Abstract
The Tuareg presently live in the Sahara and the Sahel. Their ancestors are commonly believed to be the Garamantes of the Libyan Fezzan, ever since it was suggested by authors of antiquity. Biological evidence, based on classical genetic markers, however, indicates kinship with the Beja of Eastern Sudan. Our study of mitochondrial DNA (mtDNA) sequences and Y chromosome SNPs of three different southern Tuareg groups from Mali, Burkina Faso and the Republic of Niger reveals a West Eurasian-North African composition of their gene pool. The data show that certain genetic lineages could not have been introduced into this population earlier than ∼9000 years ago whereas local expansions establish a minimal date at around 3000 years ago. Some of the mtDNA haplogroups observed in the Tuareg population were involved in the post-Last Glacial Maximum human expansion from Iberian refugia towards both Europe and North Africa. Interestingly, no Near Eastern mtDNA lineages connected with the Neolithic expansion have been observed in our population sample. On the other hand, the Y chromosome SNPs data show that the paternal lineages can very probably be traced to the Near Eastern Neolithic demic expansion towards North Africa, a period that is otherwise concordant with the above-mentioned mtDNA expansion. The time frame for the migration of the Tuareg towards the African Sahel belt overlaps that of early Holocene climatic changes across the Sahara (from the optimal greening ∼10 000 YBP to the extant aridity beginning at ∼6000 YBP) and the migrations of other African nomadic peoples in the area.
Keywords: Tuareg, genetic diversity, phylogeography
Introduction
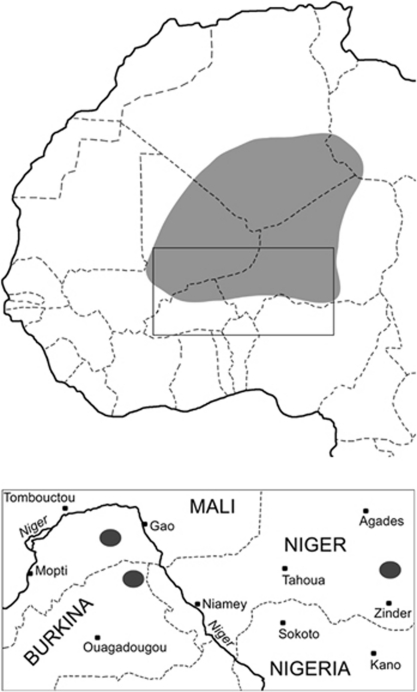
The Tuareg call themselves Kel Tamasheq (people of the Tamasheq language) or Imashaghen (free people). The Tamasheq tongue is a Berber language belonging to the Afro-Asiatic phylum. The Tuareg maintain a nomadic and/or semi-nomadic lifestyle in the Central Sahara and adjacent regions of the African Sahel, where they number about 1 262 000 in total. Their contemporary geographic distribution is shown in the upper map in Figure 1.
Figure 1.
The geographical location of southern Tuareg populations, including the ones studied here: TTan in the Republic of Niger, TGor in Burkina Faso and TGos in Mali.
The 5th century Greek historian Herodotus suggested that the ancestral homeland of the ancient Tuareg (ie Garamantes) was the Libyan Fezzan.1 It has been suggested that subsequent to the camel being adopted for Saharan trade in the 1st or 2nd century , the area of Tuareg influence expanded further to the south. Oral accounts and sparse written records in Tifinagh (the script of the Tamasheq language) date back to 14th century when the first caravan traders were documented in the Air Mountains. Hereafter, it seems that from the 17th century onwards the increasingly frequent invasions of North Africa by various Arabic tribes drove the Tuareg yet further southward to the African Sahel.
Since the beginning of European explorations of the Sahara and the Sahel, the Tuareg have been known mainly as caravan traders linking the sub-Saharan and Mediterranean cultures. Their contact with the various sub-Saharan peoples was not always peaceful and they were known to take war captives. Centuries of mutual contact led to substantial assimilation of others into the Tuareg population.
Carrying out biological or genetic investigations of the Tuareg has not always been easy because of their demanding lifestyle and their often negative attitude to the European colonists. Cavalli-Sforza et al,2 whose synthesized study of classical protein and serological markers is well known, noticed a genetic link between the Tuareg and Beja from Eastern Sudan. The fact that the genetic distances between the Tuareg and Berber/North-western Africans were larger than that between the Tuareg and Beja, provides a picture of a common origin and population separation at some point more than 5000 years ago. Interestingly, both people are also pastoralist and speak Afro-Asiatic languages, even if the Beja language (Bedawi), with its four dialects, belongs to the Cushitic branch, whereas Tamasheq belongs to the Berber branch. The fact that these two peoples today speak different languages might be explained either by the Tuareg having acquired the Berber language during their westwards migration, or possibly by the Beja coming under the influence of some Eastern African peoples as language shift is a relatively common phenomenon.
Among the first African mitochondrial DNA (mtDNA) sequences were those from data sets3, 4 obtained mostly from Tuareg living in Niger and Nigeria, and which revealed a rather sub-Saharan affinity of their population. More recently, however, a study based on 129 Tuareg samples from two villages of the Libyan Fezzan, stressed a high frequency but concomitant low diversity of the West Eurasian component, bearing only haplogroups H1, V and M1. The sub-Saharan component of the Libyan Tuareg was more diversified but predominantly represented by only two haplogroups (L2a1 and L0a1a). The Tuareg population from Libya was homogenous with very low estimates of haplotype diversity suggesting high genetic drift.5
The above-mentioned studies have thus revealed a dual influence in the genetic make-up of this African people. In this study, we provide new mtDNA and Y chromosome data sets of three unrelated Tuareg groups from three different countries (Niger, Mali and Burkina Faso). At the same time, we try to unravel the questions of their genetic origin, the mutual relationships among their sub-populations as well as possible links to neighbouring populations. The genetic heritage of the Tuareg population is analysed within the context of the West Eurasian versus sub-Saharan contributions to their gene pool.
Materials and methods
Subjects
The biological samples (buccal swabs) were obtained from three different groups of self-identified Tuareg (90 unrelated individuals in total). One population sample (n=38) was secured in Burkina Faso around the village Gorom-Gorom (further referred to as TGor). The second sample (n=31) was taken in the Republic of Niger in the vicinity of Tanut (TTan). The third sample (n=21) was collected in Mali near Gossi (TGos). The samples from Mali and Burkina Faso are geographically relatively close to each other as they are located within the bend of the Niger River, whereas the sample collected in the central part of the Republic of Niger is located some 1500 km eastward (Figure 1). The field sampling was undertaken with the collaboration of local Tuareg assistants. Of these 90 healthy and unrelated individuals, 47 were male and 43 were female. Oral informed consent was obtained from all participants in the study and research permits were obtained from the Ministries of Education and/or Health in all the three countries.
Laboratory analyses
DNA extractions and PCR amplifications of mtDNA hypervariable segments I and II (HVS-I and HVS-II) were carried out as in earlier studies.6, 7 Amplicons were purified and sequenced using forward PCR primers. In some cases the reverse primer was also used for sequencing (due to for example, the presence of poly-C stretches between nt 16184 and 16193). In some samples, SNP testing was analysed through matrix-assisted laser desorption/ionization time-of-flight mass spectrometry and minisequencing.7, 8 Whole genome sequencing of mtDNAs affiliated by D-loop to the haplogroup M1 was undertaken following the protocols reported elsewhere.7, 9, 10, 11
For the detection of Y chromosome SNP polymorphisms, the Signet Y-SNP Identification System v 2.0 (Marligen, Rockville, MD, USA) was used. All the samples were first analysed by A-R multiplex (polymorphisms M122, M168, M175, M207, M304, M343, M45, M89, M96 and M9), differentiating main evolutionary branches of Y chromosome phylogeny.12 Subsequently, samples belonging to haplogroup E were further analysed for polymorphisms DYS391, M2, M33, M35, M58, M75, M78, M81 and M123.
Haplogroup nomenclature
mtDNA classification into haplogroups L, H, U and M was carried out in accordance with the most recent phylogenetic studies of Salas et al,13 Kivisild et al14 and Behar et al15 for L; Olivieri et al16 for U and M; and Achilli et al17 for H (see also van Oven and Kayser, 2009).18 The numbering is consistent with the revised Cambridge Reference Sequence.19 The three complete mtDNA sequences have been deposited in GenBank (accession numbers GQ377749; GQ377750; and GQ377751). For the Y chromosome SNP affiliation, the nomenclature of Karafet et al12 was followed.
Statistical and phylogenetic analyses
Analysis of population structure and molecular diversity measures was calculated by using Arlequin software version 3.0.20 Two-tailed Fisher's exact test P-values of 2 × 2 contingency tables were calculated in DnaSP.21 FST genetic distances calculated by Arlequin were subsequently visualized in multidimensional scaling (MDS) by means of SPSS 10.0 software (SPSS Inc, Chicago, IL, USA). Extensive data sets of both mtDNA sequences and Y-SNPs were used to characterize the Tuareg diversity within Mediterranean and sub-Saharan population contexts (see Supplementary Material SM1 and SM2).
Phylogenetic reconstruction of mtDNA diversity was based on both HVS-I and complete sequences. The dates of the most recent common ancestor of specific subclusters in the phylogeny were estimated using ρ.22 The average number of transitions from the ancestral haplotype to all haplotypes in the cluster, for both coding (between positions 577 and 16 023) and HVS-I (between positions 16 090 and 16 365) regions, was considered with respect to mutation rate estimates of 5138 years23 and 20 180 years per transition24 within the region, respectively. Standard errors were calculated as in Saillard et al.25 Recently, updated mutation rates published by Soares et al26 were also used for the entire molecule (1 mutation every 3624 years) and synonymous substitutions (1 mutation every 7884 years). As the mutation rate determined by these authors for the HVS-I (between positions 16 090 and 16 365) region, however, is very similar to the one used above (1 transition every 20 129 years versus 20 180 years) the calculations overlapped and we present only those estimated with the original ρ mutation rate of 1 substitution every 20 180 years.
Interpolation maps
To determine and visualize the geographical distribution of haplogroups H1, H3 and V, we drew interpolation maps using the ‘Spatial Analyst Extension' of ArcView version 3.2 (http://www.esri.com/software/arcview/). Inverse distance weighted option that we used assumes that each input point has a local influence that diminishes with distance. The geographic location used is the centre of the distribution area, from where the individual samples of each population were collected. Comparative data for H1 and H3 were taken from Finnila et al,27 Herrnstadt et al,28 Pereira et al,29 Cherni et al30 and Ennafaa et al;31 and those for haplogroup V from Torroni et al,10 Pereira et al,32 Behar et al33 and Cherni et al.30
Results
The mtDNA pool of the Tuareg
The polymorphisms present in the 90 Tuareg individuals led to the identification of 53 different D-loop haplotypes (Table 1). As can be seen in the network based only on HVS-I diversity (Supplementary Material SM3), for which only 33 different haplotypes are observed, there are varying degrees of sharing of haplotypes among the analysed groups: only one belonging to haplogroup H was shared by all three groups; two haplotypes were shared by TGos–TGor and TGos–TTan; and three haplotypes were shared by TGor–TTan. Only 18 of the 33 haplotypes are unique, what is a rather low proportion when compared to most African samples.32 This is further corroborated by haplotype diversities in the three Tuareg samples, which are lower as compared with other populations – especially in the two groups of the Niger bend (0.861±0.027 in TGor; 0.910±0.037 in TGos; and 0.963±0.020 in TTan; see Supplementary Material SM4).
Table 1. HVS-I, HVS-II and other polymorphisms observed in the 90 Tuareg samples from the three populations (TGos, TGor and TTan), as well as its haplogroup (HG) affiliation.
| Haplotype ID | TGos | TGor | TTan | HV-I | HV-II | Other polymorphisms | HG |
|---|---|---|---|---|---|---|---|
| T1 | 1 | 129 183C 189 519 | 263 315.1C | 750 3010 4769 8602 | H1* | ||
| T2 | 1 | 189 519 | 073 263 315.1C | 750 3010 4769 14552 | H1a? | ||
| T3 | 2 | 189 519 | 073 263 315.1C | 750 3010 4769 8602 | H1 | ||
| T4 | 1 | 189 519 | 073 263 315.1C 328 | 750 3010 4769 8602 | H1 | ||
| T5 | 1 | 189 192C>T 519 | 073 263 315.1C | 750 3010 4769 8602 | H1 | ||
| T6 | 1 | 311 519 | 152 263 315.1C | 750 3010 4769 8602 | H1 | ||
| T7 | 1 | 311 519 | 263 315.1C | 750 | U3/H? | ||
| T8 | 1 | 311 519 | 263 315.1C | 750 3010 4769 8602 | H1 | ||
| T9 | 1 | 519 | 073 309.1 315.1C | 750 3010 4769 12308 14552 | H1a | ||
| T10 | 1 | 519 | 152 263 315.1C | 750 3010 4769 14552 | H1 | ||
| T11 | 2 | 519 | 263 309.1 315.1C | 750 3010 4769 14552T | H1 | ||
| T12 | 1 | 519 | 263 309.1C 315.1C | 750 4769 8602 | H3* | ||
| T13 | 2 | 1 | 519 | 263 315.1C | 750 3010 4769 14552 | H1 | |
| T14 | 3 | 519 | 263 315.1C | 750 3010 4769 14552T | H1 | ||
| T15 | 3 | 519 | 263 315.1C | 750 3010 4769 8602 | H1 | ||
| T16 | 1 | 093 260 343 390 519 | 073 150 263 315.1C | U3a | |||
| T17 | 1 | 234 298 | 064 072 239 263 309.1 315.1C | 750 2706 4580 4769 7028 14552 | V | ||
| T18 | 1 | 234 298 | 072 263 309.1 315.1C | 750 2706 4580 4769 7028 12308 14552 | V | ||
| T19 | 1 | 189 234 298 | 072 263 309.1 315.1C | 750 2706 4580 4769 7028 14552T | V | ||
| T20 | 1 | 234 293 298 | 072 263 309.1 315.1C | 750 2706 4580 4769 7028 12308 14552 | V | ||
| T21 | 5 | 234 298 | 072 239 263 309.1 315.1C | 750 2706 4580 4769 7028 14552 | V | ||
| T22 | 1 | 234 298 | 072 239 263 309.1 315.1C | 750 2706 4580 4769 7028 12308 14552 | V | ||
| T23 | 1 | 234 298 | 072 239 263 309.1 315.1C | 750 2706 4580 4769 7028 14552T | V | ||
| T24 | 1 | 129 148 168 172 187 188G 189 193 223 230 278 293 311 320 | 093 095C 152 185 189 236 247 263 309.1C 315.1 523–524delAC | L0a1a | |||
| T25 | 1 | 093 126 145 187 189 223 264 270 278 293 311 519 | 073 152 182 185T 195 247 263 315.1C 357 523–524 delAC | L1b | |||
| T26 | 1 | 126 172 187 189 223 264 270 278 293 311 519 | 073 146 152 182 185T 195 247 263 315.1C 357 523–524delAC | L1b1 | |||
| T27 | 1 | 126 187 189 223 264 270 278 293 311 | 073 146 152 263 315.1C 357 523–524delAC 573insCCCC | L1b1 | |||
| T28 | 1 | 126 187 189 223 264 270 278 293 311 519 | 073 152 182 185T 189 195 247 263 315.1C 357 523–524delAC | L1b1 | |||
| T29 | 1 | 126 187 189 223 264 270 278 293 311 519 | 073 152 182 185T 189 195 247 263 309.1 315.1C 357 523–524delAC | L1b1 | |||
| T30 | 1 | 129 183C 189 215 223 261 278 294 311 360 519 | 073 151 152 182 186A 189C 247 263 315.1C 316 523–524delAC | L1c | |||
| T31 | 1 | 172 213 223 261 278 318 390 | 073 146 150 152 182 195 198 263 315.1C 523–524delAC | L2* | |||
| T32 | 1 | 2 | 093 189 192 223 278 294 311 390 519 | 073 143 146 152 195 263 309.1 315.1C | L2a | ||
| T33 | 2 | 1 | 093 189 193 223 278 294 311 390 519 | 073 143 146 152 195 263 315.1C | L2a | ||
| T34 | 1 | 169 223 239 278 294 309 390 526 | 073 143 146 152 189 195 263 309.1C 315.1C 523–524delAC | L2a | |||
| T35 | 2 | 183C 189 223 278 294 390 | 073 143 146 152 189 195 263 315.1C 517 | L2a | |||
| T36 | 1 | 188 189 223 278 294 311 320 390 | 073 146 152 263 309.1 315.1C | L2a | |||
| T37 | 3 | 189 223 278 294 390 | 073 143 146 152 195 263 309.1 315.1C | L2a | |||
| T38 | 2 | 189 223 278 294 390 | 073 143 146 152 195 263 315.1C | L2a | |||
| T39 | 5 | 189 192 223 278 294 309 390 519 | 073 146 152 195 263 315.1C | L2a1 | |||
| T40 | 2 | 223 278 294 309 390 519 | 073 143 146 152 195 263 315.1 523–524delAC | L2a1 | |||
| T41 | 1 | 223 278 294 309 390 519 | 073 143 146 152 189 195 203 204 263 309.1C 315.1C | L2a1 | |||
| T42 | 1 | 223 278 294 309 390 519 | 073 146 152 195 263 315.1C 398 | L2a1 | |||
| T43 | 1 | 223 355 | 073 150 152 235 263 309.1C 315.1C 494 | L3 | |||
| T44 | 2 | 124 223 278 311 362 519 | 073 152 263 315.1 523–524delAC | L3b | |||
| T45 | 2 | 124 223 278 362 519 | 073 152 200 263 309.1C 315.1C 523–524delAC | L3b | |||
| T46 | 2 | 124 166 223 309 | 073 152 263 315.1C 523–524delAC | L3d | |||
| T47 | 1 | 209 223 261G 292 311 519 | 073 152 189 194 200 263 315.1C | L3f1 | |||
| T48 | 1 | 209 223 292 311 519 | 073 189 200 214 263 309.1C 315.1C | L3f1 | |||
| T49 | 1 | 129 174 192 218 223 256 311 362 | 073 151 152 189C 195 263 294 315.1C 523–524delAC | L3h | |||
| T50 | 1 | 093G 223 287A 293T 301 311 355 362 399 | 073 146 150 152 192 200 244 263 315.1C 513 | L4g | |||
| T51 | 7 | 129 183C 189 223 249 311 519 | 073 152 195 263 (315.1)+2C 489 513 | M1a2 | |||
| T52 | 1 | 129 182C 183C 189 223 249 311 362 399 519 | 073 195 263 315.1 489 | M1b2 | |||
| T53 | 4 | 129 185 189 223 249 311 519 | 073 195 198 263 309.1 315.1 466 489 | M1b1 | |||
| Total | 21 | 38 | 31 |
Variant positions from the rCRS are shown at least for the minimum segment between nucleotide position 16017 and 16399 in HVS-I and 064 and 340 for HVS-II. Number of variants in the HVS-I region are referred with respect to the rCRS minus 16000. Substitutions are transitions unless the base change or a deletion is explicitly indicated, while insertions in poly-C tract are indicated by ‘.1'. An heteroplasmy for position 16 192 is indicated by ‘C>T'.
A total of 48% of the mtDNA haplotypes observed in the Tuareg populations could be ascribed to sub-Saharan haplogroups. Another 39%, however, were of West Eurasian ancestry (non-L types in Table 1), which is a substantial proportion considering the sub-Saharan geographical location. In fact, it has been observed that in typical North African populations there is a gradient of increasing frequency of West Eurasian lineages ranging from around 50–75% in the northernmost locations.34 The Tuareg's neighbours, however, have a markedly smaller proportion of West Eurasian haplotypes (22% in Western Chad Arabs, 8% in Shuwa Arabs from North-eastern Nigeria, 7% in the Buduma from South-eastern Niger and 6% in the Kanuri from North-eastern Nigeria).35 The remaining 13% of Tuareg haplotypes belong to the typical East African haplogroup M1.
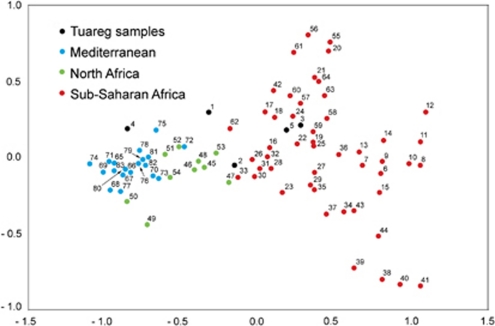
Furthermore, we noticed some differences in the distribution of West Eurasian mtDNA haplogroups between Tuareg groups. Most of the West Eurasian haplogroups (30 out of 35 sequences, amounting to 6 out of 9 HVS-I haplotypes) and the East African M1 (11 out of 12 sequences but amounting to only 2 out of 3 HVS-I haplotypes) are observed in the two Tuareg populations – TGos and TGor – located within the bend of the Niger. Tuareg from the Republic of Niger, TTan, have much higher proportion of sub-Saharan (81%) haplogroups than of West Eurasian (16%) and East African (3%) ones. These differences in haplogroup distribution led to statistically significant genetic distances when comparing HVS-I haplotypes between Tuareg from Mali (TGos) with those from the Republic of Niger (TTan) (FST=0.048; unadjusted P-value=0.009), as well as Tuareg from Burkina Faso (TGor) with those from the Republic of Niger (TTan) (FST=0.064; unadjusted P-value=0.000), whereas Tuareg from Mali (TGos) and from Burkina Faso (TGor) are not statistically different (FST=0.012; unadjusted P-value=0.234). Similarly, analysis of MDS based on FST distances and using a large database of West Eurasian and African mtDNA sequences has shown a very good separation of the sub-Saharan and West Eurasian-North African gene pools (Figure 2). Only some East African populations are closer to the West Eurasian samples, respectively, to the North African populations analysed here. This picture is a good representation of FST values as the normalized raw stress is very low (0.01165). However, the analysed Tuareg populations are divided between two gene pools: like the sample from Libya,5 the groups located within the bend of Niger (TGor and TGos) fall into the West Eurasian gene pool, whereas the Tuareg from the Republic of Niger (TTan) and the Tuareg sample from the Watson's data set3, 4 are permeated by the sub-Saharan mtDNA gene pool.
Figure 2.
MDS plot of FST genetic distances calculated from HVS-I mtDNA sequences. For numbers see Supplementary Material SM1.
The West Eurasian component observed in the Tuareg is highly interesting. A major proportion (94%) could be allocated to haplogroups H1, H3 and V, West Eurasian lineages of Iberian origin that spread to Europe7, 10, 17, 26, 29, 36 and most probably North Africa30, 31 with the improvement of the climatic conditions after the retreat of the ice sheets 15 000–13 000 years ago. The interpolation maps of these lineages across North Africa and Europe (Supplementary Material SM5) clearly place the Tuareg population in the path of the southern African edge of post-Last Glacial Maximum expansions. The H1 haplogroup (Supplementary Material SM5A and SM5B, with and without the outlier Norway, respectively) is as frequent in our southern Tuareg groups as in Libya and the centre of the dispersion within the Iberian Peninsula. The H3 haplogroup is almost vestigial in Tuareg (Supplementary Material SM5C), having the highest observed frequencies outside of Iberia in Algeria and Tunisia. Again for haplogroup V, Tuareg present frequencies as high as in the Basque country (Supplementary Material SM5D).
Both H1 and H3 commonly display rather low diversity in the D-loop region, but the Tuareg haplotypes belonging to haplogroup V have a specific diagnostic mutation – the transition at position 16 234. All the Tuareg V haplotype samples collected in Burkina Faso and the Republic of Niger (three haplotypes observed in 11 individuals) bear this mutation together with the defining substitution at position 16 298, This polymorphism is present in two of the five V haplotypes observed in the recently published Libyan Tuaregs.5 This fact seems to point to a founder effect in haplogroup V occurring in our southern Tuareg population; the further presence of two other polymorphisms in two V samples (substitutions at positions 16 189 and 16 293) allows a very preliminary estimation for the Time to the Most Recent Common Ancestor (TMRCA) of this Tuareg V sub-lineage at around 3600±2600 years ago or at a maximum of 8800 years ago if using a 95% confidence interval (see Supplementary Material SM6 for the network).
Another very interesting characteristic of the West Eurasian mtDNA pool in the Tuareg population as a whole (including Watson's and Ottoni's data sets) is the total absence of so-called Neolithic haplogroups derived from the branch JT, which are otherwise common in Near Eastern, North African, Mediterranean and even some East African populations. The virtual absence of these lineages in the Tuareg is statistically significant when comparing the frequency of these lineages in Morocco34 (18% unadjusted P-value=0.000), Tunisia30 (12% unadjusted P-value=0.000) and Egypt37 (29% unadjusted P-value=0.000). Notice also the absence of the haplogroup U6, which is present mainly in Berbers but also in several others North African groups.13, 16, 38
The sub-Saharan mtDNA pool of the Tuareg is composed of various lineages from the major L-type haplogroups including: 2.3% of L0; 14.0% of L1; 58.1% of L2; 23.3% L3; and 2.3% of L4. We assayed to search for haplotype matches in an extensive database of 7211 individuals from all over Africa (Table 2). The most ancient lineages L0a1a and L1c, characteristic of east/southeast Africa13 and the Pygmies,39 respectively, were each observed in only one individual. The highly frequent African haplogroup L2, and specifically its dominant clade L2a, is also dominant in Tuareg – it is probable that some branches of L2a were involved in the Bantu expansion towards the African south13, 40 and many matches are observed for these haplotypes all over the continent. Curiously, the two L2a lineages having substitutions at positions 16 192 and 16 193, respectively, have no match in Africa. As far as the L3 macrohaplogroup is concerned, the two L3b haplotypes observed in the Tuareg are widespread throughout the continent, but one of the L3f1 haplotypes (T47 in Table 1) has no matches. Both are included in the L3f1 sub-haplogroup, which is quite frequent and widespread, and which very probably originated in East Africa. No L3f3, a typical marker of the Chadic migration,41 has been observed in the Tuareg.
Table 2. HVS-I haplotype match for the haplotypes observed in Tuaregs samples and an extensive African database (composed of the following geographical regions: Central (AF-C), East (AF-E), North (AF-N), South (AF-S), Southeast (AF-SE), Southwest (AF-SW), West (AF-W) and West-Central (AF-WC)). Number of variants in the HVS-I region are referred with respect to the rCRS minus 16000.
| Haplotypes | Tuareg | AF-C | AF-E | AF-N | AF-S | AF-SE | AF-SW | AF-W | AF-WC | Totals | Haplogroup |
|---|---|---|---|---|---|---|---|---|---|---|---|
| rCRS | 14 | 0 | 4 | 193 | 2 | 0 | 1 | 7 | 5 | 226 | H? |
| 093 126 145 187 189 223 264 270 278 293 311 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 2 | L1b |
| 093 189 192 223 278 294 311 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 3 | L2a |
| 093 189 193 223 278 294 311 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 3 | L2a |
| 093 260 343 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | U3a |
| 093G 223 287A 293T 301 311 355 362 | 1 | 2 | 0 | 0 | 0 | 0 | 0 | 9 | 12 | L4g | |
| 124 166 223 309 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | L3d |
| 124 223 278 311 362 | 2 | 11 | 3 | 0 | 0 | 3 | 1 | 2 | 14 | 36 | L3b |
| 124 223 278 362 | 2 | 5 | 3 | 4 | 0 | 1 | 2 | 67 | 34 | 118 | L3b |
| 126 172 187 189 223 264 270 278 293 311 | 1 | 2 | 0 | 1 | 0 | 0 | 0 | 0 | 3 | 7 | L1b1 |
| 126 187 189 223 264 270 278 293 311 | 3 | 26 | 2 | 15 | 0 | 1 | 0 | 47 | 37 | 131 | L1b1 |
| 129 148 168 172 187 188G 189 193 223 230 278 293 311 320 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | L0a1a |
| 129 174 192 218 223 256 311 362 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | L3h |
| 129 182C 1831 189 223 249 311 362 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | M1b2 |
| 129 183C 189 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | H1 |
| 129 183C 189 215 223 261 278 294 311 360 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | L1c |
| 129 183C 189 223 249 311 | 7 | 0 | 0 | 10 | 0 | 0 | 0 | 0 | 1 | 18 | M1a2 |
| 129 185 189 223 249 311 | 4 | 0 | 0 | 5 | 0 | 0 | 0 | 0 | 0 | 9 | M1b1 |
| 169 223 239 278 294 309 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | L2a |
| 172 213 223 261 278 318 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | L2* |
| 183C 189 223 278 294 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 3 | L2a |
| 188 189 223 278 294 311 320 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 2 | L2a |
| 189 | 5 | 0 | 0 | 7 | 0 | 0 | 0 | 0 | 0 | 12 | H |
| 189 192 223 278 294 309 | 5 | 14 | 9 | 8 | 3 | 3 | 3 | 19 | 24 | 88 | L2a1 |
| 189 223 278 294 | 5 | 0 | 4 | 1 | 0 | 0 | 0 | 3 | 0 | 13 | L2a |
| 189 234 298 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | V |
| 209 223 261G 292 311 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | L3f1 |
| 209 223 292 311 | 1 | 1 | 12 | 4 | 0 | 0 | 0 | 4 | 18 | 40 | L3f1 |
| 223 278 294 309 | 4 | 18 | 4 | 12 | 0 | 10 | 1 | 38 | 20 | 107 | L2a1 |
| 223 355 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 3 | 1 | 5 | L3 |
| 234 293 298 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | V |
| 234 298 | 9 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 9 | V |
| 311 | 3 | 0 | 4 | 19 | 1 | 0 | 0 | 0 | 0 | 27 | U3?/H |
| Partial totals | 90 | 79 | 45 | 280 | 6 | 18 | 8 | 191 | 167 | 884 | |
| Totals (sample sizes per continental region) | 90 | 1404 | 862 | 1360 | 266 | 416 | 157 | 1454 | 1202 | 7211 |
In summary, the matches between Tuareg sub-Saharan haplotypes and the diverse African regions were, after correcting for the size for each region, 5.6% with Africa-Central; 4.3% with Africa-East; 3.4% with Africa-North; 1.1% with Africa-South; 4.3% with Africa-southeast; 4.5% with Africa-southwest; 12.7% with Africa-West; and 13.4% with Africa-westcentral. The West Africa or West-Central African lineages thus are clearly dominant in the extant Tuareg.
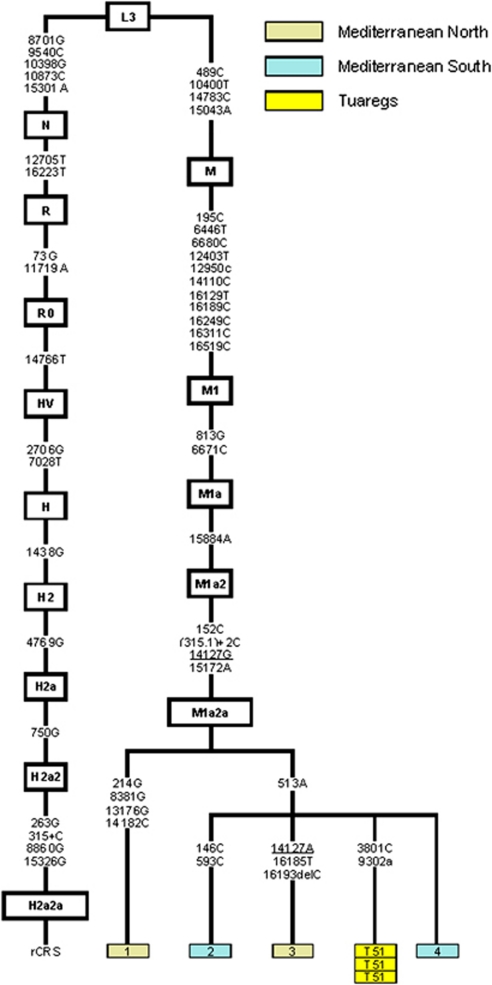
The influence of East Africa in the Tuareg can be investigated more directly through haplogroup M1.16 As concerns the finer classification of Tuareg M1 haplotypes, two of them (5 sequences out of 12) belong to M1b, which has a clear Mediterranean distribution, pointing to North Africa as its most probable gateway to the Tuareg. This finding is inconsistent with the absence of U6, which is believed to have entered Africa together with M1 in a back migration from western Eurasia around 45 000 years ago. The time estimate for M1b, based on the coding region, is 23 400±5600 years,16 placing its origin in the Early Upper Palaeolithic. More promising in ascertaining Eastern African origin is another haplotype observed in seven Tuareg individuals from Burkina Faso belonging to haplogroup M1a, which, though being considered dominant in East Africa42 also spread to the Mediterranean, and which has a total age of 28 800±4900 years.16 We performed the complete sequencing of three individuals, which despite not displaying any difference at HVS-I and HVS-II, might present some substitutions in the coding region, allowing for a better estimate of a TMRCA. These three samples, however, did not bear any difference even when sequencing the complete genome. Nonetheless, when taken together with the other M1a2a individuals (Figure 3) reported in Olivieri et al16 (sample 1 in Figure 3, accession number EF060335; sample 2, accession number EF060336), González et al43 (sample 3; accession number DQ779927) and Maca-Meyer et al44 (sample 4; accession number AF381984) allowed an age estimation for this sub-haplogroup at 8000±2400-years old based on diversity in the coding region. We checked the TMRCA using Soares et al26 mutations rates for the entire molecule and for the synonymous substitutions, obtaining, respectively, the following concordant dates: 10 400±2300 and 10 200±3400. Notice, however, that all the other four M1a2a complete sequences were observed in the Mediterranean region and in Table 2 the HVS-I motif observed in Tuareg has 10 perfect matches in the Africa-North data set and one in Africa-westcentral.
Figure 3.
Phylogeny of the complete M1a2a mtDNA sequences, including the ones from Tuaregs and the published so far. Integers represent transitions, and an upper case suffix indicates a transition while a lower case suffix indicates a transversion. Deletions are indicated by a ‘del' following the deleted nucleotide position. Underlined nucleotide positions appear more than once in the tree.
Y chromosome pool in Tuareg
From the 20 branches of the Y chromosome tree, which could be discriminated by the analyses performed, only 7 were observed in our Tuareg population sample (Supplementary Material SM7). Again, from this perspective of Y chromosome diversity, TTan is closer to sub-Saharan populations than the other two Tuareg populations, presenting 5.6% of the old AB lineages and 44.4% of E1b1a, whereas TGor and TGos have, respectively, 16.7 and 9.1% of E1b1a. Curiously, TTan also presents the highest frequency (33.3%) of West Eurasian R1b lineages whereas TGor presents only 5.6% of lineage K* (xO,P), and TGos presents none. There were no instances of the Eurasian J haplogroup in the Tuareg, which is otherwise frequent in North Africa (an average of 20% see Arredi et al45), and attains the highest frequency in the Middle East (around 50% see Semino et al)46.
The dominant haplogroup in TGor (77.8%) and TGos (81.8%) is E1b1b1b, which has a much lower frequency in TTan (11.1%). This haplogroup reaches a mean frequency of 42% in North Africa, decreasing in frequency from 76% in Morocco to ∼10% in Egypt.45 Arredi et al45 dated this haplogroup in North Africa from 2800 to 9800 YBP, associating its expansion with the Neolithic demic diffusion of Afro-Asiatic-speaking pastoralists from the Middle East.
The low level of diversity attained in the Tuareg populations (see Supplementary Material SM8) is consistent with a model of population constancy, although it can also be due in part to the ascertainment bias in the selection of a few Y-SNPs. Haplotype diversities and mean number of pairwise differences were very low in TGor and TGos, being among the lowest values observed in many populations, but TTan showed much higher levels of diversity.
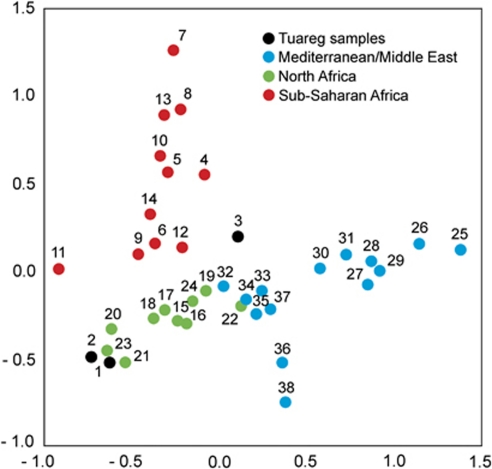
MDS of FST distances based on available Y-SNP West Eurasian and African population data sets shows, as in the case of mtDNA, separation of the West Eurasian-North African and sub-Saharan populations (Figure 4). A certain separation between the Iberian and Near Eastern groups can be explained by the absence of samples from the Central Mediterranean for the Y-NRY data set. However, though the Tuareg groups from the Niger bend (TGor and TGos) belong clearly on the West Eurasian side, the Tuareg from central Niger lean towards sub-Saharan variability.
Figure 4.
MDS plot of FST distances calculated from NRY haplogroup frequencies. Codes for numbers are as in Supplementary Material SM2.
Discussion
The Tuareg have a nomadic lifestyle and according to some demographic reports they show reduced fertility in comparison with their neighbours.47, 48, 49 The data observed here for mtDNA and Y-SNP diversities are concordant with those independent reports, especially for the Tuareg living within the bend of the Niger.
The overall West Eurasian mtDNA gene pool in the Tuareg population as a whole (H1, H3 and V) seems to favour a North African heritage.50 The only exception is the absence of the otherwise rare U5b that might have rather come to Africa through the Near East, and then drifted to higher frequencies only in some isolated populations such as in the Egyptian oasis Siwa.51 The absence of U6 can further be explained by genetic drift during the expansion of this haplogroup within North Africa.51 Note that U6 was observed at low frequencies in several population groups from the Chad Basin, such as in the Nilo-Saharan Kanuri and the Afro-Asiatic Masa.35
Relationships with the peoples of Eastern Sudan (the Beja) as pointed to by the study of classical genetic markers2 cannot yet be disregarded here as there is still no mtDNA of the Beja people available for study. However, according to historical reports, the origin of the Beja is more likely to be traceable to the Arabian Peninsula52 and the West Eurasian mtDNA lineages seen in the Tuareg have a rather Iberian affiliation in the post-LGM, and probably expanded to North Africa first.30, 31 The weak Eastern African influence in Tuareg is further supported by the M1 haplotypes belonging to the lineages characteristic of the later Mediterranean expansion (M1b and M1a2a) and the presence of very few matches for sub-Saharan L haplotypes with East Africa. The main post-LGM Eurasian and M1a2a lineages found in the Tuareg favour North African origin with migration to its southern location in the Sahel between ∼9000 and ∼3000 years ago. The upper time limit is defined by the age of the M1a2a, (estimated here from the coding region diversity observed in the three Tuareg, two North and two south Mediterranean individuals at 8000±2400), and by the upper 95% confidence interval for the Tuareg V lineages having polymorphism 16 234 (8800 years ago); the lower limit is defined by the age of the Tuareg V lineages having polymorphism 16 234 (3600 years ago).
The dates obtained from the genetic data coincide well with climatic changes in the Sahara, which resulted in repopulation during the first half of the Holocene when by ∼10 000 YBP (the Holocene climatic optimum) humid conditions and greening were established. The climatic optimum lasted until ∼6000 YBP, when the shift towards more permanent aridity occurred, culminating with the formation of the current Sahara desert. This desertification could have entrapped Tuareg populations coming from North Africa to the Sahel belt together with other pastoralists such as the Chadic speaking peoples41 coming from East Africa and Fulani nomads6 coming from West Africa. In fact, by performing complete mtDNA sequencing of the L3f3 lineage, specific for Chadic-speaking groups of the Chad Basin, Černý et al41 estimated a local demographic expansion during the Holocene period at about 8000±2500 YBP. No doubt all populations arriving to the Sahel were further enriched by various admixtures of many other sub-Saharan lineages, an effect even more pronounced in the Chadic groups who adopted a sedentary lifestyle soon after their arrival to the fertile Chad Basin than in the Tuareg who remain nomadic until present.
It is curious that, at least for the Tuareg maternal gene pool, there are no mtDNA lineages connected with the Neolithic expansion from the Near East despite being present in considerable frequencies in other North African populations. For example, the conservation of the high frequency and remarkable internal variability of T1 haplotypes within the distant and relatively isolated Egyptian oasis of el-Hayez led to an estimation of local expansion at around 5138±3633 YBP.37 There are no indications yet of the ages of local expansions in the more central and western regions of North Africa, which could contribute further insights for its absence in the Tuareg population as a whole.
Interestingly, for the Y chromosome, the dominant haplogroup in North Africa as well as the Tuareg is E1b1b1b. This haplogroup was associated with Neolithic diffusion in North Africa, with an age estimation of 2800–9800 YBP,45 but the lower resolution of the Y chromosome tree did not allow us to investigate this issue further. Nonetheless, disregarding whether they are in fact Neolithic, the ages for the mtDNA and Y chromosome lineages of North African origin observed in southern Tuareg are consistent with the same period, between 9000 and 3000 years ago.
Acknowledgments
The project was supported by the Grant Agency of the Czech Republic (Grant no. 206/08/1587), the Portuguese Fundação para a Ciência e a Tecnologia (PTDC/ANT/66275/2006 and Programa Operacional Ciência, Tecnologia e Inovação – Quadro Comunitario de Apoio III) (LP), Xunta de Galicia (Grupos Emerxentes; 2008/XA122), Fundación de Investigación Médica Mutua Madrileña (2006/CL370 and 2008/CL444) and Ministerio de Ciencia e Innovación (SAF2008-02971)(AS).
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies the paper on European Journal of Human Genetics website (http://www.nature.com/ejhg)
Supplementary Material
References
- Lhote H. Les Touaregs du Hoggar (Ahaggar) Paris: Payot,; 1955. [Google Scholar]
- Cavalli-Sforza LL, Menozzi P, Piazza A. The History and Geography of Human Genes. Princeton, NJ: Princeton University Press,; 1994. [Google Scholar]
- Watson E, Bauer K, Aman R, Weiss G, von Haeseler A, Paabo S. mtDNA sequence diversity in Africa. Am J Hum Genet. 1996;59:437–444. [PMC free article] [PubMed] [Google Scholar]
- Watson E, Forster P, Richards M, Bandelt HJ. Mitochondrial footprints of human expansions in Africa. Am J Hum Genet. 1997;61:691–704. doi: 10.1086/515503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ottoni C, Martinez-Labarga C, Loogväli EL, et al. First genetic insight into Libyan Tuaregs: a maternal perspective. Ann Hum Genet. 2009;73:438–448. doi: 10.1111/j.1469-1809.2009.00526.x. [DOI] [PubMed] [Google Scholar]
- Černý V, Hájek M, Bromová M, Čmejla R, Diallo I, Brdička R. MtDNA of Fulani nomads and their genetic relationships to neighboring sedentary populations. Hum Biol. 2006;78:9–27. doi: 10.1353/hub.2006.0024. [DOI] [PubMed] [Google Scholar]
- Álvarez-Iglesias V, Mosquera-Miguel A, Cerezo M, et al. New population and phylogenetic features of the internal variation within mitochondrial DNA macro-haplogroup R0. PLoS One. 2009;4:e5112. doi: 10.1371/journal.pone.0005112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerezo M, Cerny V, Carracedo A, Salas A. Applications of MALDI-TOF MS to large-scale human mtDNA population-based studies. Electrophoresis. 2009;30:3665–3673. doi: 10.1002/elps.200900294. [DOI] [PubMed] [Google Scholar]
- Brisighelli F, Capelli C, Álvarez-Iglesias V, et al. The Etruscan timeline: a recent Anatolian connection. Eur J Hum Genet. 2009;17:693–696. doi: 10.1038/ejhg.2008.224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torroni A, Bandelt HJ, Macaulay V, et al. A signal, from human mtDNA, of postglacial recolonization in Europe. Am J Hum Genet. 2001;69:844–852. doi: 10.1086/323485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torroni A, Rengo C, Guida V, et al. Do the four clades of the mtDNA haplogroup L2 evolve at different rates. Am J Hum Genet. 2001;69:1348–1356. doi: 10.1086/324511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karafet TM, Mendez FL, Meilerman MB, Underhill PA, Zegura SL, Hammer MF. New binary polymorphisms reshape and increase resolution of the human Y chromosomal haplogroup tree. Genome Res. 2008;18:830–838. doi: 10.1101/gr.7172008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salas A, Richards M, De la Fé T, et al. The making of the African mtDNA landscape. Am J Hum Genet. 2002;71:1082–1111. doi: 10.1086/344348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kivisild T, Reidla M, Metspalu E, et al. Ethiopian mitochondrial DNA heritage: tracking gene flow across and around the gate of tears. Am J Hum Genet. 2004;75:752–770. doi: 10.1086/425161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behar DM, Villems R, Soodyall H, et al. The dawn of human matrilineal diversity. Am J Hum Genet. 2008;82:1130–1140. doi: 10.1016/j.ajhg.2008.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olivieri A, Achilli A, Pala M, et al. The mtDNA legacy of the Levantine early Upper Palaeolithic in Africa. Science. 2006;314:1767–1770. doi: 10.1126/science.1135566. [DOI] [PubMed] [Google Scholar]
- Achilli A, Rengo C, Magri C, et al. The molecular dissection of mtDNA haplogroup H confirms that the Franco-Cantabrian glacial refuge was a major source for the European gene pool. Am J Hum Genet. 2004;75:910–918. doi: 10.1086/425590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Oven M, Kayser M. Updated comprehensive phylogenetic tree of global human mitochondrial DNA variation. Hum Mutat. 2009;30:E386–E394. doi: 10.1002/humu.20921. [DOI] [PubMed] [Google Scholar]
- Andrews RM, Kubacka I, Chinnery PF, Lightowlers RN, Turnbull DM, Howell N. Reanalysis and revision of the Cambridge reference sequence for human mitochondrial DNA. Nat Genet. 1999;23:147. doi: 10.1038/13779. [DOI] [PubMed] [Google Scholar]
- Excoffier LGL, Schneider S. Arlequin ver. 3.0: an integrated software package for population genetics data analysis. Evol Bioinform Online. 2005;1:47–50. [PMC free article] [PubMed] [Google Scholar]
- Rozas J, Rozas R. DnaSP version 3: an integrated program for molecular population genetics and molecular evolution analysis. Bioinformatics. 1999;15:174–175. doi: 10.1093/bioinformatics/15.2.174. [DOI] [PubMed] [Google Scholar]
- Bandelt HJ, Forster P, Sykes BC, Richards MB. Mitochondrial portraits of human populations using median networks. Genetics. 1995;141:743–753. doi: 10.1093/genetics/141.2.743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishmar D, Ruiz-Pesini E, Golik P, et al. Natural selection shaped regional mtDNA variation in humans. Proc Natl Acad Sci USA. 2003;100:171–176. doi: 10.1073/pnas.0136972100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forster P, Harding R, Torroni A, Bandelt HJ. Origin and evolution of Native American mtDNA variation: a reappraisal. Am J Hum Genet. 1996;59:935–945. [PMC free article] [PubMed] [Google Scholar]
- Saillard J, Forster P, Lynnerup N, Bandelt HJ, Norby S. mtDNA variation among Greenland Eskimos: the edge of the Beringian expansion. Am J Hum Genet. 2000;67:718–726. doi: 10.1086/303038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soares P, Ermini L, Thomson N, et al. Correcting for purifying selection: an improved human mitochondrial molecular clock. Am J Hum Genet. 2009;84:740–759. doi: 10.1016/j.ajhg.2009.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finnila S, Lehtonen MS, Majamaa K. Phylogenetic network for European mtDNA. Am J Hum Genet. 2001;68:1475–1484. doi: 10.1086/320591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrnstadt C, Elson JL, Fahy E, et al. Reduced-median-network analysis of complete mitochondrial DNA coding-region sequences for the major African, Asian, and European haplogroups. Am J Hum Genet. 2002;70:1152–1171. doi: 10.1086/339933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira L, Richards M, Goios A, et al. High-resolution mtDNA evidence for the late-glacial resettlement of Europe from an Iberian refugium. Genome Res. 2005;15:19–24. doi: 10.1101/gr.3182305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherni L, Fernandes V, Pereira JB, et al. Post-last glacial maximum expansion from Iberia to North Africa revealed by fine characterization of mtDNA H haplogroup in Tunisia. Am J Phys Anthropol. 2009;139:253–260. doi: 10.1002/ajpa.20979. [DOI] [PubMed] [Google Scholar]
- Ennafaa H, Cabrera VM, Abu-Amero KK, et al. Mitochondrial DNA haplogroup H structure in North Africa. BMC Genet. 2009;10:8. doi: 10.1186/1471-2156-10-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira L, Cunha C, Amorim A. Predicting sampling saturation of mtDNA haplotypes: an application to an enlarged Portuguese database. Int J Legal Med. 2004;118:132–136. doi: 10.1007/s00414-003-0424-1. [DOI] [PubMed] [Google Scholar]
- Behar DM, Garrigan D, Kaplan ME, et al. Contrasting patterns of Y chromosome variation in Ashkenazi Jewish and host non-Jewish European populations. Hum Genet. 2004;114:354–365. doi: 10.1007/s00439-003-1073-7. [DOI] [PubMed] [Google Scholar]
- Plaza S, Calafell F, Helal A, et al. Joining the pillars of Hercules: mtDNA sequences show multidirectional gene flow in the western Mediterranean. Ann Hum Genet. 2003;67:312–328. doi: 10.1046/j.1469-1809.2003.00039.x. [DOI] [PubMed] [Google Scholar]
- Černý V, Salas A, Hájek M, Žaloudková M, Brdička R. A bidirectional corridor in the Sahel-Sudan belt and the distinctive features of the Chad Basin populations: a history revealed by the mitochondrial DNA genome. Ann Hum Genet. 2007;71:433–452. doi: 10.1111/j.1469-1809.2006.00339.x. [DOI] [PubMed] [Google Scholar]
- Loogväli EL, Roostalu U, Malyarchuk BA, et al. Disuniting uniformity: a pied cladistic canvas of mtDNA haplogroup H in Eurasia. Mol Biol Evol. 2004;21:2012–2021. doi: 10.1093/molbev/msh209. [DOI] [PubMed] [Google Scholar]
- Kujanova M, Pereira L, Fernandes V, Pereira JB, Cerny V. Near eastern neolithic genetic input in a small oasis of the Egyptian Western Desert. Am J Phys Anthropol. 2009;140:336–346. doi: 10.1002/ajpa.21078. [DOI] [PubMed] [Google Scholar]
- Pereira L, Cunha C, Alves C, Amorim A. African female heritage in Iberia: a reassessment of mtDNA lineage distribution in present times. Hum Biol. 2005;77:213–229. doi: 10.1353/hub.2005.0041. [DOI] [PubMed] [Google Scholar]
- Quintana-Murci L, Quach H, Harmant C, et al. Maternal traces of deep common ancestry and asymmetric gene flow between Pygmy hunter-gatherers and Bantu-speaking farmers. Proc Natl Acad Sci USA. 2008;105:1596–1601. doi: 10.1073/pnas.0711467105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira L, Macaulay V, Torroni A, Scozzari R, Prata MJ, Amorim A. Prehistoric and historic traces in the mtDNA of Mozambique: insights into the Bantu expansions and the slave trade. Ann Hum Genet. 2001;65:439–458. doi: 10.1017/S0003480001008855. [DOI] [PubMed] [Google Scholar]
- Černý V, Fernandes V, Costa MD, Hájek M, Mulligan CJ, Pereira L. Migration of Chadic speaking pastoralists within Africa based on population structure of Chad Basin and phylogeography of mitochondrial L3f haplogroup. BMC Evol Biol. 2009;9:63. doi: 10.1186/1471-2148-9-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quintana-Murci L, Semino O, Bandelt HJ, Passarino G, McElreavey K, Santachiara-Benerecetti AS. Genetic evidence of an early exit of Homo sapiens sapiens from Africa through eastern Africa. Nat Genet. 1999;23:437–441. doi: 10.1038/70550. [DOI] [PubMed] [Google Scholar]
- González AM, Larruga JM, Abu-Amero KK, Shi Y, Pestano J, Cabrera VM. Mitochondrial lineage M1 traces an early human backflow to Africa. BMC Genomics. 2007;8:223. doi: 10.1186/1471-2164-8-223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maca-Meyer N, González AM, Larruga JM, Flores C, Cabrera VM. Major genomic mitochondrial lineages delineate early human expansions. BMC Genet. 2001;2:13. doi: 10.1186/1471-2156-2-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arredi B, Poloni ES, Paracchini S, et al. A predominantly neolithic origin for Y-chromosomal DNA variation in North Africa. Am J Hum Genet. 2004;75:338–345. doi: 10.1086/423147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semino O, Magri C, Benuzzi G, et al. Origin, diffusion, and differentiation of Y-chromosome haplogroups E and J: inferences on the neolithization of Europe and later migratory events in the Mediterranean area. Am J Hum Genet. 2004;74:1023–1034. doi: 10.1086/386295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallais J. Pasteurs et paysans du Gourma: la condition sahélienne. Paris: C.N.R.S.,; 1975. [Google Scholar]
- Hill A, Randall S. Différences géographiques et sociales dans la mortalité infantile et juvenile au Mali. Population. 1984;39:921–946. [Google Scholar]
- Randall S, Winter M. The Reluctant Spouse and the Illegitimate Slave: Marriage, Household Formation, and Demographic Behaviour Amongst Malian Tamesheq from the Niger Delta and the Gourma. London: Overseas Development Institute, Agricultural Administration Unit,; 1986. [Google Scholar]
- Torroni A, Achilli A, Macaulay V, Richards M, Bandelt HJ. Harvesting the fruit of the human mtDNA tree. Trends Genet. 2006;22:339–345. doi: 10.1016/j.tig.2006.04.001. [DOI] [PubMed] [Google Scholar]
- Coudray C, Olivieri A, Achilli A, et al. The complex and diversified mitochondrial gene pool of Berber populations. Ann Hum Genet. 2009;73:196–214. doi: 10.1111/j.1469-1809.2008.00493.x. [DOI] [PubMed] [Google Scholar]
- Paul A. A History of the Beja Tribes of the Sudan. London: F. Cass; 1971. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.