Abstract
A primary motivation for sequencing the mouse genome was to accelerate the discovery of mammalian genes by using sequence conservation between mouse and human to identify coding exons. Achieving this goal proved challenging because of the large proportion of the mouse and human genomes that is apparently conserved but apparently does not code for protein. We developed a two-stage procedure that exploits the mouse and human genome sequences to produce a set of genes with a much higher rate of experimental verification than previously reported prediction methods. RT-PCR amplification and direct sequencing applied to an initial sample of mouse predictions that do not overlap previously known genes verified the regions flanking one intron in 139 predictions, with verification rates reaching 76%. On average, the confirmed predictions show more restricted expression patterns than the mouse orthologs of known human genes, and two-thirds lack homologs in fish genomes, demonstrating the sensitivity of this dual-genome approach to hard-to-find genes. We verified 112 previously unknown homologs of known proteins, including two homeobox proteins relevant to developmental biology, an aquaporin, and a homolog of dystrophin. We estimate that transcription and splicing can be verified for >1,000 gene predictions identified by this method that do not overlap known genes. This is likely to constitute a significant fraction of the previously unknown, multiexon mammalian genes.
Complete and precise delineation of protein coding genes in mammalian genomes remains a challenging task. To produce a preliminary gene catalog for the draft sequence of the mouse (1), the Mouse Genome Sequencing Consortium relied primarily on the ensembl gene build pipeline (2). ensembl works by (i) aligning known mouse cDNAs from refseq (3), riken (4, 5), and swissprot (6, 7) to the genome, (ii) aligning known proteins from related mammalian genes to the genome, and (iii) using portions of genscan (8) predictions that are supported by experimental evidence (such as ESTs). This conservative approach yielded ≈23,600 genes. However, ensembl cannot predict genes for which there is no preexisting evidence of transcription (1). Furthermore, reliance on known transcripts may lead to a bias against predicting genes that are expressed in a restricted manner or at very low levels.
Before the production of a draft genome sequence for a second mammal, the best available methods for predicting novel mammalian genes were single-genome de novo gene-prediction programs, of which genscan (8) is one of the most accurate and most widely used. These programs work by recognizing statistical patterns characteristic of coding sequences, splice signals, and other features in the genome to be annotated. However, they tend to predict many apparently false exons caused by the occurrence of such patterns by chance. With the availability of draft sequences for both the mouse and human genomes, it is now possible to incorporate genomic sequence conservation into de novo gene prediction algorithms. However, DNA alignment programs alone are not an effective means of gene prediction because a large fraction of the mouse and human genomes is conserved but does not code for protein.
We developed a procedure that greatly reduces the false-positive rate of de novo mammalian gene prediction by exploiting mouse–human conservation in both an initial gene-prediction stage and an enrichment stage. The first stage is to run gene-prediction programs that use genome alignment in combination with statistical patterns in the DNA sequence itself. A number of such programs have been described (9–12). For these experiments, we used SGP2 (13) and twinscan (refs. 14 and 15 and http://genes.cs.wustl.edu), two such programs that we designed for efficient analysis of whole mammalian genomes. twinscan is an independently developed extension of the genscan probability model, whereas SGP2 is an extension of geneid (16, 17). The probability scores these programs assign to each potential exon are modified by the presence and quality of genome alignments. twinscan uses nucleotide alignment [blastn (18), blast.wustl.edu] and has specific models for how alignments modify the scores of coding regions, UTRs, splice sites, and translation initiation and termination signals. SGP2, in contrast, uses translated alignments [tblastx (18), blast.wustl.edu] to modify the scores of potential coding regions only. These programs predict many fewer exons than genscan with no reduction in sensitivity to the exons of known genes (13, 14).
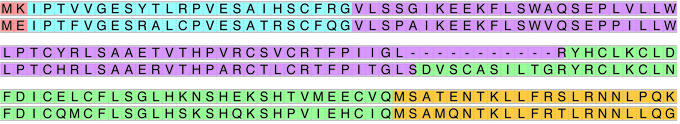
The second stage of our procedure is based on the observation that almost all mouse genes have a human counterpart with highly conserved exonic structure (1). We therefore compare all multiexon genes predicted in mouse in the first stage to those predicted in human. Predictions are retained only if the protein predicted in mouse aligns to a human protein predicted by the same program, with at least one predicted intron at the same location (aligned intron, Fig. 1). Predicted single-exon genes are always discarded by this procedure. Although there are many real single-exon genes, it is not currently possible to predict them reliably nor to verify them reliably in a cost-effective, high-throughput procedure.
Figure 1.
An example of predictions with aligned introns. RT-PCR positive predicted protein 3B1 (a novel homolog of Dystrophin) is aligned with its predicted human ortholog (N-terminal regions shown; Upper of each row: mouse, Lower of each row: human). Each color indicates one coding exon. Three of four predicted splice boundaries (color boundaries) align perfectly. Any one of these three is sufficient for surviving the enrichment step. Gaps in the alignment (shown as dashes) may indicate mispredicted regions.
In this article, we show that our two-stage process yields >1,400 predictions outside the standard annotation of the mouse genome. RT-PCR and direct sequencing of a single exon pair in a sample of these predictions indicates that the majority correspond to real spliced transcripts. Our results also show that this procedure is sensitive to genes that are hard to find by other methods. The combination of these computational and experimental techniques forms a powerful, cost-effective system for expanding experimentally supported genome annotation. This approach is therefore expected to bring the annotation of the mouse and human genomes nearer to closure.
Experimental Procedures
Genome Sequences.
The MGSCv3 assembly of the mouse genome described in ref. 1 and the December, 2001 Golden Path assembly of the human genome (National Center for Biotechnology Information Build 28) were downloaded from the University of California (Santa Cruz) genome browser (http://genome.ucsc.edu).
Genome Alignments.
twinscan was run on the mouse genome by using blastn alignments to the human genome (wu-blast, http://blast.wustl.edu). Lowercase masking in the human sequence was first converted to N masking. The result was further masked with nseg by using default parameters, all Ns were removed, and the sequence was cut into 150-kb database segments. The mouse genome sequence was divided into 1-mb query segments. blastn parameters were: M=1 N=−1Q=5 R=1 Z=3000000000 Y=3000000000 B=10000 V=100 W=8 X=20 S=15 S2=15 gapS2=30 lcmask wordmask=seg wordmask=dust topcomboN=3. twinscan was run on the human genome by using separate blastn alignments to the mouse genome, which was prepared in the same way except that Ns were not removed before creating the blast database.
SGP2 was run on the mouse and human genomes by using a single set of alignments. The masked human genome was cut into 100-kb query segments that were compared with a database of all 100-kb segments of the mouse genome with tblastx (wu-blast, parameters: B=9000 V=9000 hspmax=500 topcomboN=100 W=5 E=0.01 E2=0.01 Z=3000000000 nogap filter=xnu+seg S2=80). The substitution matrix was blosum62 modified to penalize alignments with stop codons heavily (−500).
Initial Gene Predictions.
twinscan was run on 1-mb segments of the mouse and human genomes with target genome parameters identical to the genscan parameters and the 68-set-ortholog conservation parameters (available on request). Note that the twinscan results described in ref. 14 are based on a subsequently developed set of target genome parameters that yields better results than those described here. SGP2 was run on unsegmented mouse and human chromosomes. The refseq genes (which were not tested in the experiments reported here) were incorporated directly into the SGP2 predictions, which improved the predictions outside the refseqs slightly by preventing some gene fusion errors. Note that the refseqs were not used in generating the SGP2 results described in ref. 13.
Novelty Criteria.
Mouse predictions were considered known if they overlapped ensembl predictions or had 95% nucleotide identity to a refseq mRNA or an ensembl-predicted mRNA over at least 100 bp. We used the most inclusive set of ensembl predictions available, based on the complete riken cDNA set without further filtering (1).
Enrichment Procedure.
The enrichment procedure was applied separately to predictions of twinscan and SGP2. The protein sequences predicted by each program in human and mouse were compared by using blastp (19). For each predicted mouse protein, all predicted human proteins with expect values <1 × 10−6 were called homologs. A global protein alignment was produced for the best scoring homologs (up to five) by using t-coffee (ref. 39; http://igs-server.cnrs-mrs.fr/∼cnotred/Projects_home_page/t_coffee_home_page.html) with default parameters. Exonic structure was added to the alignments by using exstral.pl (www1.imim.es/∼rcastelo/exstral.html). When both members of an aligned pair contained an intron at the same coordinate with at least 50% identity over 15 aa on both sides the corresponding mouse prediction was assigned to the “enriched” pool. Predictions with homologs but no aligned intron were assigned to the “similar” pool.
RT-PCR.
To test predictions, primers were designed in adjacent exons as described in Results and used in RT-PCR of total RNA from 12 normal mouse adult tissues. All procedures were as described (20), except that JumpStart REDTaq ReadyMix (Sigma) and primers from Sigma-Genosys were used.
Additional Details.
See supplementary information at www1.imim.es/datasets/mouse2002 for additional details of these procedures.
Results
We applied the two-stage procedure described above to the entire draft mouse and human genome sequences (see Experimental Procedures). twinscan predicted 17,271 genes with at least one aligned intron, whereas SGP2 predicted a largely overlapping set of 18,056 genes with at least one aligned intron. These predicted gene sets contain 145,734 exons and 168,492 exons, respectively. Together the two sets overlapped 90% of multiexon ensembl gene predictions.
To estimate a lower bound on the proportion of novel predictions that are transcribed and spliced, we performed a series of RT-PCR amplifications from 12 adult mouse tissues (20). We did not test genes that overlap ensembl predictions nor those that are 95% identical to ensembl predictions or refseq mRNAs over >100 bp or more. Because ensembl was the standard for annotation of the draft mouse genome, we refer to the non-ensembl genes as “novel.” A random sample of novel genes predicted by each program and containing at least one aligned intron was tested. Primer pairs were designed in adjacent exons separated by an aligned intron of at least 1,000 bp (Fig. 2). The exon pair to be tested was chosen on the basis of intron length (minimum 1,000 bp), primer design requirements, and de novo gene prediction score, with no reference to protein, EST, or cDNA databases. Amplification followed by direct sequencing of the PCR product (Fig. 3) verified the exon pair in 133 unique predicted genes of 214 tested (62%, enriched pool, see Table 1 and www1.imim.es/datasets/mouse2002). Mouse genes predicted by both programs were verified at a much higher rate than those predicted by just one program (76% vs. 27%). Extrapolating from the success rates in Table 1, testing the entire pool of 1,428 enriched predictions in this way is expected to yield a total of 788 (±48) predictions with confirmed splices, none of which overlap ensembl predictions.
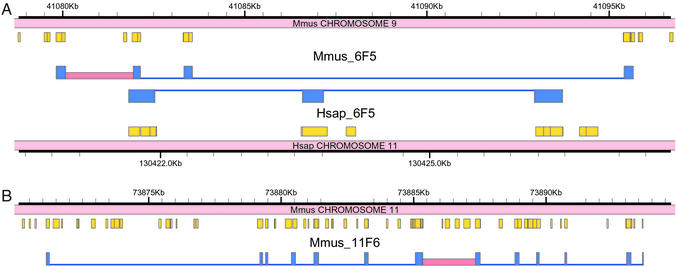
Figure 2.
Two examples of predicted gene structures (blue) with introns verified by RT-PCR from primers located in exons flanking the introns indicated in red. Mouse–human genomic alignments (orange) correlate with predicted exons but do not match them exactly. (A) Verified mouse prediction 6F5, a novel homolog of Drosophila brain-specific homeobox protein (bsh), with matching human prediction. (B) Verified mouse prediction 11F6, a homolog of rat vanilloid receptor type 1-like protein 1. No matching human gene was predicted. A cDNA (GenBank accession no. AF510316) that matches the predicted protein over four protein-coding exons was deposited in GenBank subsequent to our analysis.
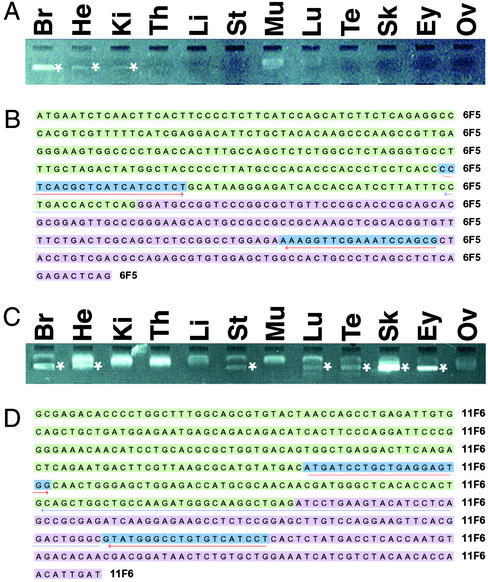
Figure 3.
Verification of gene predictions by RT-PCR analysis. (A and B) Test of prediction 6F5, a homolog of Drosophila brain-specific homeobox protein (bsh). (C and D) Test of prediction 11F6, a homolog of rat vanilloid receptor type 1-like protein. Gel analysis of amplimers (*) with the source of the cDNA pool indicated above is shown in A and C. Primers (blue) and the region to which the amplimer sequence aligned (underlining) are shown in B and D. The indicated forward primers were used to generate the amplimer sequences (brain amplimer, B; skin amplimer, D). Br, brain; Ey, eye; He, heart; Ki, kidney; Li, liver; Lu, lung; Mu, muscle; Ov, ovary; Sk, skin; St, stomach; Te, testis; Th, thymus.
Table 1.
Predicted novel gene sets and RT-PCR verification rates
| Pool | Programs* | No. of predictions | No. tested | No. positive | Success rate, % | Expected successes | Standard error |
|---|---|---|---|---|---|---|---|
| Enriched† | Both | 827 | 154 | 117 | 75.97 | 628 | |
| One | 601 | 60 | 16 | 26.67 | 160 | ||
| Total | 1,428 | 214 | 133 | 62.15 | 788 | 48 | |
| Similar‡ | Both | 505 | 16 | 4 | 25.00 | 126 | |
| One | 1,620 | 22 | 0 | 0.00 | 0 | ||
| Total | 2,125 | 38 | 4 | 10.53 | 126 | 105 | |
| Other§ | Both | 234 | 5 | 1 | 20.00 | 46 | |
| One | 3,425 | 58 | 1 | 1.72 | 59 | ||
| Total | 3,659 | 63 | 2 | 3.17 | 105 | 83 | |
| All | Total | 7,212 | 315 | 139 | N/A | 1,019 |
N/A, not applicable.
Both, Genes predicted at least partially by both twinscan and sgp2 programs. One, Genes predicted by one program that are not overlapped by predictions of the other program. N/A, not applicable.
†Mouse gene predictions containing an intron whose flanking exonic regions align with flanking exonic regions predicted by the same program in human.
‡Mouse gene predictions that fail the enrichment step but show regions of strong similarity to a gene predicted by the same program in human.
§Mouse gene predictions without regions of strong similarity to any gene predicted by the same program in human.
Considered in isolation, genes predicted by twinscan had a higher verification rate than those predicted by SGP2 (83% vs. 44%), but that difference is skewed by the fact that twinscan predicted fewer exons per gene, and hence its predictions were less likely to overlap ensembl predictions. We corrected for this by clustering overlapping twinscan and SGP2 predictions to ensure that both were counted as positive if either was verified experimentally. For each program, the predictions belonging to a given cluster were counted only once, even if more than one was RT-PCR positive. After this correction, the confirmation rates were much closer (76% for twinscan vs. 62% for SGP2). The results shown in Table 1 include the correction. The twinscan verification rate is similar to the verification rate for genes predicted by both programs because the exons predicted by twinscan are largely a subset of those predicted by SGP2.
Before the enrichment procedure, the combined predictions of SGP2 and twinscan overlap 98% of multiexon ensembl genes, as compared with 90% for the enriched pool. This finding suggests that the enrichment procedure reduces sensitivity by a small but noticeable degree. To investigate the potential loss of sensitivity further, we applied the same RT-PCR procedure to two samples of gene predictions that were excluded by the enrichment criterion and did not overlap ensembl predictions. One sample had one or more regions of strong similarity to a predicted human gene but did not satisfy the aligned intron criterion (similar pool) whereas the other lacked any strong similarity to a human prediction by the same program (other pool). The verification rates for the similar and other pools were 25% and 20%, respectively, for genes predicted by both programs, and 0% and 2%, respectively, for genes predicted by only one program (Table 1 and www1.imim.es/datasets/mouse2002). This finding shows that the enrichment procedure increases specificity greatly and, consistent with the ensembl overlap analysis, reduces sensitivity only slightly. If all predictions in the similar and other pools were tested the expected numbers of successes are 126 (±105) and 105 (±83), respectively, with the large standard errors resulting from the small number of successful amplifications in these pools.
As a control, we also tested 113 predictions from the enriched pool that did overlap ensembl predictions. In 66 of the predictions the splice boundary we tested was predicted identically in ensembl, and 64 of these tests (97%) were positive. In 47 of the predictions the splice boundary we tested was not predicted identically in ensembl, and 21 of these tests (45%) were positive, despite the fact that ensembl predictions are based on transcript evidence. This verification rate may reflect alternative splices identified by our method but not by ensembl.
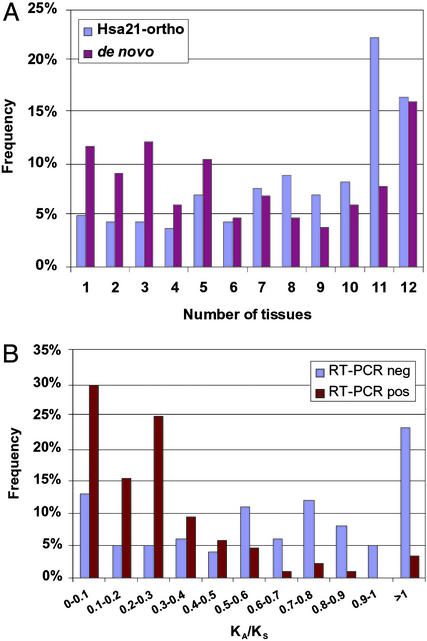
To determine whether tissue-restricted expression could explain the absence of the predictions we verified from the transcript-based annotation, we compared the expression patterns of our RT-PCR positive predictions to those of the complete set of mouse orthologs of genes mapping to human chromosome 21 (Hsa21). These genes were chosen for comparison because they had been previously subjected to the same protocol with the same cDNA pools in the same laboratory (20). Our verified novel gene predictions showed a significantly more restricted pattern of expression (Fig. 4A). The mean number of tissues for our positive predictions was 6.3, and 33% of the positive predictions showed expression in three or fewer tissues; the corresponding numbers for the mouse orthologs of human chromosome 21 genes are 8.2 tissues on average and 14% showing expression in three or fewer tissues. This difference in expression specificity was statistically significant (ANOVA, F = 23.22, df = 1, P < 0.001).
Figure 4.
Characteristics of verified predictions. (A) Expression specificity. Percentages of RT-PCR positive de novo predictions (red) and Hsa21 mouse orthologs (blue) expressed in 1–12 tissues, tested in the same cDNA pools. (B) Distributions of the ratio of nonsynonymous to synonymous substitution rate (KA/KS) in 83 RT-PCR positive (red) vs. 98 RT-PCR negative (blue) mouse predictions with reciprocal best blast matches among the human predictions.
To determine whether prediction of pseudogenes by our method could explain some of the RT-PCR negatives, we computed the ratio of nonsynonymous to synonymous substitution rates (KA/KS) (21) for the subset of tested mouse predictions with unique putative human orthologs (Fig. 4B). The mean for PCR-positive predictions was 0.29 whereas for PCR-negative predictions it was 0.72. The difference was statistically significant (ANOVA, F = 34.86, df = 1, P < 0.001), suggesting that (i) some of the negative predictions may be pseudogenes, and (ii) KA/KS can be efficiently incorporated in the enrichment protocol to increase specificity (22).
Among the predictions with confirmed splices, 112 had significant homology to known genes and/or domains. A few of these genes, which were not represented in databases at the beginning of our gene survey, were submitted to databases and/or published in the literature in the intervening months. For example, we correctly predicted the first four protein coding exons of TRPV3, a heat-sensitive TRP channel in keratinocytes (23), and both exons of RLN3 (preprorelaxin 3), an insulin-like prohormone (24). The verified predictions with the most notable homologies are shown in Table 2, including a novel homolog of dystrophin that is discussed in the mouse genome paper (1). Table 2 includes two noncanonical homeobox genes, one that is most similar to fruitfly brain-specific homeobox protein (Figs. 2 and 3 A and B) (25) and another that is a Not-class homeobox, likely to be involved in notochord development (26). Four predicted genes were found to be expressed in the brain and are likely to have neuronal functions, including one paralog each of: Nna1, which is expressed in regenerating motor neurons (27); an N-acetylated-α-linked-acidic dipeptidase, which hydrolyses the neuropeptide N-acetyl-aspartyl-glutamate to terminate its neurotransmitter activity (28); a novel γ-aminobutyric acid type B receptor, which regulates neurotransmitter release (29); and an Ent2-like nucleoside transporter, which modulates neurotransmission by altering adenosine concentrations (30). Other verified genes are likely to be important in muscle contraction (myosin light chain kinase homolog), degradation of cell cycle proteins (fizzy/CDC20 homolog), Wnt-dependent vertebrate development (Dapper/frodo homolog), and solute and steroid transport in the liver (solute transporter β). Homologs of two further genes predicted in our studies are associated with disease. ATP10C, an aminophospholipid translocase, is absent from Angelman syndrome patients with imprinting mutations (31), and otoferlin, which is mutated in a nonsyndromic form of deafness (32).
Table 2.
Novel mouse genes, their tissue expression, and their homologs
| Code | B | H | K | Y | V | S | M | L | T | K | E | O | %Id | Ln | Homology |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 3B1 | + | + | 38 | 134 | Dystrophin-like; with ZZ domain | ||||||||||
| 3B3 | + | + | + | + | + | 25 | 184 | Novel aquaporin; similar to Drosophila CG12251 | |||||||
| 3C3 | + | + | + | + | + | 25 | 260 | TEP1 (telomerase associated); probable ATPase | |||||||
| 3C5 | + | + | 47 | 198 | Voltage-dependent calcium channel γ subunit | ||||||||||
| 4B3 | + | + | + | 34 | 74 | IFN-induced/fragilis transmembrane family | |||||||||
| 4C6 | + | + | + | + | + | 30 | 134 | IL-22-binding protein CRF2-10 | |||||||
| 4G4 | + | + | + | + | 64 | 109 | Nna1p, nuclear ATP/GTP-binding protein | ||||||||
| 5B5 | + | + | + | 43 | 111 | Likely aminophospholipid flippase (transporting ATPase) | |||||||||
| 1E3 | + | + | + | + | + | 40 | 106 | N-acetylated-α-linked-acidic dipeptidase (NAALADase) | |||||||
| 6C4 | + | + | 42 | 117 | Not-type homeobox; poss. involved in notochord development | ||||||||||
| 6F5 | + | + | + | 66 | 102 | Drosophila brain-specific homeobox protein (bsh) | |||||||||
| 11F2 | + | + | + | + | + | 29 | 216 | Human γ-aminobutyric acid type B receptor 2, neurotransmitter release regulator | |||||||
| 5A2 | + | + | + | + | 41 | 36 | Skate liver organic solute transporter β | ||||||||
| 11B6 | + | + | + | 55 | 116 | IFN-activatable protein 203; nuclear protein | |||||||||
| 12B3 | + | + | + | + | + | + | + | + | 25 | 229 | Fatty acid desaturase; maintains membrane integrity | ||||
| 11F6 | + | + | + | + | + | + | + | 44 | 494 | Rat vanilloid receptor type 1 like protein 1 | |||||
| 12E3 | + | + | 52 | 175 | Fizzy/CDC20; modulates degradation of cell-cycle proteins | ||||||||||
| 12F1 | + | + | + | + | + | 43 | 355 | Otoferlin (mutated in DFNB9, nonsyndromic deafness) | |||||||
| 12H1 | + | + | + | 45 | 116 | Fruitfly additional sex combs; a Polycomb group protein | |||||||||
| 12C4 | + | + | + | 43 | 133 | Caenorhabditis elegans C15C8.2; single-minded-like; HLH and PAS domains | |||||||||
| 12D2 | + | 41 | 397 | Cytosolic phospholipase A2, group IVB | |||||||||||
| 12A5 | + | 38 | 415 | Fruitfly GH15686p; Ent2-like nucleoside transporter | |||||||||||
| 12E5 | + | + | + | + | 32 | 111 | Relaxin 3 preproprotein; prohormone of the insulin family | ||||||||
| 11A1 | + | + | + | + | + | 89 | 75 | Mouse BET3, involved in ER to Golgi transport | |||||||
| 11A2 | + | + | + | + | + | + | 70 | 207 | Vacuolar ATP synthase subunit S1 | ||||||
| 11B2 | + | + | + | + | + | + | 54 | 271 | Myosin light chain kinase, skeletal muscle | ||||||
| 11G2 | + | + | + | + | + | + | + | + | + | + | 36 | 179 | Dapper/frodo (transduces Wnt signals by interacting with Dsh) |
Code, Coding name of tested gene model. B, brain; H, heart; K, kidney; Y, thymus; V, liver; S, stomach; M, muscle; L, lung; T, testis; K, skin; E, eye; O, ovary. %Id, Percentage amino acid identity. Ln, Number of amino acids in the local alignment between the prediction and the homolog.
Discussion
We have demonstrated a remarkably efficient mammalian gene discovery system. This system exploits the draft mouse and human genome sequences in both an initial gene-prediction stage and an enrichment stage. The first stage consists of SGP2 and twinscan, gene-prediction programs that use genome alignment in combination with statistical patterns in the DNA sequence. We have shown elsewhere that both programs have greater sensitivity and specificity than single-genome de novo predictors, such as genscan (13, 14). In this article, we have demonstrated the effectiveness of the enrichment stage, in which predictions are retained only if the protein predicted in mouse aligns to a human protein predicted by the same program, with at least one predicted intron at the same location (aligned intron, Fig. 1). In our pool of predictions, the aligned intron filter is expected to eliminate 24 times more RT-PCR negatives than RT-PCR positives. This enrichment procedure can be applied to predictions from any program.
Our goal was to develop a low-cost, high-throughput system for finding and verifying coding regions that are missed by annotation systems that require existing transcript evidence. ensembl was chosen as the representative of such systems because the Mouse Genome Sequencing Consortium judged it to be the most suitable tool for timely, cost-effective, reliable annotation of the mouse genome sequence. Thus, we evaluated our system by investigating genes that do not overlap ensembl predictions. Our system is not designed to find genes that would be missed by expert manual annotators, who can effectively integrate information such as the predictions of genscan (8) and genomescan (33), percent-identity plots (34), comparison to fish genomes (35, 36), alignment of weakly homologous proteins, and alignment of EST sequences. As a result, we did not exclude gene predictions from our evaluation based on these indicators.
Our two-stage system identified a highly reliable pool of 827 predicted genes not overlapping the standard annotation, of which we tested 154 for expression by using RT-PCR and direct sequencing. Primers designed for a single pair of adjacent exons in each predicted gene yielded a spliced PCR product whose sequence closely matched that of the predicted exons in 76% of these tests. In the only other published report of high-throughput verification of gene predictions of which we are aware, 14% of predictions not overlapping the standard annotation yielded spliced products (37). These numbers cannot be compared directly because of differences in the sampling criteria, but the magnitude of the difference suggests our method provides new levels of efficiency in experimental confirmation of genes outside the standard annotation set.
The sensitivity of our method also appears to be high. Predictions in our enriched pool overlap 90% of multiexon genes predicted by ensembl. However, it has been estimated that >4,000 ensembl predictions comprising 12,000 predicted exons are in fact pseudogenes (1). Although the precise number of multiexon pseudogenes in the ensembl annotation is unknown, this estimate suggests that our enriched pool may overlap a much larger fraction of the functional genes identified by ensembl. Further, RT-PCR tests of twinscan and SGP2 predictions outside the enriched pool indicate that a relatively small number of these predictions are transcribed and spliced in the 12 tissues tested. Thus, the enrichment procedure is sensitive to both ensembl predictions and verifiable predictions by twinscan and SGP2.
Using our system, we confirmed one intron of 139 predicted genes that do not overlap any gene in the standard mouse genome annotation (1). Ninety-two of the RT-PCR positive introns (66%) did not align to any mouse EST, and these might have posed difficulties even for human annotators. Furthermore, seven of the RT-PCR negative introns (4%) did align to mouse ESTs and six of these were in the enriched pool, suggesting that the true percentage of transcribed and spliced predictions in this pool may be even higher than the RT-PCR positive percentage.
Among RT-PCR positive predictions, 24 had homologies to known proteins that we found particularly interesting (Table 2). The positive identification of these homologs is expected to impact numerous research programs devoted to genes of developmental and medical importance. In general, these genes were probably missed in the ensembl annotation because the length and percent identity of the homologies were not sufficient to support a protein-based gene prediction (Table 2). In many cases, such as the predicted homolog of a brain-specific homeobox protein, the expression patterns we found were consistent with what would be expected from the function of the known homolog (Fig. 3 A and B).
The confirmed 139 genes also showed a relatively restricted expression pattern, on average. Because all mouse orthologs of genes on human chromosome 21 had already been tested by using the same experimental protocol and the same cDNA pools, we were able to directly compare expression patterns. To the extent that the known genes on chromosome 21 are no more tissue specific than the complete set of known genes, the results (Fig. 4) suggest that our system may be particularly sensitive to genes with tissue-restricted expression. Qualitatively similar restricted expression patterns were reported for novel genscan predictions on chromosome 22 (37), lending further support to the value of de novo prediction for identifying genes with tissue-restricted expression.
Of the RT-PCR positive novel predictions, only 33% have identifiable homologs in the sequenced fish (Fugu/Tetraodon/zebrafish) genomes. Comparing this finding to the recent estimate that three-quarters of all human genes can be recognized in the Fugu genome (36) suggests that our system may be particularly sensitive to genes that are not ubiquitous in the vertebrate lineage. Genes with relatively restricted expression patterns and species distribution can be difficult to find by using transcript-based methods like genewise (38) and compact-genome methods like exofish (35), but they appear to be tractable for our system.
Extrapolating from the success rates in all categories, the expected total number of gene predictions that could be successfully RT-PCR amplified in the cDNA pools we tested is 1,019 (Table 1), adding ≈5% to the number of functional mouse genes identified by ensembl (1). The number of distinct genes verifiable in this way may be slightly smaller, because the effect of fragmentation in ensembl and in our predictions is not readily testable. However, the number of predictions that are transcribed and spliced is likely to be >1,019, because (i) we tested only one exon pair from each prediction and (ii) we used only 12 adult mouse tissues (20).
The relatively low success rate in the pools failing the enrichment step suggests that the number of real, multiexon genes whose existence has been predicted but not yet confirmed is in the range of 1,000–2,000 (including those predictions in the enriched pool that have not been confirmed). Because we have used only two prediction programs, twinscan and SGP2, it is possible that other programs might yield a large additional set of predictions that pass the enrichment step. However, genscan yields only 49 additional predictions that pass enrichment and novelty criteria and do not overlap the 1,428 “aligned intron” novel predictions from twinscan and SGP2 (3%). These 49 are worth testing, and adding more prediction programs will yield at least a few more predictions with aligned introns. Nonetheless, the data presented here suggest that the 1,428 predictions in the enriched pool may overlap a significant fraction of the previously unannotated, multiexon mouse genes.
Using the draft sequences of the mouse and human genomes, we have developed a cost-effective, high-throughput system for predicting genes and verifying the existence of corresponding spliced transcripts. Applying this system to the entire mouse genome, we showed that an automated system can produce a large set of experimentally supported mammalian gene predictions outside the standard annotation. Further, the average cost per verified exon pair is less than two primer pairs and sequencing reactions. We expect that testing the remaining predictions in the enriched pool will locate most multiexon mouse genes that are currently unannotated, bringing us significantly closer to identification of the complete mammalian gene set.
As more mammalian genomes are sequenced, the need for experimentally validated high-throughput annotation will continue to grow, as will the data available for methods such as ours. Using the sequences of more genomes, it may be possible to extend this approach to single-exon and lineage-specific genes. In combination with methods like ensembl and refinement by expert annotators, these developments may bring complete, experimentally supported genome annotation within reach.
Acknowledgments
We are grateful to the Mouse Genome Sequencing Consortium for providing the mouse genome sequence as well as support throughout the analysis process. We are particularly grateful to Eric Lander, Robert Waterston, Ewan Birney, Adam Felsenfeld, and Ross Hardison for advice and encouragement. Thanks are also due to Marc Vidal, Lior Pachter, Kerstin Lindblad-Toh, and Gwen Acton for participation in pilot experiments and Tamara Doering for helpful comments on the manuscript. Research at Institut Municipal d'Investigació Mèdica/Universitat Pompeu Fabra/Centre de Regulació Genòmica is supported by a grant from the Spanish Plan Nacional de Investigacion y Desarrollo. J.F.A. is supported by a fellowship from the Instituto de Salud Carlos III. The Division of Medical Genetics is supported by the Swiss National Science Foundation, National Centres of Competence in Research Frontiers in Genetics, and the Childcare and J. Lejeune Foundations. Research at Washington University was supported by Grant DBI-0091270 from the National Science Foundation (to M.R.B.) and Grant HG02278 from the National Institutes of Health (to M.R.B.).
References
- 1.Mouse Genome Sequencing Consortium. Nature. 2002;420:520–562. doi: 10.1038/nature01262. [DOI] [PubMed] [Google Scholar]
- 2.Hubbard T, Barker D, Birney E, Cameron G, Chen Y, Clark L, Cox T, Cuff J, Curwen V, Down T, et al. Nucleic Acids Res. 2002;30:38–41. doi: 10.1093/nar/30.1.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pruitt K D, Maglott D R. Nucleic Acids Res. 2001;29:137–140. doi: 10.1093/nar/29.1.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kawai J, Shinagawa A, Shibata K, Yoshino M, Itoh M, Ishii Y, Arakawa T, Hara A, Fukunishi Y, Konno H, et al. Nature. 2001;409:685–690. doi: 10.1038/35055500. [DOI] [PubMed] [Google Scholar]
- 5.The FANTOM Consortium and The RIKEN Genome Exploration Research Group Phase II Team. Nature. 2002;420:563–571. [Google Scholar]
- 6.Bairoch A, Apweiler R. Nucleic Acids Res. 2000;28:45–48. doi: 10.1093/nar/28.1.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gasteiger E, Jung E, Bairoch A. Curr Issues Mol Biol. 2001;3:47–55. [PubMed] [Google Scholar]
- 8.Burge C, Karlin S. J Mol Biol. 1997;268:78–94. doi: 10.1006/jmbi.1997.0951. [DOI] [PubMed] [Google Scholar]
- 9.Wiehe T, Gebauer-Jung S, Mitchell-Olds T, Guigó R. Genome Res. 2001;11:1574–1583. doi: 10.1101/gr.177401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pachter L, Alexandersson M, Cawley S. J Comput Biol. 2002;9:389–399. doi: 10.1089/10665270252935520. [DOI] [PubMed] [Google Scholar]
- 11.Batzoglou S, Pachter L, Mesirov J P, Berger B, Lander E S. Genome Res. 2000;10:950–958. doi: 10.1101/gr.10.7.950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bafna V, Huson D H. Proc Int Conf Intell Syst Mol Biol. 2000;8:3–12. [PubMed] [Google Scholar]
- 13.Parra G, Agarwal P, Abril J F, Wiehe T, Fickett J W, Guigó R. Genome Res. 2003;13:108–117. doi: 10.1101/gr.871403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Flicek P, Keibler E, Hu P, Korf I, Brent M R. Genome Res. 2003;13:46–54. doi: 10.1101/gr.830003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Korf I, Flicek P, Duan D, Brent M R. Bioinformatics. 2001;17, Suppl. 1:S140–S148. doi: 10.1093/bioinformatics/17.suppl_1.s140. [DOI] [PubMed] [Google Scholar]
- 16.Parra G, Blanco E, Guigó R. Genome Res. 2000;10:511–515. doi: 10.1101/gr.10.4.511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guigó R, Knudsen S, Drake N, Smith T. J Mol Biol. 1992;226:141–157. doi: 10.1016/0022-2836(92)90130-c. [DOI] [PubMed] [Google Scholar]
- 18.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 19.Altschul S F, Madden T L, Schaffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Reymond A, Marigo V, Yaylaoglu M B, Leoni A, Ucla C, Scamuffa N, Caccioppoli C, Dermitzakis E T, Lyle R, Banfi S, et al. Nature. 2002;420:582–586. doi: 10.1038/nature01178. [DOI] [PubMed] [Google Scholar]
- 21.Hughes A L, Nei M. Nature. 1988;335:167–170. doi: 10.1038/335167a0. [DOI] [PubMed] [Google Scholar]
- 22.Nekrutenko A, Makova K D, Li W H. Genome Res. 2002;12:198–202. doi: 10.1101/gr.200901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Peier A M, Reeve A J, Andersson D A, Moqrich A, Earley T J, Hergarden A C, Story G M, Colley S, Hogenesch J B, McIntyre P, et al. Science. 2002;296:2046–2049. doi: 10.1126/science.1073140. [DOI] [PubMed] [Google Scholar]
- 24.Bathgate R A, Samuel C S, Burazin T C, Layfield S, Claasz A A, Reytomas I G, Dawson N F, Zhao C, Bond C, Summers R J, et al. J Biol Chem. 2002;277:1148–1157. doi: 10.1074/jbc.M107882200. [DOI] [PubMed] [Google Scholar]
- 25.Jones B, McGinnis W. Development (Cambridge, UK) 1993;117:793–806. doi: 10.1242/dev.117.2.793. [DOI] [PubMed] [Google Scholar]
- 26.Talbot W S, Trevarrow B, Halpern M E, Melby A E, Farr G, Postlethwait J H, Jowett T, Kimmel C B, Kimelman D. Nature. 1995;378:150–157. doi: 10.1038/378150a0. [DOI] [PubMed] [Google Scholar]
- 27.Harris A, Morgan J I, Pecot M, Soumare A, Osborne A, Soares H D. Mol Cell Neurosci. 2000;16:578–596. doi: 10.1006/mcne.2000.0900. [DOI] [PubMed] [Google Scholar]
- 28.Pangalos M N, Neefs J M, Somers M, Verhasselt P, Bekkers M, van der Helm L, Fraiponts E, Ashton D, Gordon R D. J Biol Chem. 1999;274:8470–8483. doi: 10.1074/jbc.274.13.8470. [DOI] [PubMed] [Google Scholar]
- 29.Billinton A, Ige A O, Bolam J P, White J H, Marshall F H, Emson P C. Trends Neurosci. 2001;24:277–282. doi: 10.1016/s0166-2236(00)01815-4. [DOI] [PubMed] [Google Scholar]
- 30.Crawford C R, Patel D H, Naeve C, Belt J A. J Biol Chem. 1998;273:5288–5293. doi: 10.1074/jbc.273.9.5288. [DOI] [PubMed] [Google Scholar]
- 31.Meguro M, Kashiwagi A, Mitsuya K, Nakao M, Kondo I, Saitoh S, Oshimura M. Nat Genet. 2001;28:19–20. doi: 10.1038/ng0501-19. [DOI] [PubMed] [Google Scholar]
- 32.Yasunaga S, Grati M, Cohen-Salmon M, El-Amraoui A, Mustapha M, Salem N, El-Zir E, Loiselet J, Petit C. Nat Genet. 1999;21:363–369. doi: 10.1038/7693. [DOI] [PubMed] [Google Scholar]
- 33.Yeh R F, Lim L P, Burge C B. Genome Res. 2001;11:803–816. doi: 10.1101/gr.175701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schwartz S, Zhang Z, Frazer K A, Smit A, Riemer C, Bouck J, Gibbs R, Hardison R, Miller W. Genome Res. 2000;10:577–586. doi: 10.1101/gr.10.4.577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Roest Crollius H, Jaillon O, Bernot A, Dasilva C, Bouneau L, Fischer C, Fizames C, Wincker P, Brottier P, Quetier F, et al. Nat Genet. 2000;25:235–238. doi: 10.1038/76118. [DOI] [PubMed] [Google Scholar]
- 36.Aparicio S, Chapman J, Stupka E, Putnam N, Chia J M, Dehal P, Christoffels A, Rash S, Hoon S, Smit A, et al. Science. 2002;297:1301–1310. doi: 10.1126/science.1072104. [DOI] [PubMed] [Google Scholar]
- 37.Das M, Burge C B, Park E, Colinas J, Pelletier J. Genomics. 2001;77:71–78. doi: 10.1006/geno.2001.6620. [DOI] [PubMed] [Google Scholar]
- 38.Birney E, Durbin R. Genome Res. 2000;10:547–548. doi: 10.1101/gr.10.4.547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Notre dame C, Higgins D G, Heringa J. J Mol Biol. 2000;302:205–217. doi: 10.1006/jmbi.2000.4042. [DOI] [PubMed] [Google Scholar]