Abstract
Stickler syndrome is an autosomal dominant connective tissue disorder caused by mutations in different collagen genes. The aim of our study was to define more precisely the phenotype and genotype of Stickler syndrome type 1 by investigating a large series of patients with a heterozygous mutation in COL2A1. In 188 probands with the clinical diagnosis of Stickler syndrome, the COL2A1 gene was analyzed by either a mutation scanning technique or bidirectional fluorescent DNA sequencing. The effect of splice site alterations was investigated by analyzing mRNA. Multiplex ligation-dependent amplification analysis was used for the detection of intragenic deletions. We identified 77 different COL2A1 mutations in 100 affected individuals. Analysis of the splice site mutations showed unusual RNA isoforms, most of which contained a premature stop codon. Vitreous anomalies and retinal detachments were found more frequently in patients with a COL2A1 mutation compared with the mutation-negative group (P<0.01). Overall, 20 of 23 sporadic patients with a COL2A1 mutation had either a cleft palate or retinal detachment with vitreous anomalies. The presence of vitreous anomalies, retinal tears or detachments, cleft palate and a positive family history were shown to be good indicators for a COL2A1 defect. In conclusion, we confirm that Stickler syndrome type 1 is predominantly caused by loss-of-function mutations in the COL2A1 gene as >90% of the mutations were predicted to result in nonsense-mediated decay. On the basis of binary regression analysis, we developed a scoring system that may be useful when evaluating patients with Stickler syndrome.
Keywords: COL2A1, Stickler syndrome, genotype–phenotype correlation, type II collagenopathies, splice site mutation
INTRODUCTION
Stickler syndrome (MIM no. 108300) is a connective tissue disorder first described by Stickler et al1, 2 in 1965. It is characterized by ocular, orofacial, auditory and skeletal manifestations with considerable intra- and inter-familial variability. The incidence is estimated to range between 1 in 7500 and 1 in 9000 newborns (http://ghr.nlm.nih.gov/condition%3Dsticklersyndrome; US Department of Health). The most characteristic ocular features include congenital myopia, vitreous alterations, cataract, glaucoma and a high risk of spontaneous retinal detachments. The orofacial changes include cleft palate, midfacial hypoplasia, low nasal bridge and micrognathia. Joint pain is common in childhood and osteoarthrosis may be apparent from the third or fourth decade onward. Radiographs may show signs of a spondylo-epiphyseal dysplasia congenita (SEDC). Mild sensorineural hearing loss, mainly for the high tones, can be present in Stickler syndrome type 1 (COL2A1 gene); more severe sensorineural hearing loss is usually found in other types of Stickler syndrome.3, 4
At present, at least three types of autosomal dominant Stickler syndrome have been discerned. A correlation between these different types and their accompanying vitreous anomalies has been suggested.5 ‘Membranous' or type 1 vitreous has been associated with Stickler syndrome type 1 caused by heterozygous mutations in the COL2A1 gene (MIM no. 108300).6 Type 2 or ‘beaded' vitreous is mainly found in patients with Stickler syndrome type 2, which is due to a heterozygous mutation in the COL11A1 gene (MIM no. 604841).3, 7 Stickler syndrome type 3 or ‘non-ocular Stickler syndrome' refers to the phenotype of patients with a mutation in the COL11A2 gene that is not expressed in the eye (MIM no. 184840).8 In addition to the different types of autosomal dominant Stickler syndrome, recently a recessive form of Stickler syndrome, caused by a mutation in the COL9A1 gene, has also been described (MIM no. 120210).9
Stickler syndrome type 1 is the most common form. The majority of COL2A1 mutations identified in patients with Stickler syndrome type 1 are predicted to result in nonsense-mediated decay (NMD). On the other hand, missense mutations (usually glycine substitutions) in COL2A1 usually result in short-stature disorders, such as achondrogenesis type II/hypochondrogenesis, SEDC, Kniest dysplasia, spondyloperipheral dysplasia and Torrance dysplasia (MIM nos 200610, 183900, 156550, 271700, 151210, respectively).5, 10, 11
The aim of this study was to define more precisely the phenotype and genotype of Stickler syndrome type 1 by investigating a large series of Stickler syndrome patients with a heterozygous mutation in the COL2A1 gene.
MATERIALS AND METHODS
Evaluation of phenotype
Over the past 10 years, blood or DNA samples obtained from 278 individuals were referred for mutation analysis of the COL2A1 gene in order to confirm or exclude the clinical diagnosis of Stickler syndrome.
Information on clinical and radiographic features of each patient was requested by using a specific questionnaire (Supplementary Table 1). A total of 90 patients were excluded from the study because insufficient clinical data were available (in 11 of those patients, a COL2A1 mutation was identified). Each patient in the group of 188 remaining subjects had 2 or more of the following features reminiscent of Stickler syndrome: myopia, spontaneous retinal detachment, cleft palate, sensorineural hearing loss and arthropathy. Informed consent was obtained from each enrolled patient.
Analysis of genomic DNA
Genomic DNA was extracted from blood samples using standard procedures, followed by touchdown PCR amplification of the COL2A1 gene using forward and reverse primers located in the flanking introns. The PCR products were analyzed by gel electrophoresis and visualized by ethidium bromide staining on 2% agarose gels.
Mutation screening was performed by SSCP and CSGE (period 1997–2002) or by DHPLC analysis (period 2003–2006) using the WAVE DNA fragment analysis system (Transgenomic, Crewe, UK).12, 13, 14 All fragments showing an aberrant pattern were directly sequenced on the ABI PRISM 3730 automated sequencer (Applied Biosystems, Foster City, CA, USA) using the BigDye terminator cycle sequencing chemistry. From 2007 onward, direct sequencing of all 54 exons was performed. These obtained sequences were compared with the wild-type sequence as submitted to GenBank accession number NM_001844. The nucleotides were numbered starting from the first base of the start codon (ATG) of the cDNA reference sequence. Amino-acid residues were numbered from the first methionine (start codon for translation) of the procollagen α1(II)-chain (GenBank accession number L10347).
RNA studies
In patients with a splice site mutation, an EBV cell line or skin biopsy was requested for analysis of mRNA splicing. To stabilize mutant COL2A1 mRNA, cycloheximide (Sigma, St Louis, MO, USA; http://www.sigmaaldrich.com) was added to the cultures, followed by mRNA isolation and cDNA preparation. Nested PCR was used to obtain sufficient PCR fragments for direct sequencing.
MLPA analysis
Multiplex ligation-dependent amplification (MLPA) was set up, following the directions provided by the manufacturer (MRC Holland, Amsterdam, The Netherlands) (http://www.MPLA.com).15 The probe set for COL2A1 (SALSA MLPA kit P214) covering exons 1, 4, 6, 8, 10, 16, 17, 19, 20, 24, 27, 29, 31, 35, 39, 43, 46, 49, 51 and 54 was used.
Binary logistic regression analysis/statistics
The formula for the proposed scoring system was developed using binary (mutation positive or not) logistic regression analysis.16, 17, 18 The parameters tested in the model comprised vitreous abnormalities, retinal abnormalities, flat face, micrognathia, retinal tear and/or detachment, cataract, low nasal bridge, cleft palate, positive family history, myopia, conductive hearing loss, premature arthropathy, hypermobility, epiphyseal dysplasia on X-rays and sensorineural hearing loss. The weight (score) for each characteristic in the scoring system was proportional to its regression coefficient in the model. To simplify the scoring system, the scores were rounded to positive integers and the scores of the characteristics with lowest significant regression coefficients were conventionally given a score value of one, and the intercept of the linear predictor was neglected. Otherwise, no recalibration, shrinkage factor or model revision or extension seemed to be required to study the whole study population. Calibration of the scoring system was further evaluated with the Hosmer–Lemeshow test. The clinical applicability of the obtained score was evaluated for several thresholds using conventional receiver-operating characteristics (such as positive and negative predictive value, sensitivity and specificity). All statistical analyses were performed using SPSS 15.0 for Windows (SPSS Inc., Chicago, IL, USA).
RESULTS
In 100 of 188 individuals referred with a potential diagnosis of Stickler syndrome, a heterozygous COL2A1 mutation was identified. This panel of 77 different mutations included 1 deletion of the entire gene,19 13 nonsense mutations, 21 deletions, 1 insertion, 9 duplications, 2 combinations of an insertion and a deletion, 22 splice site alterations, 1 synonymous mutation, 2 missense mutations resulting in an arginine-to-cysteine substitution20 and 5 missense mutations substituting a glycine residue in the triple helical domain of the protein. The mutations were distributed over the entire gene and no hot spot regions were apparent (Table 1). Thirteen mutations were observed in >1 proband: c.625C>T, p.Arg209X and c.1833+1G>A, p.GlyfsX619 were found in 4 patients each; c.3106C>T, p.Arg1036X occurred five times (Table 1).
Table 1. Summary of 77 different COL2A1 mutations identified in a series of 100 affected individuals.
| Patient ID | Age (years) | Score | Exon/intron | cDNA | Protein | Mutation type | Mutation effect |
|---|---|---|---|---|---|---|---|
| 1 | 46 | 15 | del COL2A1 | del COL2A1 | Large deletion | Deletion[19] | |
| 2 | 54 | 13 | 02 | c.211_233dup | p.Glu79ThrfsX2 | Duplication | Frameshift |
| 3 | 58 | 17 | 02 | c.264_276del | p.Cys89SerfsX24 | Deletion | Frameshift |
| 4 | 4 | 6 | IVS 04 | c.342+1G>A | p.Asp114_Ile115insIleSerAlaAsnTyrSerHisProValLeuGlnLeuLeuX14 | RNA processing | Insertion with premature stop codon |
| 5 | 42 | 17 | IVS 06 | c.430-1G>C | p.Gly144ValfsX54; p.Gln125_Gly126insArgGluGlyGluAsnLeuPheLeuArgProPheLeuAlaAlaGlnValThrAspLeuX20; p.Lys143_Asn178delExon7a | RNA processing | Frameshift; insertion with premature stop codon; exon deletion |
| 6 | 6 | 6 | 07 | c.492delT | p.Gly165ValfsX34 | Deletion | Frameshift |
| 7 | 3 | 6 | 09 | c.625C>T | p.Arg209X | Nonsense | Premature stop codon |
| 8 | 6 | 11 | 09 | c.625C>T | p.Arg209X | Nonsense | Premature stop codon |
| 9 | 19 | 13 | 09 | c.625C>T | p.Arg209X | Nonsense | Premature stop codon |
| 10 | 12 | 14 | 09 | c.625C>T | p.Arg209X | Nonsense | Premature stop codon |
| 11 | 22 | 15 | 09 | c.647G>A | p.Gly216Asp | Missense | Glycine substitution |
| 12 | 8 | 8 | IVS 09 | c.654+1G>A | ND | RNA processing | ND |
| 13 | 34 | 10 | 10 | c.655G>C | p.Gly219Arg | Missense | Glycine substitution |
| 14 | 27 | 14 | 10 | c.665G>T | p.Gly222Val | Missense | Glycine substitution |
| 15 | 24 | 11 | 11 | c.744delT | p.Gly249GlufsX59 | Deletion | Frameshift |
| 16 | 45 | 18 | 12 | c.793delG | p.Glu265fsX43 | Deletion | Frameshift |
| 17 | 37 | 14 | IVS 13 | c.870+5 G> A | ND | RNA processing | ND |
| 18 | 30 | 9 | IVS 14 | c.925-1G>A | p.Lys308_Gly309insGluPheAlaGlyGlyGlnGluTrpGlyProArgHisX13 | RNA processing | Insertion with premature stop codon |
| 19 | 67 | 12 | 17 | c.1030C>T | p.Arg344X | Nonsense | Premature stop codon |
| 20 | 9 | 9 | 17 | c.1030C>T | p.Arg344X | Nonsense | Premature stop codon |
| 21 | 62 | 21 | IVS 18 | c.1123-1G>A | p.Gly375ValfsX253 | RNA processing | Frameshift |
| 22 | 6 | 7 | 19 | c.1172delC | p.Pro391LeufsX238 | Deletion | Frameshift |
| 23 | 11 | 11 | IVS 19 | c.1221+1G>A | ND | RNA processing | ND |
| 24 | 43 | 12 | 21 | c.1311_1313delinsCA | p.Gly438ThrfsX191 | Deletion/insertion | Frameshift |
| 25 | 33 | 12 | 23 | c.1428_1429insTGGC | p.Gly477TrpfsX12 | Insertion | Frameshift |
| 26 | 13 | 8 | 23 | c.1475G>A | p.Gly492Asp | Missense | Glycine substitution |
| 27 | 40 | 13 | 25 | c.1597C>T | p.Arg533X | Nonsense | Premature stop codon |
| 28 | 10 | 10 | 25 | c.1597C>T | p.Arg533X | Nonsense | Premature stop codon |
| 29 | 12 | 15 | IVS 25 | c.1680+2delGTinsAA | ND | RNA processing | ND |
| 30 | 24 | 10 | 26 | c.1693C>T | p.Arg565Cys | Missense | Arginine-to-cysteine substitution[20] |
| 31 | 20 | 8 | 26 | c.1693C>T | p.Arg565Cys | Missense | Arginine-to-cysteine substitution[20] |
| 32 | 9 | 7 | 26 | c.1693C>T | p.Arg565Cys | Missense | Arginine-to-cysteine substitution[20] |
| 33 | 11 | 19 | 27 | c.1777C>T | p.Gln593X | Nonsense | Premature stop codon |
| 34 | 14 | 14 | 27 | c.1828delG | p.Ala610ProfsX19 | Deletion | Frameshift |
| 35 | 11 | 10 | IVS 27 | c.1833+1G>A | ND | RNA processing | ND |
| 36 | 36 | 12 | IVS 27 | c.1833+1G>A | p.Gly609GlyfsX1 | RNA processing | Frameshift |
| 37 | 40 | 14 | IVS 27 | c.1833+1G>A | ND | RNA processing | ND |
| 38 | 17 | 17 | IVS 27 | c.1833+1 G>A | p.Gly609GlyfsX1 | RNA processing | Frameshift |
| 39 | 14 | 10 | IVS 28 | c.1888-2A>G | p.Gly630MetfsX53 | RNA processing | Frameshift |
| 40 | 13 | 6 | 29 | c.1931delC | p.Pro644LeufsX144 | Deletion | Frameshift |
| 41 | 40 | 19 | 30 | c.1957C>T | p.Arg653X | Nonsense | Premature stop codon |
| 42 | 41 | 12 | 30 | c.1957C>T | p.Arg653X | Nonsense | Premature stop codon |
| 43 | 4 | 6 | IVS 32 | c.2094+1G>A | ND | RNA processing | ND |
| 44 | 35 | 9 | IVS 32 | c.2095-1G>A | ND | RNA processing | ND |
| 45 | 40 | 8 | 33 | c.2101C>T | p.Arg701X | Nonsense | Premature stop codon |
| 46 | 31 | 10 | 33 | c.2101C>T | p.Arg701X | Nonsense | Premature stop codon |
| 47 | 8 | 11 | IVS 33 | c.2193+2T>C | ND | RNA processing | ND |
| 48 | 43 | 14 | 34 | c.2257_2264delGGCGAGAG | p.Glu754SerfsX13 | Deletion | Frameshift |
| 49 | 5 | 5 | 34 | c.2263_2264delAG | p.Arg755GlyfsX14 | Deletion | Frameshift |
| 50 | 9 | 11 | 35 | c.2353C>T | p.Arg785X | Nonsense | Premature stop codon |
| 51 | 14 | 10 | 35 | c.2353C>T | p.Arg785X | Nonsense | Premature stop codon |
| 52 | 37 | 7 | 35 | c.2353C>T | p.Arg785X | Nonsense | Premature stop codon |
| 53 | 33 | 8 | IVS 35 | c.2355+5G>A | ND | RNA processing | ND |
| 54 | 38 | 13 | IVS 35 | c2355+5G>A | p.Arg785_Gly786insValAsnGluCysGlyLeuLeuAspCysTrpAlaPheGlySerX15 | RNA processing | Insertion with premature stop codon |
| 55 | 11 | 11 | 36 | c.2381dupC | p.Gly795TrpfsX6 | Duplication | Frameshift |
| 56 | 5 | 10 | 36 | c.2382delT | p.Gly795Alafs86 | Deletion | Frameshift |
| 57 | 41 | 12 | 36 | c.2382delT | p.Gly795Alafs86 | Deletion | Frameshift |
| 58 | 14 | 9 | 38 | c.2467G>T | p.Glu823X | Nonsense | Premature stop codon |
| 59 | 44 | 9 | 38 | c.2493dupA | p.Pro832ThrfsX11 | Duplication | Frameshift |
| 60 | 66 | 13 | IVS 38 | c.2517+2T>G | ND | RNA processing | ND |
| 61 | 24 | 15 | IVS 38 | c.2518-1 G>A | p.Gly840ValfsX41 | RNA processing | Frameshift |
| 62 | 41 | 17 | 39 | c.2588-2604delCTGG TCCTCAGGGCCCC | p.Pro863LeufsX16 | Deletion | Frameshift |
| 63 | 39 | 17 | 40 | c.2659C>T | p.Arg887X | Nonsense | Premature stop codon |
| 64 | 12 | 12 | 40 | c.2673dupC | p.Ala895SerfsX49 | Duplication | Frameshift |
| 65 | 33 | 9 | 40 | c.2673delC | p.Pro893ArgfsX135 | Deletion | Frameshift |
| 66 | 9 | 14 | 41 | c.2710C>T | p.Arg904Cys | Missense | Arginine-to-cysteine substitution[20] |
| 67 | 18 (8 at examination) | 0 | 41 | c.2710C>T | p.Arg904Cys | Missense | Arginine-to-cysteine substitution[20] |
| 68 | 70 | 9 | 41 | c.2715dupT | p.Gly906TrpfsX38 | Duplication | Frameshift |
| 69 | 40 | 17 | 41 | c.2719dupC | p.Gly909ArgfsX35 | Duplication | Frameshift |
| 70 | 44 | 9 | 42 | c.2813delC | p.Pro938LeufsX90 | Deletion | Frameshift |
| 71 | 58 | 9 | 42 | c.2839C>T | p.Gln947X | Nonsense | Premature stop codon |
| 72 | 12 | 10 | 42 | c.2862C>T | p.Gly954Glyb | Synonymous | Frameshift |
| 73 | 11 | 8 | IVS 43 | c.3003+1G>A | ND | RNA processing | ND |
| 74 | 20 | 13 | IVS 43 | c.3003+5G>A | p.Gly966_Ser1001del; p.Gly990GlyfsX1a | RNA processing | Deletion; frameshift |
| 75 | 32 | 16 | 44 | c.3081_3087delGACGGT insCCTGG | p.Thr1028LeufsX100 | Deletion/insertion | frameshift |
| 76 | 18 | 17 | 44 | c.3106C>T | p.Arg1036X | Nonsense | Premature stop codon |
| 77 | 39 | 14 | 44 | c.3106C>T | p.Arg1036X | Nonsense | Premature stop codon |
| 78 | 10 | 11 | 44 | c.3106C>T | p.Arg1036X | Nonsense | Premature stop codon |
| 79 | 47 | 13 | 44 | c.3106C>T | p.Arg1036X | Nonsense | Premature stop codon |
| 80 | 45 | 10 | 44 | c.3106C>T | p.Arg1036X | Nonsense | Premature stop codon |
| 81 | 49 | 13 | IVS 44 | c.3111+1G>T | p.Glu1033LysfsX4 | RNA processing | Frameshift |
| 82 | 8 | 11 | IVS 44 | c.3112-1G>A | p.Gly1038GlufsX92 | RNA processing | Frameshift |
| 83 | 29 | 12 | 45 | c.3137delC | p.Pro1046LeufsX84 | Deletion | Frameshift |
| 84 | 8 | 12 | 45 | c.3137dupC | p.Gly1047TrpfsX11 | Duplication | Frameshift |
| 85 | 17 | 13 | 45 | c.3138delT | p.Gly1047AlafsX83 | Deletion | Frameshift |
| 86 | 20 | 12 | 46 | c.3228delT | p.Gly1077AlafsX53 | Deletion | Frameshift |
| 87 | 35 | 18 | 46 | c.3258_3261delAGAC | p.Asp1087GlufsX42 | Deletion | Frameshift |
| 88 | 42 | 9 | 47 | c.3325delC | p.Gln1109ArgfsX21 | Deletion | Frameshift |
| 89 | 18 | 11 | 48 | c.3392G>C | p.Gly1131Ala | Missense | Glycine substitution |
| 90 | 8 | 11 | 50 | c.3574C>T | p.Arg1192X | Nonsense | Premature stop codon |
| 91 | 47 | 16 | 50 | c.3574C>T | p.Arg1192X | Nonsense | Premature stop codon |
| 92 | 40 | 18 | 50 | c.3574C>T | p.Arg1192X | Nonsense | Premature stop codon |
| 93 | 42 | 11 | 51 | c.3623delC | p.Pro1208LeufsX19 | Deletion | Frameshift |
| 94 | 33 | 12 | 51 | c.3641dupC | p.Gly1215TrpfsX38 | Duplication | Frameshift |
| 95 | 41 | 10 | 51 | c.3864-3865delCT | p.Cys1289ProfsX3 | Deletion | Frameshift |
| 96 | 11 | 10 | 51 | c.3878G>A | p.Trp1293X | Nonsense | Premature stop codon |
| 97 | 55 | 12 | 52 | c.3891_3898dupCTACTGGA | p.Ile1300ThrfsX15 | Duplication | Frameshift |
| 98 | 53 | 12 | 52 | c.3903delC | p.Asn1303ThrfsX9 | Deletion | Frameshift |
| 99 | 52 | 17 | IVS 52 | c.4074+1 G>T | p.Gln1238_Leu1411del; p.Trp1348CysfsX17a | RNA processing | Deletion; frameshift |
| 100 | 8 | 10 | IVS 53 | c.4317+2T>C | ND | RNA processing | ND |
| Indication N-propeptide (p.26–181) – triple helical domain (p.201–1214) – C-propeptide (p.1242–1487) | |||||||
IVS, intervening sequence; ND, not determined.
Items in italics represent recurrent mutations.
Exons are numbered 1–54.
cDNA mutations are numbered starting from the first base of the start codon (ATG) of the cDNA reference sequence (GenBank accession number NM_001844).
Amino-acid mutations were numbered from the first Methionine of the α1(II) collagen chain (GenBank L10347).
Score as calculated by the proposed scoring system in Table 2.
Splice site mutations with multiple isoforms: see Supplementary Figure 1.
Synonymous mutation: see Supplementary Figure 2.
Two mutations were located in the alternatively spliced exon 2. The first mutation, a duplication of 23 nucleotides (c.211_233dup; p.Glu79ThrfsX2) causes a frameshift that leads to a premature stop codon within the exon itself. The patient with this mutation only had ocular features (retinal detachment) of Stickler syndrome as expected, as exon 2 is retained in the eye but spliced out in the cartilage.21, 22, 23 The second patient had the deletion c.264_276del; p.Cys89SerfsX24 that causes a frameshift with a premature stop codon in exon 3. However, this patient had both ocular and extraocular manifestations of Stickler syndrome, including vitreal abnormalities, a retinal detachment, flat face, sensorineural hearing loss, arthropathy and epiphyseal changes on radiographs.
In the skin fibroblasts or the EBV cell line available from 13 patients with 12 different splice site alterations, cDNA analysis showed that each splice site alteration resulted in a premature stop codon (data not shown). For the three splice site mutations, multiple isoforms of mRNA were detected. In each case, at least one isoform harbored a premature stop codon (Supplementary Figure 1: isoforms A3, B2 and C2). In the additional isoforms A1, B1 and C1, only skipping of the adjacent exon was observed. In the C1 isoform, skipping of all three consecutive exons (51–53) had occurred. These exons constitute the carboxypropeptide of the procollagen α1(II)-chain, which is necessary for chain association and initiation of the triple helix formation.24 Consequently, the resulting truncated protein will most likely be lost and not incorporated into the collagen trimer. In the isoform A2, exon 7 was deleted but intron 5 retained, the latter containing an in-frame stop codon. Both patients harboring the c.430-1G>C and c.4074+1G>T splice site mutation suffered from myopia, vitreoretinal abnormalities and spontaneous retinal detachments. They also showed a flat face. The individual with the c.3003+5G>A splice site mutation was born with a Pierre Robin anomaly and had myopia, a retinal detachment and cataract. He also suffered from conductive hearing loss. His affected father had a history of spontaneous bilateral retinal detachments in childhood.
One patient was heterozygous for a synonymous mutation (c.2862C>T; p.Gly954Gly) in exon 42. As this mutation was cosegregating with Stickler syndrome in the affected family, the pathogenic effect was further explored at the mRNA level. cDNA analysis showed that this mutation generated a cryptic splice site 35 nucleotides upstream of the normal donor splice site in intron 42, resulting in a frameshift with a premature stop codon (Supplementary Figure 2).
The nonsense (p.Trp1293X) and frameshift mutations (p.Cys1289ProfsX3, p.Ile1300ThrfsX15, p.Asn1303ThrfsX9) residing in the carboxypropeptide were predicted to result in NMD as they occur before the last 50 nucleotides of the last exon–exon junction (Table 1).25 In the patient with the splice site alteration in intron 53 (c.4317+2T>C), the splice site prediction program (http://www.fruitfly.org/seq_tools/splice.html) computed an insertion of a part of the intron 53 containing an in-frame stop codon.
As only 100 mutations were identified in a series of 188 patients, we decided to expand the molecular analysis with MLPA to explore the possibility of missed intragenic deletions. For this analysis, we selected 20 patients in whom we strongly suspected the diagnosis of Stickler syndrome because of the presence of severe myopia, retinal detachment and/or cleft palate. However, no additional mutations were identified in these affected individuals.
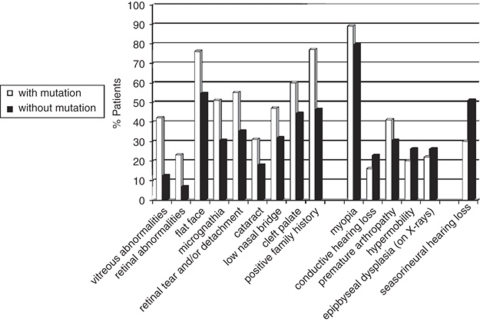
In the next step, we evaluated the clinical and radiographic features in our series of 188 patients and looked for differences between the mutation-positive (n=100) and mutation-negative (n=88) groups. The results are summarized in Figure 1. A positive family history, orofacial anomalies (cleft palate, low nasal bridge, flat face, micrognathia) and vitreoretinal changes were more frequently (P-value ≤0.05) present in the mutation-positive group. On the other hand, sensorineural hearing loss was observed more frequently in the mutation-negative group (P<0.005). Overall, 20 of 23 of the sporadic patients with a COL2A1 mutation had either a cleft palate or retinal detachment(s) with vitreous anomalies and myopia.
Figure 1.
Frequency of clinical and radiographic characteristics in patients with a COL2A1 mutation (white bars) and patients without a COL2A1 mutation (black bars). From left to right: the first nine characteristics have a P-value ≤0.05, the next five characteristics are not statistically significant and the remaining characteristic (sensorineural hearing loss) shows reverse significance with P-value <0.005.
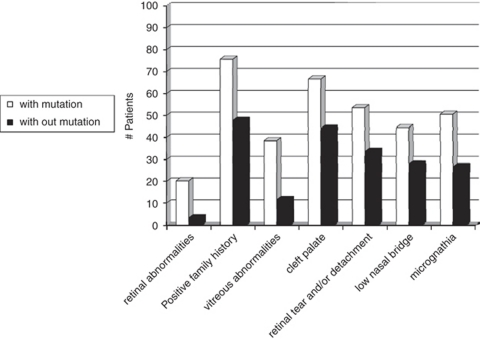
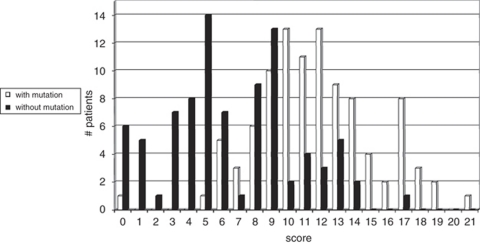
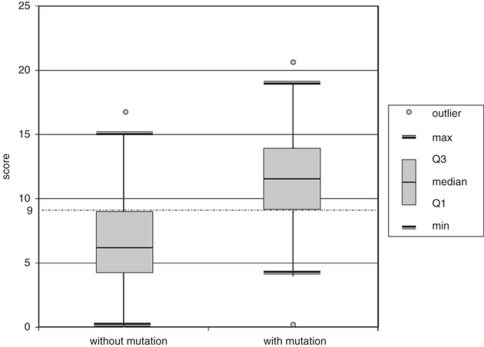
To determine the discriminating power of these features, we conducted a binary logistic regression analysis. The following characteristics were most distinguishing between both groups: (1) vitreous abnormalities, (2) retinal abnormalities, (3) history of retinal tear and/or detachment, (4) low nasal bridge, (5) cleft palate, (6) micrognathia and (7) positive family history (Figure 2). On the basis of the regression coefficient of each distinguishing characteristic, a specific scoring system was proposed. The highest score (score 5) was attributed to retinal abnormalities and positive family history, a score of 4 was assigned to the cleft palate and vitreous abnormalities, a retinal tear and/or detachment represented a score of 3, whereas low nasal bridge and micrognathia received the lowest score (score 1) (Table 2). When applying this scoring system to each patient, we observed a higher median score for patients with a COL2A1 mutation compared with those without a mutation (11.5 versus 6). The calculated score ranged from 0 to 21 with a theoretical maximum of 23. The distribution of the score for mutation-positive and mutation-negative cases is shown in Figure 3. Overall, 75% of the patients with a COL2A1 mutation had a total score ≥9 (Figure 4). The presence of vitreoretinal anomalies and a retinal detachment yields a total score of 12, illustrating the importance of a thorough ophthalmological evaluation in patients with Stickler syndrome.
Figure 2.
Frequency of the seven most distinguishing characteristics in both mutation-negative and mutation-positive groups.
Table 2. Proposed scoring system.
| Characteristics | Score |
|---|---|
| Retinal abnormalities | 5 |
| Positive family history | 5 |
| Vitreous abnormalities | 4 |
| Cleft palate | 4 |
| Retinal tear and/or detachment | 3 |
| Low nasal bridge | 1 |
| Micrognathia | 1 |
| Total score | 23 |
Figure 3.
Overlap in total score between mutation-positive and mutation-negative groups of patients.
Figure 4.
Box plot presentation of the total scores in both patient groups with Q1 representing the first quartile or 25th centile and Q3 representing the third quartile or 75th centile. Max indicates the maximum score, and min the minimum score, that is not an outlier or that is within 1.5 times the interquartile range (Q1–Q3). Overall, 75% of the patients with a COL2A1 mutation had a total score ≥9.
DISCUSSION
In the past decade, we have identified a large series of COL2A1 mutations in a group of patients referred with the diagnosis of Stickler syndrome. The availability of these data prompted us to retrospectively analyze both genotype and phenotype of these patients. In this study, we aimed to define more precisely the phenotype of Stickler syndrome type 1 and were interested in identifying discriminating features between patients with and without a COL2A1 mutation. In addition, we wanted to investigate in what respect Stickler syndrome type 1 mutations were different from other COL2A1 mutations causing the type II collagenopathies with short stature. More precisely, we wanted to learn whether all Stickler syndrome mutations were predicted to exert a loss-of-function effect on the procollagen α1(II)-chain.
Sufficient clinical and radiographic data were available on 188 probands, and in 100 of these individuals, a heterozygous COL2A1 mutation was identified. The 77 different mutations were distributed over the entire gene, and no regions of mutation clustering were found. In all, 13 mutations were observed in more than 1 proband, with 10 involving a CpG dinucleotide. One patient was heterozygous for a deletion of the entire gene and details have been published earlier.19 The 34 smaller and intragenic deletions, insertions, duplications and insertion–deletions were all out of frame and therefore predicted to result in NMD. A similar effect was shown for the synonymous mutation (p.Gly954Gly), which created a cryptic splice site (Supplementary Figure 2). This mutation is the second example of an apparently silent COL2A1 mutation that alters RNA splicing, illustrating the importance of studying the effect of so-called synonymous mutations at the mRNA level.26 Analysis of cDNA also allowed us to study the effect of 12 different splice site alterations. In addition, it provided us with more insights into the complexity of mRNA splicing of the COL2A1 gene. Each splice site mutation was shown to create at least one isoform with a frameshift and premature stop codon as a consequence (Supplementary Figure 1). In addition, some unexpected splice site outcomes were observed with skipping of one or more consecutive exons and even retention of introns more remote from the mutation. As shown before for collagen types I and V, introns are not consecutively removed in a 5′–3′ direction which may explain some unusual RNA isoforms observed in our patients.27, 28
In addition to the above-mentioned hypomorphic mutations, seven different missense mutations were also identified in this series of patients. Five mutations (p.Gly216Asp, p.Gly219Arg, p.Gly222Val, p.Gly492Asp and p.Gly1131Ala) were predicted to result in a glycine substitution. Glycine substitutions in the triple helical domain usually have a dramatic effect by hampering proper triple helix formation of the collagen trimer. They usually result in a type II collagen disorder with either lethal outcome (achondrogenesis type 2/hypochondrogenesis) or severe short stature (SEDC, Kniest dysplasia). After review of the literature and our own data, it was observed that glycine substitutions causing these short-stature phenotypes never seem to occur amino-terminal to the glycine residue at position 303.29 Glycine substitutions upstream of this residue seem to exert a less deleterious effect on collagen trimer formation and function which may explain Stickler syndrome phenotype in our patients with the p.Gly216Asp, p.Gly219Arg or p.Gly222Val substitution. For the more carboxyterminally located missense mutations, there is a less clear correlation between the location of the glycine substitution and the phenotypic outcome. The nature of the substituting amino acid may also have a role as is exemplified by the p.Gly492Val mutation that causes SEDC 30 and the Gly492Asp mutation that results in Stickler syndrome (our series).
Not only missense mutations substituting a glycine residue were identified but also two different missense mutations changing an arginine for a cysteine residue (Arg565Cys and Arg904Cys) were found in a group of five patients. These substitutions involve an arginine residue in the X position of the Gly-X-Y triplet.20, 31 As we reported earlier, substituting an arginine in the X position seems to cause Stickler syndrome, whereas substituting an arginine in the Y position rather causes a type II collagenopathy without ocular involvement.20 Cysteine residues are normally not present in the triple helical domain of the procollagen α1(II)-chain.24 The insertion of such a residue may generate aberrant disulfide bonds between mutant procollagen chains and as such may hamper proper chain alignment and trimer formation. Under these circumstances, the mutation may exert a loss-of-function effect on the protein.
The second major goal of this study was to delineate the phenotype of Stickler syndrome type 1 and to try and identify distinguishing characteristics between patients with and without a COL2A1 mutation. In the group of 100 patients with a mutation, 89% had myopia and 55% suffered from at least one episode of spontaneous retinal detachment. Vitreous abnormalities were identified in 42% of the affected individuals, but it proved difficult for most referring ophthalmologists to classify these anomalies into either a type 1 or type 2 vitreous anomaly. Overall, 60% of the mutation-positive patients presented with a cleft palate at birth. Binary logistic regression analysis showed that the ocular and orofacial features were the most distinguishing clinical characteristics between both groups. An affected first-degree relative, the presence of vitreoretinal anomalies and cleft palate were good indicators for Stickler syndrome type 1. Their presence in a patient with Stickler syndrome increases the likelihood of finding a COL2A1 mutation on molecular analysis. On the other hand, severe sensorineural hearing loss was more frequently observed in the mutation-negative group (Figure 1). The latter confirms the findings of previous studies indicating that hearing loss is more prevalent and pronounced in type 2 Stickler syndrome.3 Some features (such as myopia) were not included in the scoring system because they were frequently reported in both groups and thus only had a weak discriminating power. Interestingly, there was no statistical difference in the occurrence of early-onset osteoarthrosis and spondyloepiphyseal anomalies between the group with and without a COL2A1 mutation. When applying the proposed score system, a higher total score was found in the group of patients with a COL2A1 mutation (Figures 3 and 4), which is in contrast to previous studies in which no differences were observed.32 Nevertheless, a considerable overlap between both groups was present. This overlap is most likely due to an age-of-onset effect in the mutation-positive group and genetic heterogeneity in the mutation-negative group. In the latter group, individuals with a COL11A1 mutation may be present (especially those with severe hearing loss), as well as patients with an undetected COL2A1 mutation (false-negative patients). Indeed, samples referred at the beginning of the study were analyzed with less sensitive mutation screening techniques, such as SSCP and CSGE. In addition, deletions involving one particular amplicon will be missed by sequencing analysis. However, MLPA analysis in a selected group of patients failed to unravel new mutations. In addition, regions outside the coding sequences such as the promoter were not analyzed in this study. Lower scores in the mutation-positive group may be attributed to the young age of the affected individuals not yet showing all features (such as retinal detachments) of Stickler syndrome type 1. Of the 16 cases with a score of ≤8, 14 were <14 years of age (Table 1).
In conclusion, this study conducted in a large series of patients, confirms that Stickler syndrome type 1 is predominantly caused by loss-of-function mutations in the COL2A1 gene. Only 10% of the gene alterations were missense mutations residing in the triple helical domain, some of which may still exert a hypomorphic effect (such as the arginine-to-cysteine substitutions). Vitreoretinal abnormalities including the occurrence of a retinal tear or detachment were statistically more frequent in Stickler syndrome patients with a COL2A1 mutation compared with those without a mutation. Together with cleft palate and a positive family history, these characteristics were shown to be good indicators for a type II collagen defect (in contrast to severe sensorineural hearing loss). Finally, binary regression analysis allowed us to develop a scoring system that highlighted the importance of a thorough vitreoretinal assessment when evaluating individuals suspected with Stickler syndrome type 1.
Acknowledgments
We are grateful to the patients and their families for their cooperation. We thank the following clinicians for the referral of samples: M Ausems, M Baumgartner, K Becker, S Bertok, F Betis, AM Bisgaard, K Bouman, H Brunner, O Calabrese, K Chandler, S De Almeida, T De Ravel, K Devriendt, M Drolenga, I Feenstra, JP Fryns, H Fryssira, F Goodman, BCJ Hamel, JM Hertz, T Homfray, J Hurst, S Janssens, D Johnson, J Kamphoven, WS Kerstjens-Frederikse, K Keymolen, I Liebaers, M Maas, F Malfait, H Malmgren, S Mancini, S Mansour, I Mathijssen, T McDevitt, EJ Meijers, F Meire, A Mendicino, N Mignone, A Muellner-Eidenbock, R Newbury-Ecob, A Nordgren, C Postma, EM Ruiter, P Schmidt, C Schrander-Stumpel, F Stanzial, A Superti-Furga, K Ten Berg, P Terhal, S Tinschert, A Tzschach, D van den Boogaard, I Van Der Burgt, P Van Kerrebroeck, L Van Maldergem, N Van Regemorter, J Vigneron, AMC Vos, M Wright and A Zankl. This study was made possible by Grant no. G.0331.03 from the Research Foundation-Flanders (FWO) and GOA grant no. 12051203 from the Ghent University. Geert R Mortier is senior clinical investigator at the Research Foundation – Flanders (FWO). The corresponding author has the right to grant on behalf of all authors and does grant on behalf of all authors, an exclusive license (or non-exclusive for government employees) on a worldwide basis to the BMJ Publishing Group Ltd and its Licensees to permit this article (if accepted) to be published in the Journal of Medical Genetics and any other BMJPGL products to exploit all subsidiary rights, as set out in our license.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies the paper on European Journal of Human Genetics website (http://www.nature.com/ejhg)
Supplementary Material
References
- Stickler GB, Belau PG, Farrell FJ, et al. Hereditary progressive arthro-ophthalmopathy. Mayo Clin Proc. 1965;40:433–455. [PubMed] [Google Scholar]
- Stickler GB, Hughes W, Houchin P. Clinical features of hereditary progressive arthro-ophthalmopathy (Stickler syndrome): a survey. Genet Med. 2001;3:192–196. doi: 10.1097/00125817-200105000-00008. [DOI] [PubMed] [Google Scholar]
- Majava M, Hoornaert KP, Bartholdi D, et al. A report on 10 new patients with heterozygous mutations in the COL11A1 gene and a review of genotype-phenotype correlations in type XI collagenopathies. Am J Med Genet A. 2007;143:258–264. doi: 10.1002/ajmg.a.31586. [DOI] [PubMed] [Google Scholar]
- Snead MP, Yates JR. Clinical and molecular genetics of Stickler syndrome. J Med Genet. 1999;36:353–359. [PMC free article] [PubMed] [Google Scholar]
- Richards AJ, Laidlaw M, Whittaker J, et al. High efficiency of mutation detection in type 1 Stickler syndrome using a two-stage approach: vitreoretinal assessment coupled with exon sequencing for screening COL2A1. Hum Mutat. 2006;27:696–704. doi: 10.1002/humu.20347. [DOI] [PubMed] [Google Scholar]
- Ahmad NN, Ala-Kokko L, Knowlton RG, et al. Stop codon in the procollagen II gene (COL2A1) in a family with the Stickler syndrome (arthro-ophthalmopathy) Proc Natl Acad Sci USA. 1991;88:6624–6627. doi: 10.1073/pnas.88.15.6624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards AJ, Yates JR, Williams R, et al. A family with Stickler syndrome type 2 has a mutation in the COL11A1 gene resulting in the substitution of glycine 97 by valine in alpha 1 (XI) collagen. Hum Mol Genet. 1996;5:1339–1343. doi: 10.1093/hmg/5.9.1339. [DOI] [PubMed] [Google Scholar]
- Vikkula M, Mariman EC, Lui VC, et al. Autosomal dominant and recessive osteochondrodysplasias associated with the COL11A2 locus. Cell. 1995;80:431–437. doi: 10.1016/0092-8674(95)90493-x. [DOI] [PubMed] [Google Scholar]
- Van Camp G, Snoeckx RL, Hilgert N, et al. A new autosomal recessive form of Stickler syndrome is caused by a mutation in the COL9A1 gene. Am J Hum Genet. 2006;79:449–457. doi: 10.1086/506478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spranger J, Winterpacht A, Zabel B. The type II collagenopathies: a spectrum of chondrodysplasias. Eur J Pediatr. 1994;153:56–65. doi: 10.1007/BF01959208. [DOI] [PubMed] [Google Scholar]
- Zankl A, Neumann L, Ignatius J, et al. Dominant negative mutations in the C-propeptide of COL2A1 cause platyspondylic lethal skeletal dysplasia, torrance type, and define a novel subfamily within the type 2 collagenopathies. Am J Med Genet. 2005;133:61–67. doi: 10.1002/ajmg.a.30531. [DOI] [PubMed] [Google Scholar]
- Ganguly A, Rock MJ, Prockop DJ. Conformation-sensitive gel electrophoresis for rapid detection of single-base differences in double-stranded PCR products and DNA fragments: evidence for solvent-induced bends in DNA heteroduplexes. Proc Natl Acad Sci USA. 1993;90:10325–10329. doi: 10.1073/pnas.90.21.10325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orita M, Iwahana H, Kanazawa H, et al. Detection of polymorphisms of human DNA by gel electrophoresis as single-strand conformation polymorphisms. Proc Natl Acad Sci USA. 1989;86:2766–2770. doi: 10.1073/pnas.86.8.2766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao W, Oefner PJ. Denaturing high-performance liquid chromatography: a review. Hum Mutat. 2001;17:439–474. doi: 10.1002/humu.1130. [DOI] [PubMed] [Google Scholar]
- Schouten JP, McElgunn CJ, Waaijer R, et al. Relative quantification of 40 nucleic acid sequences by multiplex ligation-dependent probe amplification. Nucleic Acids Res. 2002;30:e57. doi: 10.1093/nar/gnf056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrell FE, Jr, Lee KL, Mark DB. Multivariable prognostic models: issues in developing models, evaluating assumptions and adequacy, and measuring and reducing errors. Stat Med. 1996;15:361–387. doi: 10.1002/(SICI)1097-0258(19960229)15:4<361::AID-SIM168>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- Laupacis A, Sekar N, Stiell IG. Clinical prediction rules. A review and suggested modifications of methodological standards. JAMA. 1997;277:488–494. [PubMed] [Google Scholar]
- Spiegelhalter DJ. Probabilistic prediction in patient management and clinical trials. Stat Med. 1986;5:421–433. doi: 10.1002/sim.4780050506. [DOI] [PubMed] [Google Scholar]
- Van Der Hout AH, Verlind E, Beemer FA, et al. Occurrence of deletion of a COL2A1 allele as the mutation in Stickler syndrome shows that a collagen type II dosage effect underlies this syndrome. Hum Mutat. 2002;20:236. doi: 10.1002/humu.9061. [DOI] [PubMed] [Google Scholar]
- Hoornaert KP, Dewinter C, Vereecke I, et al. The phenotypic spectrum in patients with arginine to cysteine mutations in the COL2A1 gene. J Med Genet. 2006;43:406–413. doi: 10.1136/jmg.2005.035717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donoso LA, Edwards AO, Frost AT, et al. Clinical variability of Stickler syndrome: role of exon 2 of the collagen COL2A1 gene. Surv Ophthalmol. 2003;48:191–203. doi: 10.1016/s0039-6257(02)00460-5. [DOI] [PubMed] [Google Scholar]
- McAlinden A, Majava M, Bishop PN, et al. Missense and nonsense mutations in the alternatively spliced exon 2 of COL2A1 cause the ocular variant of Stickler syndrome. Hum Mutat. 2008;29:83–90. doi: 10.1002/humu.20603. [DOI] [PubMed] [Google Scholar]
- Richards AJ, Martin S, Yates JR, et al. COL2A1 exon 2 mutations: relevance to the Stickler and Wagner syndromes. Br J Ophthalmol. 2000;84:364–371. doi: 10.1136/bjo.84.4.364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kielty C, Grant M.The collagen family: structure, assembly, and organization in the extracellular matrixin Royce PM, Steinmann B, (eds).: Connective Tissue and its Heritable Disorders Wiley Liss Inc.: New York; 2002159–222. [Google Scholar]
- Schell T, Kulozik AE, Hentze MW. Integration of splicing, transport and translation to achieve mRNA quality control by the nonsense-mediated decay pathway. Genome Biol. 2002;3:REVIEWS1006. doi: 10.1186/gb-2002-3-3-reviews1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards AJ, Laidlaw M, Meredith SP, et al. Missense and silent mutations in COL2A1 result in Stickler syndrome but via different molecular mechanisms. Hum Mutat. 2007;28:639. doi: 10.1002/humu.9497. [DOI] [PubMed] [Google Scholar]
- Takahara K, Schwarze U, Imamura Y, et al. Order of intron removal influences multiple splice outcomes, including a two-exon skip, in a COL5A1 acceptor-site mutation that results in abnormal pro-alpha1(V) N-propeptides and Ehlers-Danlos syndrome type I. Am J Hum Genet. 2002;71:451–465. doi: 10.1086/342099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarze U, Starman BJ, Byers PH. Redefinition of exon 7 in the COL1A1 gene of type I collagen by an intron 8 splice-donor-site mutation in a form of osteogenesis imperfecta: influence of intron splice order on outcome of splice-site mutation. Am J Hum Genet. 1999;65:336–344. doi: 10.1086/302512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkin DJ, Bogaert R, Lachman RS, et al. A single amino acid substitution (G103D) in the type II collagen triple helix produces Kniest dysplasia. Hum Mol Genet. 1994;3:1999–2003. doi: 10.1093/hmg/3.11.1999. [DOI] [PubMed] [Google Scholar]
- Tiller GE, Polumbo PA, Weis MA, et al. Dominant mutations in the type II collagen gene, COL2A1, produce spondyloepimetaphyseal dysplasia, Strudwick type. Nat Genet. 1995;11:87–89. doi: 10.1038/ng0995-87. [DOI] [PubMed] [Google Scholar]
- Richards AJ, Baguley DM, Yates JR, et al. Variation in the vitreous phenotype of Stickler syndrome can be caused by different amino acid substitutions in the X position of the type II collagen Gly-X-Y triple helix. Am J Hum Genet. 2000;67:1083–1094. doi: 10.1016/s0002-9297(07)62938-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zechi-Ceide RM, Jesus Oliveira NA, Guion-Almeida ML, et al. Clinical evaluation and COL2A1 gene analysis in 21 Brazilian families with Stickler syndrome: identification of novel mutations, further genotype/phenotype correlation, and its implications for the diagnosis. Eur J Med Genet. 2008;51:183–196. doi: 10.1016/j.ejmg.2007.12.008. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.