Abstract
Rearrangements between tandemly repeated DNA sequences are a common source of genetic instability. Such rearrangements underlie several human genetic diseases. In many organisms, the mismatch-repair (MMR) system functions to stabilize repeats when the repeat unit is short or when sequence imperfections are present between the repeats. We show here that the action of single-stranded DNA (ssDNA) exonucleases plays an additional, important role in stabilizing tandem repeats, independent of their role in MMR. For perfect repeats of ≈100 bp in Escherichia coli that are not susceptible to MMR, exonuclease (Exo)-I, ExoX, and RecJ exonuclease redundantly inhibit deletion. Our data suggest that >90% of potential deletion events are avoided by the combined action of these three exonucleases. Imperfect tandem repeats, less prone to rearrangements, are stabilized by both the MMR-pathway and ssDNA-specific exonucleases. For 100-bp repeats containing four mispairs, ExoI alone aborts most deletion events, even in the presence of a functional MMR system. By genetic analysis, we show that the inhibitory effect of ssDNA exonucleases on deletion formation is independent of the MutS and UvrD proteins. Exonuclease degradation of DNA displaced during the deletion process may abort slipped misalignment. Exonuclease action is therefore a significant force in genetic stabilization of many forms of repetitive DNA.
Rearrangements between homologous tandem DNA sequences are a common source of genetic variation. Such rearrangements underlie a number of human genetic diseases, including those involving trinucleotide repeat arrays (as in Huntington's disease or fragile X syndrome; ref. 1) or larger repeated DNA sequences such as tandem globin genes or Alu sequences (2, 3). Sequence divergence between repeated sequences serves to stabilize the repeats. The reduced level of rearrangements between homeologous (imperfectly homologous) DNA strands has been attributed to the antirecombination effect of mismatch repair (MMR) (4). In Escherichia coli, MMR aborts rearrangements between both dispersed (5) and tandemly arranged (6) repeats.
In E. coli, rearrangements between tandemly repeated DNA sequences have been deduced to occur primarily by slipped misalignment during DNA replication (reviewed in ref. 7). Such rearrangements occur at high frequency, even in the absence of the RecA protein that is required for general homologous recombination (8–12). The rate of deletion of a 101-bp perfect duplication within the tetA gene carried on a circular plasmid is significantly increased by mutations in many components of the replication machinery (ref. 13; V.V.F. and S.T.L., unpublished results), but unaffected by mutations in recA or any component of MMR. When the perfect duplication within tetA is replaced by an imperfect duplication with four base substitutions, the rate of deletion plummets several-hundred-fold (6). This reduction is caused in part by MMR, because mutations blocking MMR (dam, mutH, mutL, mutS, or uvrD) restore deletion to levels approaching that seen for perfect repeats (6).
During MMR in E. coli, the MutHLS complex of proteins mediates recognition of mispairs and incision of duplex DNA (reviewed in ref. 14). Excision of one strand is accomplished by the helicase activity of UvrD, followed by DNA degradation by one of several single-stranded DNA (ssDNA) exonucleases (ssExos). The four exonucleases (Exos) I, VII, X, and RecJ act in a redundant fashion both in vivo and in vitro (15, 16); the presence of any one exonuclease is sufficient to support MMR to the extent that no elevation of spontaneous mutation is evident. This redundancy occurs despite the fact that the exonucleases have distinct polarities of degradation. ExoI and ExoX specifically degrade DNA from a 3′ end (17, 18), whereas RecJ initiates DNA cleavage from a 5′ ssDNA end (19). ExoVII is the only exonuclease that can attack both 3′ and 5′ ends (20). ExoI, ExoVII, and RecJ are highly specific for ssDNA (17, 19, 21), and ExoX has a preference for ssDNA, but can also degrade duplex DNA (18).
Our expectation was that the ssExos would also act redundantly in the inhibition of homeologous recombination by MMR. However, we show here that these exonucleases inhibit homeologous rearrangements by a mechanism distinct from their role in MMR; these exonucleases affect fully homologous rearrangements as well.
Materials and Methods
Bacterial Strains, Plasmids, and Media.
All strains were grown at 37°C, in liquid LB medium, or minimal media consisting of 56/2 salts (22), 0.2% glucose, and 50 μg/ml required amino acids. Plate media contained 1.5% agar. LCG media, used for P1 lysates and transductions, consisted of LB medium supplemented with 2 mM calcium chloride, 50 μg/ml thymine, and 0.2% glucose. Antibiotics were used at the following concentrations: ampicillin, 100 μg/ml; tetracycline, 15 μg/ml; chloramphenicol, 30 μg/ml; and kanamycin, 60 μg/ml.
Isogenic E. coli K-12 strains used in this study were constructed by P1 vir transduction (23) and derived from strain AB1157 (24) with the genotype F− thi-1 hisG4 argE Δ(gpt-proA)62 thr-1 leuB6 kdgK51 rfbD1 ara-14 lacY1 galK2 xyl-5 mtl-1 tsx-33 supE44 rpsL31 rac. Details of strain constructions are available from the authors by request and are briefly described below. The mutS458∷miniTn10kan allele used was derived from GM4799 (M. Marinus, University of Massachusetts Medical Center, Worcester), and the uvrD254∷Tn5 allele was derived from STL1526 (25). The recA allele used in this study, recA∷cat, was transduced from strain STL3817. (GM4799, STL1526, and STL3817 are also isogenic with AB1157.) The lexA3 allele, which blocks induction of the SOS response, was introduced by linkage with malF3180∷Tn10kan, selecting kanamycin resistance, and scoring for UV sensitivity. These alleles were moved into the quadruple exonuclease-deficient strain STL6283 (described below) by P1 transduction, producing strains STL6377 (ssExo− MutS−), STL6381 (ssExo− UvrD−), STL6525 (ssExo− RecA−), and STL7610 (ssExo− LexAInd−). RecA− MutS− derivatives were constructed by the transduction of recA∷cat into GM4799 (yielding STL7534, ssExo+) and STL6377 (yielding STL6644, also ssExo−).
Precise deletion of the exonuclease-encoding regions in strains STL5908 (recJ), STL6064 (xonA), STL6001 (xseA), and STL6229 (exoX) was accomplished by λ Red-mediated recombination (26). PCR products containing the kan gene and 40 bp of homology to sequences flanking the target gene were generated by using appropriate primers listed below and pKD4 as the template. All upstream disruption primers terminated in the common sequence 5′-TGTAGGCTGG AGCTGCTTCG, and downstream primers terminated in the sequence 5′-CATATGAATA TCCTCCTTAG, which served to amplify the kan gene flanked by directly repeated FRT (FLP recombinase) sites. In addition, primers contained the following ORF-specific 40-base sequences at their 5′ ends: 5′-TCGACGAACA CCAAAAAATG ACCAGCGGTA AATAATTCGC (recJ upstream) and 5′-TTGTACCCAA TCCACGCTCT TTTTTATAGA GAAGATGACG (recJ downstream); 5′-GACTGAATAA CCTGCTGATT TAGAATTTGA TCTCGCTCAC (xseA upstream) and 5′-ATGGCTTGAT ATCGAAAAAA CGCGTTGAAT TCGTGCTGGC (xseA downstream); 5′-TGATACTCTG GCAGACAGCA GAAATAACGG ATTTAACCTA (xonA upstream) and 5′-CAACATTGTC CTCCGCCGTA CCAGCGGCGG AGGCTTCAAA (xonA downstream); and 5′-TCATTCCATT ACGCTAGGC TTTTTCGGCC TGGAGCATGCC (exoX upstream) and 5′-CGCTGGCGCA GGGAACATTA CCCGCTACGC CTGCGGACTA (exoX downstream). The PCR products were introduced into strain BW26308 by electroporation (27). Recombination between the PCR product and the E. coli chromosome was mediated by the λ Red recombinase, which was expressed from plasmid pKD46 in the presence of 1 mM l-arabinose. Kanamycin-resistant recombinants were selected at 37°C and gene disruptions were confirmed by PCR analysis. The disrupted allele was introduced into AB1157 by P1 transduction to kanamycin-resistance. Excision of the kan gene was later accomplished by the transformation of temperature-sensitive plasmid pCP20, which expresses FLP recombinase, into the appropriate strain at 30°C. Growth at 42°C resulted in the loss of pCP20, yielding kanamycin-sensitive and ampicillin-sensitive deletion derivatives.
Multiple combinations of exonuclease-deletion mutations were constructed by the transduction of a kan-marked allele into the various deletion strains, followed by FLP-mediated excision as described above. The resulting strains are double mutants STL5998 (ΔxseA ΔrecJ), STL6108 (ΔxonA ΔxseA), STL6125 (ΔxseA ΔexoX), STL6230 (ΔxonA ΔexoX), STL6232 (ΔexoX ΔrecJ), and STL6234 (ΔxonA ΔrecJ), triple mutants STL6103 (ΔxonA ΔxseA ΔrecJ), STL6127 (ΔxonA ΔxseA ΔexoX), STL6139 (ΔxseA ΔexoX ΔrecJ), and STL6255 (ΔxonA ΔexoX ΔrecJ), and quadruple mutant STL6283 (ΔxonA ΔxseA ΔexoX ΔrecJ).
The xseA complementation plasmid was produced by PCR of AB1157 chromosomal DNA with the primers 5′-CAATATCTCG ACCAGAGTGG and 5′-ATTACTTCTG TCAGCACGGG. The fragment was digested with EcoRI and SacII and ligated into vector pBS SK+ (Stratagene), producing plasmid pSTL340. The correct xseA sequence was confirmed by DNA-sequence analysis. By EcoRI and SacII digestion, the xseA fragment was introduced into the low-copy vector conferring kanamycin resistance, pWKS130 (28), producing plasmid pSTL341.
Deletion Assays.
Plasmid pSTL57 (11) is derived from pBR322 and contains an intact copy of the bla gene and a copy of the tetA gene with a perfect internal 101-bp duplication. Plasmid pSTL113 (6) is an imperfect duplication, differing by four bases that are located at 21-bp intervals. Plasmids were introduced into various AB1157 derivatives by electroporation (27) or by treatment with polyethylene glycol, DMSO, and MgCl2 (29). Deletion was measured as the ratio of tetracycline-resistant colonies to ampicillin-resistant colonies in at least eight independent cultures as described (10). The method of the median (30) was used to calculate deletion rates by the following formula: M/N = deletion rate, where M is the calculated number of deletion events, and N is the final average number of ampicillin-resistant cells in the 1-ml culture. M is solved by interpolation from experimental determination of r0, where r0 is the median number of tetracycline-resistant cells as determined by the formula r0 = M(1.24 + lnM). Confidence intervals of 95% were determined as described (31). As a control, the plasmid copy number was determined for pSTL57 in strains AB1157 and STL6255 (32). The presence of rolling-circle replication intermediates, long linear multimers, was determined by Southern blot analysis of extracted total cellular DNA by using the MasterPure DNA purification kit (Epicentre Technologies, Madison, WI) from the above strains with pBR322 DNA probe, and by using the Renaissance random primer fluorescein kit (Perkin–Elmer Life Sciences, Boston), and the methods supplied by the manufacturer.
Results
Using the model organism E. coli, we examined the effect of ssExos (RecJ and ExoI, -VII, and -X) on deletion of perfect and imperfect tandem repeats. These studies used E. coli K-12 mutants carrying all combinations (single, double, triple, and quadruple) of precise deletion of the exonuclease-coding regions. Deletion of internal 101-bp tetA tandem repeats was scored by using plasmids pSTL57 and pSTL113 (6, 11), selecting acquisition of tetracycline resistance (Fig. 1). Plasmid pSTL57 carries perfectly homologous 101-bp repeats. The downstream repeat of pSTL113 carries four coding-silent base-substitution transition mutations that generate homeologous 101-bp tandem repeats. Previous work has shown that deletion of homeologous repeats of pSTL113 occurs at a rate about two to three orders of magnitude lower than the perfect repeats of pSTL57; this lower rate is partially alleviated by mutations in components of the MMR pathway (6).
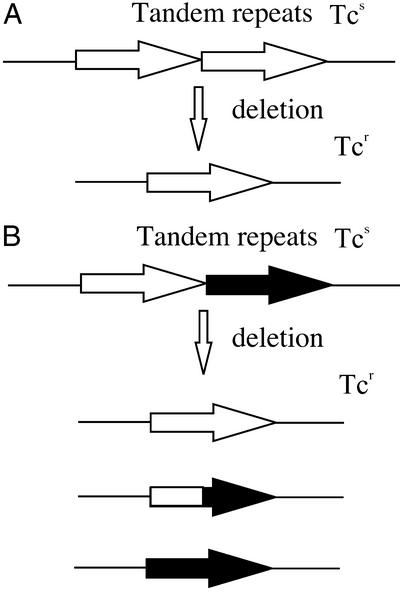
Figure 1.
Assay of tandem repeat deletion. Perfect 101-bp repeats (A) or imperfect repeats carrying four-base heterologies (B) render the tetA gene nonfunctional (Tcs). Deletion of one repeat yields a functional tetA gene that confers tetracycline resistance to the cell (Tcr). Deletion products of homeologous repeats (B) may carry the tetA segment derived from either of the parental repeats, or a hybrid fusion of the two repeats.
The loss of any one ssExo had only modest effects on deletion of perfectly homologous tandem repeats (Table 1). Significant enhancement of deletion rates was seen when three or more exonucleases are deficient, with the deletion rate about 10 times higher in the absence of all four ssExos (Table 1). The maximal increase of 27-fold over wild-type levels was seen when ExoI, ExoX, and RecJ exonuclease were simultaneously inactivated. Control experiments confirmed that plasmid copy number was normal in this strain, and that no rolling circle replication intermediates were evident.
Table 1.
Deletion rate of perfect (in pSTL57) 101-bp tandem repeats in ssExo-deficient strains
| Strain | Exonuclease
|
Deletion rate | Confidence interval | Relative rate | |||
|---|---|---|---|---|---|---|---|
| I | VII | X | RecJ | ||||
| AB1157 | + | + | + | + | 3.0 × 10−5 | 1.8–4.5 × 10−5 | 1.0 |
| STL6064 | − | + | + | + | 7.0 × 10−5 | 5.4–13 × 10−5 | 2.3 |
| STL6001 | + | − | + | + | 2.6 × 10−5 | 1.9–6.5 × 10−5 | 0.9 |
| STL6229 | + | + | − | + | 5.4 × 10−5 | 2.6–7.8 × 10−5 | 1.8 |
| STL5908 | + | + | + | − | 5.8 × 10−5 | 1.7–7.8 × 10−5 | 1.9 |
| STL6108 | − | − | + | + | 6.6 × 10−5 | 2.9–8.9 × 10−5 | 2.2 |
| STL6230 | − | + | − | + | 1.1 × 10−4 | 0.8–2.1 × 10−4 | 3.7 |
| STL6234 | − | + | + | − | 7.7 × 10−5 | 4.7–17 × 10−5 | 2.6 |
| STL6125 | + | − | − | + | 5.7 × 10−5 | 3.9–9.7 × 10−5 | 1.9 |
| STL5998 | + | − | + | − | 4.8 × 10−5 | 3.5–13 × 10−5 | 1.6 |
| STL6232 | + | + | − | − | 7.7 × 10−5 | 4.1–10 × 10−5 | 2.6 |
| STL6127 | − | − | − | + | 1.2 × 10−4 | 0.7–2 × 10−4 | 4.0 |
| STL6103 | − | − | + | − | 3.3 × 10−4 | 2.5–5.3 × 10−4 | 11.0 |
| STL6255 | − | + | − | − | 8.2 × 10−4 | 2.4–13 × 10−4 | 27.0 |
| STL6139 | + | − | − | − | 1.4 × 10−4 | 0.5–3.2 × 10−4 | 4.7 |
| STL6283 | − | − | − | − | 2.9 × 10−4 | 1.5–6.9 × 10−4 | 9.7 |
| STL6283/ pSTL341 | − | + | − | − | 8.0 × 10−4 | 3.3–14 × 10−4 | 27.0 |
Deletion between imperfect tandem repeats of pSTL113 occurred at lower rates and were more strongly regulated by ssExos (Table 2). ExoI played a prominent role in aborting imperfect repeat deletion. Mutants in ExoI exhibited an almost two orders of magnitude increase in deletion rate as compared with that of wild type. Other single mutants showed either a modest increase or no increase in the deletion rate.
Table 2.
Deletion rate of imperfect (in pSTL113) 101-bp tandem repeats in ssExo-deficient strains
| Strain | Exonuclease
|
Deletion rate | Confidence interval | Relative rate | |||
|---|---|---|---|---|---|---|---|
| I | VII | X | RecJ | ||||
| AB1157 | + | + | + | + | 1.0 × 10−7 | 0.6–2.6 × 10−7 | 1.0 |
| STL6064 | − | + | + | + | 9.1 × 10−6 | 4.8–10.3 × 10−6 | 91.0 |
| STL6001 | + | − | + | + | 9.0 × 10−8 | 5.2–18 × 10−8 | 0.9 |
| STL6229 | + | + | − | + | 1.9 × 10−7 | 1.0–2.6 × 10−7 | 1.9 |
| STL5908 | + | + | + | − | 1.5 × 10−7 | 0.4–3.1 × 10−7 | 1.5 |
| STL6108 | − | − | + | + | 1.9 × 10−6 | 1.2–2.9 × 10−6 | 19.0 |
| STL6230 | − | + | − | + | 1.1 × 10−5 | 0.5–3.1 × 10−5 | 110.0 |
| STL6234 | − | + | + | − | 6.0 × 10−6 | 3.9–11 × 10−6 | 60.0 |
| STL6125 | + | − | − | + | 3.0 × 10−7 | 1.7–4.4 × 10−7 | 3.0 |
| STL5998 | + | − | + | − | 5.2 × 10−7 | 3.2–10 × 10−7 | 5.2 |
| STL6232 | + | + | − | − | 2.9 × 10−7 | 1.2–3.4 × 10−7 | 2.9 |
| STL6127 | − | − | − | + | 4.0 × 10−6 | 1.7–6.1 × 10−6 | 40.0 |
| STL6103 | − | − | + | − | 8.9 × 10−6 | 5.9–12 × 10−6 | 89.0 |
| STL6255 | − | + | − | − | 2.3 × 10−5 | 1.5–3.2 × 10−5 | 230.0 |
| STL6139 | + | − | − | − | 4.6 × 10−7 | 2.1–7.1 × 10−7 | 4.6 |
| STL6283 | − | − | − | − | 4.0 × 10−6 | 2.6–7.1 × 10−6 | 40.0 |
| STL6283/ pSTL341 | − | + | − | − | 1.4 × 10−5 | 0.8–2.3 × 10−5 | 140.0 |
In contrast to the inhibitory role of ExoI on deletion, ExoVII was found to stimulate deletion in some instances. The elevation of homeologous deletion by ExoI deficiency required, to some extent, functional ExoVII activity. The loss of ExoVII diminished the homeologous deletion rates by 3- to 6-fold in three of the four combinations lacking ExoI. For example, the highest homeologous deletion rate, elevated 260-fold relative to wild type, was seen in the triple mutant deleted for ExoI, ExoX, and RecJ. Additional loss of ExoVII reduced the deletion rate ≈6-fold, to a level elevated ≈40-fold relative to wild-type. ExoVII may be required for deletions between perfect repeats as well, because inactivation of it reduced the elevated deletion rate in ExoI− ExoX− RecJ− strains ≈3-fold. Introduction of the xseA+ gene on a low-copy plasmid in the recJ xonA xseA exoX mutant strain elevated deletion rates of both perfect and imperfect repeats, confirming that ExoVII alone is responsible for this effect.
Epistasis analysis suggests that the inhibitory effect of exonucleases on homeologous deletion rates was independent of their role in MMR. Although deficiency for MMR (mutS, uvrD) or ssExos each elevate homeologous deletion rates from 20- to 40-fold, the deletion rate in the MutS− ssExo− strain was 260-fold greater than wild type (Table 3), indicating genetic synergy between MMR and exonuclease deficiency. The MutS-independent effect of the exonucleases does not appear to require UvrD; the effect of uvrD was no greater than that of mutS and was, likewise, comparably synergistic with the loss of the exonucleases. Neither mutS nor uvrD significantly increased the rate of perfect repeat deletion (Table 4), as we have reported (6). In the absence of all four ssExos, a mutation in uvrD lowered deletion rates of the perfect repeats ≈2-fold (Table 4), suggesting that UvrD helicase action may promote slipped misalignment in the absence of ssExos.
Table 3.
Effect of RecA and MMR on deletion of imperfect tandem repeats (pSTL113) in ssExo-deficient strains
| Strain | MutS | UvrD | ssExos* | RecA | LexA | Deletion rate | Confidence interval | Relative rate |
|---|---|---|---|---|---|---|---|---|
| AB1157 | + | + | + | + | + | 1.0 × 10−7 | 6.2–26 × 10−8 | 1.0 |
| GM4799 | − | + | + | + | + | 2.3 × 10−6 | 1.7–5.2 × 10−6 | 23.0 |
| STL1526 | + | − | + | + | + | 2.1 × 10−6 | 1.4–3.6 × 10−6 | 21.0 |
| STL6283 | + | + | − | + | + | 4.0 × 10−6 | 2.6–7.1 × 10−6 | 40.0 |
| STL6377 | − | + | − | + | + | 2.6 × 10−5 | 1.8–4.2 × 10−5 | 260.0 |
| STL6381 | + | − | − | + | + | 1.9 × 10−5 | 1.0–4.9 × 10−5 | 190.0 |
| STL3817 | + | + | + | − | + | 2.0 × 10−7 | 0.9–4.5 × 10−7 | 2.0 |
| STL7534 | − | + | + | − | + | 2.9 × 10−6 | 1.7–7.6 × 10−6 | 29.0 |
| STL6525 | + | + | − | − | + | 3.2 × 10−5 | 1.5–4.5 × 10−5 | 320.0 |
| STL7610 | + | + | − | + | − | 8.4 × 10−6 | 2.7–18 × 10−6 | 84.0 |
| STL2172 | − | + | + | − | + | 2.4 × 10−5 | 2.0–4.7 × 10−5 | 240.0 |
| STL6644 | − | + | − | − | + | 7.6 × 10−5 | 6–10 × 10−5 | 760.0 |
+, Proficiency for all four ssExos; −, deficiency in all four ssExos.
Table 4.
Effect of RecA and MMR on deletion of perfect tandem repeats (pSTL57) in ssExo-deficient strains
| Strain | MutS | UvrD | ssExos* | RecA | LexA | Deletion rate | Confidence interval | Relative rate |
|---|---|---|---|---|---|---|---|---|
| AB1157 | + | + | + | + | + | 3.0 × 10−5 | 1.8–4.5 × 10−5 | 1.0 |
| GM4799 | − | + | + | + | + | 3.8 × 10−5 | 2.5–6.9 × 10−5 | 1.3 |
| STL1526 | + | − | + | + | + | 4.0 × 10−5 | 3.1–7.1 × 10−4 | 1.3 |
| STL6283 | + | + | − | + | + | 2.9 × 10−4 | 1.5–6.9 × 10−4 | 9.7 |
| STL6377 | − | + | − | + | + | 4.2 × 10−4 | 3.1–5.2 × 10−4 | 14.0 |
| STL6381 | + | − | − | + | + | 1.2 × 10−4 | 0.7–2.3 × 10−4 | 4.0 |
| STL3817 | + | + | + | − | + | 3.8 × 10−5 | 2.9–4.4 × 10−5 | 1.3 |
| STL6525 | + | + | − | − | + | 4.2 × 10−4 | 1.4–8.2 × 10−4 | 14.0 |
| STL7610 | + | + | − | + | − | 1.2 × 10−4 | 0.8–2 × 10−4 | 4.0 |
| STL6644 | − | + | − | − | + | 3.2 × 10−4 | 0.7–8 × 10−4 | 11.0 |
+, Proficiency for all four ssExos; −, deficiency in all four ssExos.
Deletion in wild-type strains occurs primarily by slipped misalignment during replication, and it therefore does not require the RecA homologous recombination protein (reviewed in ref. 7). We tested the recA-dependence of the deletion process in the absence of ssExos and found a surprising role for RecA in the avoidance of deletion. A recA mutation had little effect on the deletion rates of either the perfect or the imperfect repeats in an Exo+ background. However, addition of the recA mutation to the ssExo− background led to an 8-fold increase in the homeologous deletion rate relative to the recA+ strain, and a 320-fold increase relative to the wild-type strain (Exo+; Table 3). In combination with MutS− and ssExo−, recA− further increased the deletion rate of imperfect repeats to >700-fold relative to wild type, to a level comparable to that seen for deletion of perfect repeats. A lexA3 mutation, specifying the noncleavable form of LexA repressor (33), enhanced deletion of homeologous repeats only 2-fold, suggesting that the effect of RecA was not primarily because of its role in regulation of the SOS response by means of promoting cleavage of the LexA repressor. The recA mutations have little effect on deletion rates of perfect repeats (11, 13), even when the ssExos are deleted (Table 4). These results suggest that the RecA strand exchange protein acts in some way to prevent deletion between homeologous repeats, which is particularly evident when ssExos fail to abort deletion.
Discussion
ssExos Inhibit Deletion of Tandem Repeats.
Our results demonstrate that the action of ssExos plays a major role in promoting genetic stability of tandemly repeated DNA sequences. For perfectly homologous 101-bp repeats, the vast majority of potential deletion events are avoided by the redundant action of RecJ, ExoI, and ExoX of E. coli. The antideletion effect of ssExos was most pronounced when the repeated sequences were imperfectly homologous. ExoI, among the ssExos, plays the most important role in controlling homeologous deletion formation.
Genetic analysis with mutS and uvrD suggests that the antideletion properties of ssExos are independent of their proposed role in MMR (15, 16). The ssExos therefore most likely abort deletion by degradation of displaced ssDNA during slipped misalignment of repeated sequences (Fig. 2). For the perfect repeats, both 5′-end ssDNA degradation (by RecJ) and 3′ end degradation (by ExoI and ExoX) contribute to deletion avoidance. For the imperfect repeats, 3′ degradation through the strongly processive ExoI aborts most events. This surveillance by ssExos does not absolutely require the UvrD helicase because a uvrD mutant did not phenotypically mimic the ssExo-deficient strain. Rather, in the absence of ssExos, UvrD may actively promote some slipped misalignments because a uvrD mutation reduced deletion rates of the ssExo− strain >2-fold. UvrD helicase activity may unwind replication intermediates to promote strand dislocation, as shown in the first step of Fig. 2. UvrD's polarity of translocation on ssDNA, 3′ to 5′ (34), would promote 5′ strand displacement susceptible to RecJ exonuclease and the slipped misalignment as in Fig. 2B.
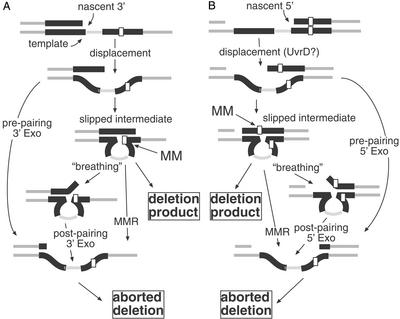
Figure 2.
Slipped misalignment and the stabilizing role of ssExos. During replication of a tandem repeat, replication may arrest, allowing the nascent strand to displace and mispair with the downstream repeat, producing the slipped intermediate. This nascent strand end could be the 3′ end of the polymerized strand (A) or a 5′ end of an Okazaki fragment (B). Unwinding of the nascent strand may be mediated by transient thermal denaturation or by active unwinding of the end by helicases, such as UvrD. Exonuclease digestion of the 3′ or 5′ displaced strand preceding mispairing can abort deletion. After slipped mispairing, if heterologies exist between the two tandem repeats, the slipped intermediate may include a mismatched base pair (MM). It is this mispair that is potentially recognized by the MMR system, leading to degradation of the nascent strand, thereby destroying the slipped intermediate. If not processed, this slipped intermediate will resolve to a deletion product after another round of DNA replication. Alternatively, in a mispair repair-independent fashion, transient unwinding of the mispaired intermediate (“breathing”) reveals a single strand that may be digested by ssExos (“postpairing”). Sequence heterologies between the mispaired strands of the intermediate may favor this unwinding.
The ssExos may have a more pronounced effect on slipped misalignment of imperfect repeats because these pairing imperfections lead to a greater probability that the DNA strands “breathe” to become unpaired at some point during the deletion process (Fig. 2). Once a ssDNA end presents itself, it may be quickly degraded by these exonucleases, thereby aborting the deletion process. Alternatively, DNA helicases may actively unwind mispaired DNA. (However, if this is true, our experiments suggest that UvrD is not the only helicase that can reveal ssDNA to the ssExos.)
ExoVII, with dual 3′ and 5′ polarity, in some backgrounds appeared to promote rather than discourage deletion formation. Loss of ExoVII alone did not influence deletion formation. However, in strains lacking ExoI, loss of ExoVII reduced deletion rates from 3- to 6-fold. This finding may mean that some ssDNA processing is necessary during deletion formation. Why ExoVII alone, of the ssExos, has this property is curious. ExoVII has dual polarity on ssDNA, and ExoVII degradation produces oligonucleotide products (20). This result might suggest its favored substrate is a single-strand flap, either 5′ or 3′. If polymerization has arrested outside the repeats, subsequent slipped misalignment at the repeats will produce a nonpaired 3′ tail (Fig. 3). Cleavage of the 3′ end by ExoI or flap cleavage by ExoVII could be required to mature these intermediates into paired 3′ ends that can prime DNA synthesis.
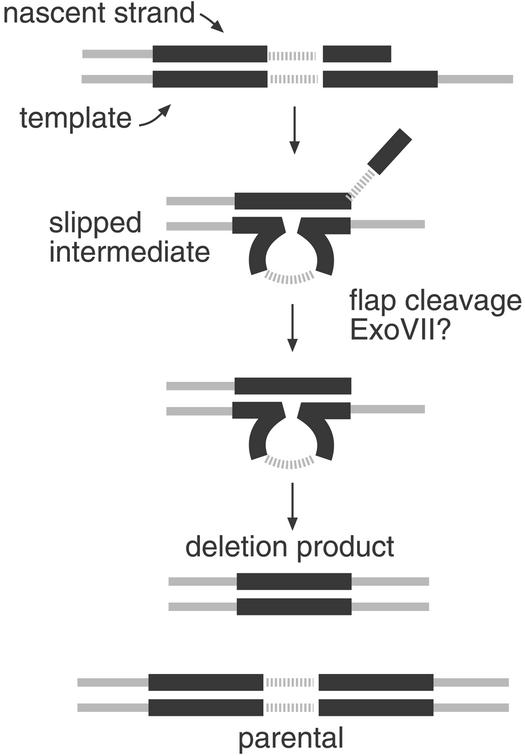
Figure 3.
Slipped mispairing and the role of trimming enzymes. If replication of tandem repeats arrests in the second repeat or in regions outside the repeat, after slipped misalignment at the repeats, an unpaired 3′ tail would be produced. ExoVII may promote deletion in the absence of ExoI by digestion of this unpaired flap. Removal of this tail produces a base-paired 3′ end that can prime DNA synthesis.
Perfect Versus Imperfect Repeat Deletion.
It has long been known that sequence imperfections between repeats reduce rearrangements that occur between them (reviewed in ref. 35). In human trinucleotide repeat arrays, such mismatches cause a significant reduction in the likelihood and severity of repeat expansion-associated diseases (36–39). The MMR system reduces incremental deletion or expansion of arrays of short repeats (less than about five nucleotides), because the MMR system detects heteroduplex loops of four nucleotides or less that would be produced by slipped misalignment. Even larger repeats are stabilized by MMR (5, 6), presumably because mismatches present in the mispaired heteroduplex elicit incision and destruction of one strand of this intermediate.
Our work confirms that a few mismatches dramatically stabilize tandem repeats. Our constructs employ 4% evenly spaced heterologies; we have seen that a single mismatch in 100 bp stabilizes repeats by an order of magnitude (V.V.F. and S.T.L., unpublished results). We show here that this effect is due only in part to the MMR system. The action of ssExos is essential not only as part of the MMR system (15, 16) but also in MMR-independent modes of stabilization.
Imperfect-repeat deletion was much more dramatically affected by exonucleases, and ExoI in particular, than the comparable deletion of perfect repeats. Mispaired intermediates of deletion may be spontaneously prone or actively promoted to unwind, revealing an unpaired 3′ end (Fig. 2). ExoI, with its potent and processive 3′ exonuclease action on ssDNA, may scavenge any such displaced DNA, thereby aborting deletion. Because exonucleases can inhibit deletion by degrading displaced DNA either before mispairing or afterward, the differences of particular exonucleases on perfect versus imperfect deletion may reflect whether they are more likely to act pre- or postmispairing. Prepairing avoidance mechanisms should affect homologous and homeologous repeat deletion equally. Effects specific for mismatched repeats must be postpairing by necessity because mispairs are generated only after annealing of DNA from the two repeats in the slipped intermediate. According to this reasoning, perfectly homologous repeats may be aborted preceding mispairing by both 3′ and 5′ end degradation by means of RecJ, ExoI, or ExoX. Postmispairing heterologies in the repeats may promote 3′ end fraying, and thereby provide more potential for 3′ end-scavenging by ExoI, such that ExoI has a predominant effect on homeologous deletion.
RecA Inhibits Slipped Misalignment of Homeologous Repeats.
Somewhat surprising was the finding that the addition of a recA mutation to the quadruple ssExo mutant resulted in a significant increase in the deletion rate of the imperfect repeat, but not of the perfect-repeat construct. This effect was not seen in Exo+ strains nor was it produced by lexA3, which impairs induction of the SOS response. This finding suggests that RecA may act directly to reverse misalignment of homeologous repeats. In the absence of ssExos, the looped nascent strand may be a substrate for RecA binding and subsequent strand pairing to reverse the reaction (Fig. 4). Binding of RecA may be aided by the action of MMR on the imperfectly paired repeats, or by destabilization of pairing by the mispairs themselves.
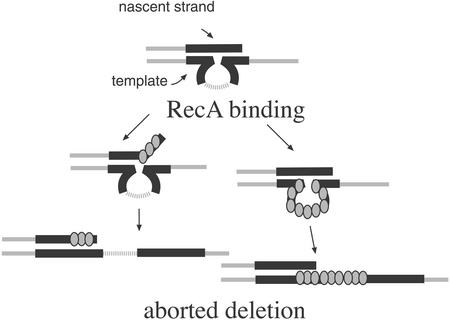
Figure 4.
Stabilization of tandem repeats by RecA in the absence of ssExos. In the absence of ssExos, RecA may stabilize repeats by binding to transiently displaced ssDNA and promoting annealing to the unslipped conformation. Alternatively, RecA may bind the looped region of the slipped intermediate to promote reannealing to the correctly paired configuration.
Exonuclease Inhibition of Genomic Instability.
Exonuclease regulation of genetic instability involves a number of different mechanisms, each of which may have in common the destruction of ssDNA-ended intermediates. This important role of ssExos has been obscured by their redundancy. A number of mutational events and genetic rearrangements are inhibited by cohort of ssExos used in this study. ExoI and ExoVII redundantly inhibit certain frameshift mutations (40) and base substitution mutations templated from quasipalindrome replication (41). Here we show that ExoI, ExoX, and RecJ play an important role in the avoidance of rearrangement between tandem-repeated sequences. Other unpublished work (V. A. Sutera, Jr., R. A. Hurley, and S.T.L.) suggests that all four exonucleases contribute to the prevention of intermolecular recombination between short (25–51 bp) sequence homologies. Paradoxically, in addition to the antirecombination activity of the ssExos, they may also promote recombination. During recombination of longer sequence homologies (as in transduction, conjugation, and bacteriophage λ recombination reactions), RecJ and ExoI redundantly promote RecABCD-mediated recombination (40, 42, 43). In this case, postsynaptic degradation of displaced DNA strands by either RecJ or ExoI may stabilize recombination intermediates, an idea supported by in vitro effects of these exonucleases during RecA-mediated strand transfer reactions (44, 45).
Acknowledgments
We thank Martin Marinus, Barry Wanner (Purdue University, West Lafayette, IN), and Mary Berlyn (Coli Genetic Stock Center, Yale University, New Haven, CT) for strains, and Vincent A. Sutera, Jr., for assistance in the construction of the exonuclease deletion strains and the xseA plasmid. This work was supported by National Institutes of Health Grants RO1 GM43889, GM51753, and F31 GM19179 (to L.A.R.) from the National Institute of General Medical Sciences.
Abbreviations
- MMR
mismatch repair
- ssDNA
single-stranded DNA
- Exo
exonuclease
- ssExo
ssDNA Exo
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.McMurray C T. Chromosoma. 1995;104:2–13. doi: 10.1007/BF00352220. [DOI] [PubMed] [Google Scholar]
- 2.Hu X, Worton R G. Hum Mutat. 1992;1:3–12. doi: 10.1002/humu.1380010103. [DOI] [PubMed] [Google Scholar]
- 3.Krawczak M, Cooper D N. Hum Genet. 1991;86:425–441. doi: 10.1007/BF00194629. [DOI] [PubMed] [Google Scholar]
- 4.Rayssiguier C, Thaler D S, Radman M. Nature. 1989;342:396–401. doi: 10.1038/342396a0. [DOI] [PubMed] [Google Scholar]
- 5.Petit M A, Dimpfl J, Radman M, Echols H. Genetics. 1991;129:327–332. doi: 10.1093/genetics/129.2.327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lovett S T, Feschenko V V. Proc Natl Acad Sci USA. 1996;93:7120–7124. doi: 10.1073/pnas.93.14.7120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bzymek M, Lovett S T. Proc Natl Acad Sci USA. 2001;98:8319–8325. doi: 10.1073/pnas.111008398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bi X, Liu L F. J Mol Biol. 1994;235:414–423. doi: 10.1006/jmbi.1994.1002. [DOI] [PubMed] [Google Scholar]
- 9.Dianov G L, Kuzminov A V, Mazin A V, Salganik R I. Mol Gen Genet. 1991;228:153–159. doi: 10.1007/BF00282460. [DOI] [PubMed] [Google Scholar]
- 10.Lovett S T, Drapkin P T, Sutera V A, Jr, Gluckman-Peskind T J. Genetics. 1993;135:631–642. doi: 10.1093/genetics/135.3.631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lovett S T, Gluckman T J, Simon P J, Sutera V A, Jr, Drapkin P T. Mol Gen Genet. 1994;245:294–300. doi: 10.1007/BF00290109. [DOI] [PubMed] [Google Scholar]
- 12.Mazin A V, Kuzminov A V, Dianov G L, Salganik R I. Mol Gen Genet. 1991;228:209–214. doi: 10.1007/BF00282467. [DOI] [PubMed] [Google Scholar]
- 13.Bzymek M, Saveson C J, Feschenko V V, Lovett S T. J Bacteriol. 1999;181:477–482. doi: 10.1128/jb.181.2.477-482.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Modrich P. Annu Rev Genet. 1991;25:229–253. doi: 10.1146/annurev.ge.25.120191.001305. [DOI] [PubMed] [Google Scholar]
- 15.Burdett V, Baitinger C, Viswanathan M, Lovett S T, Modrich P. Proc Natl Acad Sci USA. 2001;98:6765–6770. doi: 10.1073/pnas.121183298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Viswanathan M, Burdett V, Baitinger C, Modrich P, Lovett S T. J Biol Chem. 2001;276:31053–31058. doi: 10.1074/jbc.M105481200. [DOI] [PubMed] [Google Scholar]
- 17.Lehman I R, Nussbaum A L. J Biol Chem. 1964;239:2628–2636. [PubMed] [Google Scholar]
- 18.Viswanathan M, Lovett S T. J Biol Chem. 1999;274:30094–30100. doi: 10.1074/jbc.274.42.30094. [DOI] [PubMed] [Google Scholar]
- 19.Lovett S T, Kolodner R D. Proc Natl Acad Sci USA. 1989;86:2627–2631. doi: 10.1073/pnas.86.8.2627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chase J W, Richardson C C. J Biol Chem. 1974;249:4553–4561. [PubMed] [Google Scholar]
- 21.Chase J W, Richardson C C. J Biol Chem. 1974;249:4545–4552. [PubMed] [Google Scholar]
- 22.Willetts N S, Clark A J, Low B. J Bacteriol. 1969;97:244–249. doi: 10.1128/jb.97.1.244-249.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Miller J H. A Short Course in Bacterial Genetics. Plainview, NY: Cold Spring Harbor Lab. Press; 1992. [Google Scholar]
- 24.Bachmann B J. In: Escherichia coli and Salmonella: Cellular and Molecular Biology. Neidhardt F C, Curtis R III, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaecter A, Umbarger H E, editors. Vol. 2. Washington, DC: Am. Soc. Microbiol.; 1996. pp. 2460–2488. [Google Scholar]
- 25.Lovett S T, Sutera V A., Jr Genetics. 1995;140:27–45. doi: 10.1093/genetics/140.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Datsenko K A, Wanner B L. Proc Natl Acad Sci USA. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dower W J, Miller J F, Ragsdale C W. Nucleic Acids Res. 1988;16:6127–6145. doi: 10.1093/nar/16.13.6127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang R F, Kushner S R. Gene. 1991;100:195–199. [PubMed] [Google Scholar]
- 29.Chung C T, Niemela S L, Miller R H. Proc Natl Acad Sci USA. 1989;86:2172–2175. doi: 10.1073/pnas.86.7.2172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lea D E, Coulson C A. J Genet. 1949;49:264–285. doi: 10.1007/BF02986080. [DOI] [PubMed] [Google Scholar]
- 31.Saveson C J, Lovett S T. Genetics. 1997;146:457–470. doi: 10.1093/genetics/146.2.457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lovett S T, Hurley R L, Sutera V A, Jr, Aubuchon R H, Lebedeva M A. Genetics. 2002;160:851–859. doi: 10.1093/genetics/160.3.851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Little J W, Edmiston S H, Pacelli L Z, Mount D W. Proc Natl Acad Sci USA. 1980;77:3225–3229. doi: 10.1073/pnas.77.6.3225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Matson S W. J Biol Chem. 1986;261:10169–10175. [PubMed] [Google Scholar]
- 35.Modrich P, Lahue R. Annu Rev Biochem. 1996;65:101–133. doi: 10.1146/annurev.bi.65.070196.000533. [DOI] [PubMed] [Google Scholar]
- 36.Eichler E E, Holden J J, Popovich B W, Reiss A L, Snow K, Thibodeau S N, Richards C S, Ward P A, Nelson D L. Nat Genet. 1994;8:88–94. doi: 10.1038/ng0994-88. [DOI] [PubMed] [Google Scholar]
- 37.Snow K, Tester D J, Kruckeberg K E, Schaid D J, Thibodeau S N. Hum Mol Genet. 1994;3:1543–1551. doi: 10.1093/hmg/3.9.1543. [DOI] [PubMed] [Google Scholar]
- 38.Rolfsmeier M L, Dixon M J, Lahue R S. Mol Cell. 2000;6:1501–1507. doi: 10.1016/s1097-2765(00)00146-5. [DOI] [PubMed] [Google Scholar]
- 39.Choudry S, Mukerji M, Srivastava A K, Jain S, Brahmachari S K. Hum Mol Genet. 2001;10:2437–2446. doi: 10.1093/hmg/10.21.2437. [DOI] [PubMed] [Google Scholar]
- 40.Viswanathan M, Lovett S T. Genetics. 1998;149:7–16. doi: 10.1093/genetics/149.1.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Viswanathan M, Lacirignola J J, Hurley R L, Lovett S T. J Mol Biol. 2000;302:553–564. doi: 10.1006/jmbi.2000.4088. [DOI] [PubMed] [Google Scholar]
- 42.Miesel L, Roth J R. J Bacteriol. 1996;178:3146–3155. doi: 10.1128/jb.178.11.3146-3155.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Razavy H, Szigety S K, Rosenberg S M. Genetics. 1996;142:333–339. doi: 10.1093/genetics/142.2.333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Corrette-Bennett S E, Lovett S T. J Biol Chem. 1995;270:6881–6885. doi: 10.1074/jbc.270.12.6881. [DOI] [PubMed] [Google Scholar]
- 45.Bedale W A, Inman R B, Cox M M. J Biol Chem. 1993;268:15004–15016. [PubMed] [Google Scholar]