 0.09 | 0.06 | ||||
| rs6659942* | 0.1 | 0.2 | ||||
| rs7543964* | 0.09 | 0.07 | ||||
| 6. rs6680578 | rs6680578 | T | 0.38 | 0.0005 | 1.11 (1.04–1.17) | GFI1-EVI5- |
| rs2811600 | C | 0.37 | 0.0022 | 1.09 (1.03–1.16) | RPL5-FAM69 | |
| rs4847267 | C | 0.38 | 0.0036 | 1.09 (1.03–1.15) | ||
| rs17380908 | T | 0.36 | 0.00026 | 1.11 (1.05–1.18) | ||
| rs2255723 | G | 0.37 | 0.0094 | 1.08 (1.02–1.14) | ||
| rs10874746 | T | 0.35 | 0.0002 | 1.11 (1.05–1.18) | ||
| rs7536563 | A | 0.38 | 0.000009 | 1.12 (1.06–1.18) | ||
| rs11164607 | A | 0.38 | 0.011 | 1.08 (1.02–1.14) | ||
| rs9651257 | C | 0.37 | 0.00049 | 1.11 (1.04–1.17) | ||
| rs12745968 | G | 0.37 | 0.00065 | 1.1 (1.04–1.17) | ||
| rs10735781 | G | 0.38 | 0.00033 | 1.11 (1.05–1.18) | ||
| rs11164835 | A | 0.37 | 0.00097 | 1.10 (1.05–1.24) | ||
| rs4970700 | A | 0.37 | 0.018 | 1.07 (1.01–1.14) | ||
| 7. rs7514280 | rs7514280 | T | 0.28 | 0.00007 | 1.13 (1.06–1.2) | FAM69 |
| rs6604026 | C | 0.29 | 0.000008 | 1.15 (1.08–1.22) | ||
| 8. rs11164838 | rs11164838 | C | 0.43 | 0.00019 | 1.11 (1.05–1.18) | FAM69-EVI5 |
Abbreviations: CI, confidence interval; EVI5, ecotropic viral integration site 5; FAM69, family with sequence similarity 69; GFI1, growth factor independent 1; IMSGC-GWAS, international multiple sclerosis genetics consortium-genome wide association study; OR, odds ratio; RPL5, ribosomal protein L5; SNP single-nucleotide polymorphism.
Data from IMSGC study and IMSGC Supplementary material,8 combining the analysis of 1540 family trios; 2322 cases and 5418 control subjects.
Figure 1.
The LD structure of Tag-SNPs at the GFI1-EVI5-RPL5-FAM69A locus and schematic illustration of the genes in the locus with the positions in chromosome 1 (Ch1). Pairwise r2 values are shown for the Spanish population.
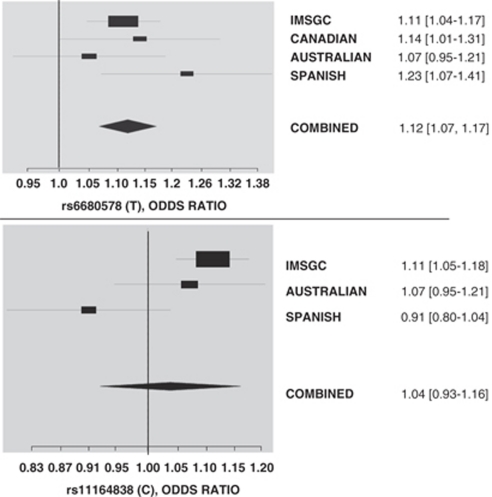
SNPs rs6680578 and rs11164838 have been analyzed in previous studies with discordant results. To address this issue, we performed a meta-analysis with the data obtained from IMSGC,8 Canadian,11 Australian18 and Spanish (this study) cohorts, as shown in Figure 2. The summary effect estimate for rs6680578 is significant with a combined OR=1.12, 95% CI (1.07–1.17) and an estimated heterogeneity variance of no significance (P=0.395). On the other hand, rs11164838 showed heterogeneity among results (P=0.025) with a combined effect estimate of no significance (OR=1.04, 95% CI (0.93–1.16)).
Figure 2.
Summary effect estimate for SNPs rs6680578 (upper panel) and rs11164838 (lower panel) using information from four different studies: IMSGC8 (combined analysis of 1540 family trios and replicated in 2322 cases and 5418 controls), CANADIAN11 (Canadian Collaborative Project on the Genetic Susceptibility to MS performed on 756 families, which included 1318 individuals with definite MS and 1507 of their unaffected first-degree relatives), AUSTRALIAN18 (cohort of 1134 cases and 1265 controls), SPANISH, (this study consisted of 732 patients with clinically defined MS and 974 healthy controls). Individual effect estimates reported by the authors and their confidence intervals have been used. A random effects model was used when heterogeneity was observed (http://cran.r-project.org/web/packages/rmeta/index.html). The odds ratio (OR) estimate of each study is marked with a square, which is also indicated in the figure. The size of the square represents the weight that the corresponding study exerts in the meta-analysis. Estimated heterogeneity variance for rs6680578=0 (P=0.395), and for rs11164838=0.0071 (P=0.025).
DISCUSSION
In this study, we analyzed the GFI-EVI5-RPL5-FAM69A locus to test, in a Spanish MS cohort, the association found in a genome-wide association study by the IMSGC.8 A fine mapping of this genomic region was performed by a Tag-SNP approach that points to the EVI5 as the most probable risk gene for MS. Our results replicate those obtained by the IMSGC, except for rs7514280 and rs11164838, which were not associated with MS in our cohort.
SNPs rs6680578 and rs10735781, located in the EVI5 gene in nearly complete LD (D′=0.98), have been previously tested in an independent set of 756 Canadian families containing 1318 MS patients.11 Both SNPs had a weak but significant contribution in this population. However, a replication study of four SNPs in the GFI-EVI5-RPL5-FAM69A locus, conducted on 1134 Australian MS patients,18 showed no association. The meta-analysis that we performed for rs6680578 using data obtained from the IMSGC,8 Canadian,11 Australian18 and Spanish (this study) cohorts showed significant results, with a combined odds ratio of 1.12 (95% CI (1.07–1.17)). On the other hand, rs11164838, which has been associated with MS in the IMSGC genome-wide study,8 did not show an association in either Spanish or Australian18 cohorts. This SNP is in low LD (D′=0.27, r2=0.014) with rs11804321, the most associated Tag-SNPs in this study; therefore, the power to capture the association with MS due to rs11804321 with rs11164838 would be very low for the Australian and Spanish cohorts. Thus, the lack of power seems to be the cause of the different results among cohorts. In fact, a recent genome-wide study performed by the Australia and New Zealand Multiple Sclerosis Genetics Consortium (ANZgene)19 in 1618 cases and 3413 controls has found association of rs6604026 with MS in the EVI5-RPL5 locus. This polymorphism is in high LD with rs11804321 (D′=0.72). Altogether, these data support the association of the locus with MS in all the populations studied.
The logistic regression analysis to determine the primarily associated marker showed that the association within this region could best be explained by one disease locus tagged by rs11804321. The two SNPs tagged from the HapMap database by rs11804321 are located in the EVI5 gene. We note, however, that until a more complete set of polymorphisms is identified and genotyped in a large collection of cases and control subjects, we cannot exclude another variant in LD, with SNP rs11804321 being the causal variant. Future resequencing of the locus may provide as-yet-undiscovered variants that will need to be assessed for disease susceptibility.
The EVI-5 protein contains a TBC (TRE2/BUB/CDC16 homology) motif located in the N-terminal 200 amino-acid region (EVI-5N) that binds and activates the guanosine triphosphate-bound form of Rab11 (GTPase-activating protein).20 This heterodimer complex may coordinate vesicular trafficking, cytokinesis and cell-cycle control independent of GTPase-activating protein function.21 EVI-5 is also a nuclear zinc-finger protein that functions as a transcriptional repressor that is possibly involved in centrosome stability and dynamics.22 Interestingly, SNP4 located in exon 15 of the EVI5 gene is a nonsynonymous polymorphism (612 His/Gln), but the potential effect on the EVI5N domain is unknown. Statistically, SNP4 did not survive Benjamini–Hochberg correction. SNP5 at exon 7 of the EVI5 gene produces a 316 Val-Ile change, but it did not show association with MS. None of the SNPs analyzed seems to interfere with any splice site or transcription factor recognition site at the promoter of the genes in the locus.
In conclusion, these findings suggest that EVI5 is the most likely candidate in the GF1I-EVI5-RPL5-FAM69A locus as a risk gene for MS in a cohort of Caucasians from the south of Spain. We confirm the data of the GWAS of IMSGC,8 leaving open the question of the determination of the causal polymorphism.
Acknowledgments
We thank the patients with multiple sclerosis and the persons who acted as controls for making this study possible. Financial support for the study was provided by the Ministerio de Ciencia e Innovación-Fondos Feder (Grant SAF2009–11491) and Junta de Andalucía (P07-CVI-02551) to A. Alcina, and by Fondo de Investigación Sanitaria (PI081636) to F Matesanz. María Fedetz is a holder of a fellowship from Fundación IMABIS. Dorothy Ndagire is a holder of AECI-Ministerio de Asuntos Exteriores fellowship.
The authors declare no conflict of interest.
References
- Compston A, Coles A. Multiple sclerosis. Lancet. 2002;359:1221–1231. doi: 10.1016/S0140-6736(02)08220-X. [DOI] [PubMed] [Google Scholar]
- Fernández O, Fernández V, Martinez-Cabrera V, et al. Multiple sclerosis in Gypsies from southern Spain: prevalence, mitochondrial DNA haplogroups and HLA class II association. Tissue Antigens. 2008;71:426–433. doi: 10.1111/j.1399-0039.2008.01016.x. [DOI] [PubMed] [Google Scholar]
- Alonso A, Jick SS, Olek MJ, Hernán MA. Incidence of multiple sclerosis in the United Kingdom: findings from a population-based cohort. J Neurol. 2007;254:1736–1741. doi: 10.1007/s00415-007-0602-z. [DOI] [PubMed] [Google Scholar]
- Lyndsay JW. Familiar recurrence rates and genetic models of multiple sclerosis. Am J Med Genet. 2005;135:53–58. doi: 10.1002/ajmg.a.30702. [DOI] [PubMed] [Google Scholar]
- McElroy JP, Oksenberg JR. Multiple sclerosis genetics. Curr Top Microbiol Immunol. 2008;318:45–72. doi: 10.1007/978-3-540-73677-6_3. [DOI] [PubMed] [Google Scholar]
- Lundmark F, Duvefelt K, Iacobaeus E, et al. Variation in interleukin 7 receptor alpha chain (IL7R) influences risk of multiple sclerosis. Nat Genet. 2007;39:1108–1113. doi: 10.1038/ng2106. [DOI] [PubMed] [Google Scholar]
- Gregory SG, Schmidt S, Seth P, et al. Interleukin 7 receptor alpha chain (IL7R) shows allelic and functional association with multiple sclerosis. Nat Genet. 2007;39:1083–1091. doi: 10.1038/ng2103. [DOI] [PubMed] [Google Scholar]
- The International Multiple Sclerosis Genetics Consortium Risk alleles for multiple sclerosis identified by a genome wide study. N Engl J Med. 2007;357:851–862. doi: 10.1056/NEJMoa073493. [DOI] [PubMed] [Google Scholar]
- Kristjansdottir G, Sandling JK, Bonetti A, et al. Interferon regulatory factor 5 (IRF5) gene variants are associated with multiple sclerosis in three distinct populations. J Med Genet. 2008;45:362–369. doi: 10.1136/jmg.2007.055012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matesanz F, Caro-Maldonado A, Fedetz M, et al. IL2RA/CD25 polymorphisms contribute to multiple sclerosis susceptibility. J Neurol. 2007;254:682–684. doi: 10.1007/s00415-006-0416-4. [DOI] [PubMed] [Google Scholar]
- Hoppenbrouwers IA, Aulchenko YS, Ebers GC, et al. EVI5 is a risk gene for multiple sclerosis. Genes Immun. 2008;9:334–337. doi: 10.1038/gene.2008.22. [DOI] [PubMed] [Google Scholar]
- Poser CM, Paty DW, Scheinberg L, et al. New diagnostic criteria for multiple sclerosis: guidelines for research protocols. Ann Neurol. 1983;13:227–231. doi: 10.1002/ana.410130302. [DOI] [PubMed] [Google Scholar]
- Wigginton JE, Cutler DJ, Abecasis GR. A note on exact tests of Hardy–Weinberg equilibrium. Am J Hum Genet. 2005;76:887–893. doi: 10.1086/429864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- González JR, Carrasco JL, Dudbridge F, Armengol L, Estivill X, Moreno V. Maximizing association statistics over genetic models. Genet Epidemiol. 2008;32:246–254. doi: 10.1002/gepi.20299. [DOI] [PubMed] [Google Scholar]
- González JR, Armengol L, Solé X, et al. SNPassoc: an R package to perform whole genome association studies. Bioinformatics. 2007;23:644–645. doi: 10.1093/bioinformatics/btm025. [DOI] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J Roy Stat Soc Ser B. 1995;57:289–300. [Google Scholar]
- Sarkar SK. False discovery and false nondiscovery rates in single-step multiple testing procedures. Ann Stat. 2006;34:394–415. [Google Scholar]
- Rubio JP, Stankovich J, Field J, et al. Replication of KIAA0350, IL2RA, RPL5 and CD58 as multiple sclerosis susceptibility genes in Australians. Genes Immun. 2008;9:624–630. doi: 10.1038/gene.2008.59. [DOI] [PubMed] [Google Scholar]
- Australia and New Zealand Multiple Sclerosis Genetics Consortium (ANZgene): Genome-wide association study identifies new multiple sclerosis susceptibility loci on chromosomes 12 and 20. Nat Genet. 2009;41:824–828. doi: 10.1038/ng.396. [DOI] [PubMed] [Google Scholar]
- Westlake CJ, Junutula JR, Simon GC, et al. Identification of Rab11 as a small GTPase binding protein for the Evi5 oncogene. Proc Natl Acad Sci USA. 2007;104:1236–1241. doi: 10.1073/pnas.0610500104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson PM, Zon LI. Molecular cloning of a cDNA with a novel domain present in the tre-2 oncogene and the yeast cell cycle regulators BUB2 and cdc16. Oncogene. 1995;11:1139–1148. [PubMed] [Google Scholar]
- Faitar SL, Dabbeekeh JT, Ranalli TA, Cowell JK. EVI5 is a novel centrosomal protein that binds to alpha- and gamma-tubulin. Genomics. 2005;86:594–605. doi: 10.1016/j.ygeno.2005.06.002. [DOI] [PubMed] [Google Scholar]