Abstract
Holoprosencephaly is a severe malformation of the brain characterized by abnormal formation and separation of the developing central nervous system. The prevalence is 1:250 during early embryogenesis, the live-born prevalence is 1:16 000. The etiology of HPE is extremely heterogeneous and can be teratogenic or genetic. We screened four known HPE genes in a Dutch cohort of 86 non-syndromic HPE index cases, including 53 family members. We detected 21 mutations (24.4%), 3 in SHH, 9 in ZIC2 and 9 in SIX3. Eight mutations involved amino-acid substitutions, 7 ins/del mutations, 1 frame-shift, 3 identical poly-alanine tract expansions and 2 gene deletions. Pathogenicity of mutations was presumed based on de novo character, predicted non-functionality of mutated proteins, segregation of mutations with affected family-members or combinations of these features. Two mutations were reported previously. SNP array confirmed detected deletions; one spanning the ZIC2/ZIC5 genes (approx. 100 kb) the other a 1.45 Mb deletion including SIX2/SIX3 genes. The mutation percentage (24%) is comparable with previous reports, but we detected significantly less mutations in SHH: 3.5 vs 10.7% (P=0.043) and significantly more in SIX3: 10.5 vs 4.3% (P=0.018). For TGIF1 and ZIC2 mutation the rate was in conformity with earlier reports. About half of the mutations were de novo, one was a germ line mosaic. The familial mutations displayed extensive heterogeneity in clinical manifestation. Of seven familial index patients only two parental carriers showed minor HPE signs, five were completely asymptomatic. Therefore, each novel mutation should be considered as a risk factor for clinically manifest HPE, with the caveat of reduced clinical penetrance.
Keywords: Holoprosencephaly, SHH, SIX3, ZIC2, TGIF, genotype–phenotype
Introduction
Holoprosencephaly (HPE; MIM236100) is a structural anomaly of the human brain that occurs in early embryonic life. The disorder originates around the fourth week of pregnancy when the prosencephalon (forebrain) fails to cleave or incompletely cleaves into diencephalon and telencephalon, into distinct left and right hemispheres and into olfactory and optic bulb tracts. The estimated prevalence of HPE is 1 in 16 000 after birth, but the prevalence of HPE in early embryogenesis is much higher (1 in 250).1 Classification is based on severity of the brain malformation and ranges from the most severe alobar form (single primitive ventricle, >60% of the cases), to the semi-lobar (incomplete separation of the ventricles, around 25% of the cases) and the lobar form (complete separation with continuity across the cortex).2, 3 Another mild form of HPE, called middle inter-hemispheric variant is characterized by failure of separation of the posterior frontal and parietal lobes.4 In many but not all cases the cerebral defects or classifications correlate very well with the facial appearance, better known as ‘the face predicts the brain'.5 These facial appearances range from severe anomalies such as cyclopia, anophthalmia and proboscis to mild such as midline cleft lip/palate, hypotelorism, flat nose, microcephaly and single maxillary central incisor. In addition, craniofacial malformations, neurological features, endocrine disorders, oromotor dysfunction and dysautonomic dysfunction can be observed.1
The etiology of HPE is very heterogeneous and both environmental as well as genetic causes are known to be involved. Environmental factors, including maternal insulin-dependent diabetes mellitus, alcoholism and exposure to drugs or infections are the primary cause for only a very small percentage of patients.1 Approximately 25% of patients comprise monogenic syndromes in which HPE is one of the malformations and about 40% of HPE patients harbor a chromosomal aberration.6 Non-syndromic HPE in the remaining 35% of patients is caused by single-gene defects with reduced penetrance. Several genes have been identified to cause HPE including SHH,7 SIX3,8 ZIC2,9 TGIF,10 GLI2,11 PTCH112 and DISP1.13 In this study, we screened the four major genes known to be involved in HPE namely SHH, ZIC2, SIX3 and TGIF by sequence and MLPA analysis in a Dutch cohort of 86 sporadic and familial non-syndromic HPE cases with a variable clinical phenotype. When a large deletion was detected, an additional SNP-array was performed to determine the exact position and size of the deleted region.
Materials and methods
Patient population
Between September 2001 and December 2008 a total of 86 index patients with a normal karyotype were referred to us for holoprosencephaly (HPE) genetic testing. These patients included 18 fetuses, 18 neonatal deceased children, 42 children and 8 adults. The department of Clinical Genetics Maastricht is the only center in the Netherlands performing genetic testing for HPE. Referral for HPE screening was based on the presence of clinical features belonging to the HPE spectrum (see Table 1). If the index patient carried a mutation, parents and family members were tested (54 in total). Informed consent was obtained from all patients (or parents) and family members, in accordance with guidelines of the local medical ethical committee. All families with a mutation were contacted and informed consent for publication of data and photos was obtained.
Table 1. Mutations detected in the Dutch population.
| Nr | Gene | Fam nr. | Mutation | Amino-acid change | Mode of inheritance | Age at testing (Sex) | Picture (Figure 2) |
|---|---|---|---|---|---|---|---|
| 1 | SHH | 10304 | c.148_166del19 | p.N50X | Paternal | Fetus (M) | A/B |
| 2 | 10959 | c.304 T>C | p.C102R | De novo | Fetus (F) | ||
| 3 | 14317 | c.1145 T>C | p.L382P | Paternal | 46 (F) | ||
| 4 | ZIC2 | 8774 | c.479delC | p.P159RfsX58 | Germline | 8 (M) | |
| 5 | 9004 | c.1090 C>T | p.Q364X | De novo | 0 (F) | C | |
| 6 | 14914 | c.1325delC | p.S442SfsX112 | De novo | Fetus (F) | ||
| 7 | 7430 | c.1377_1406dup30 | p.A461_470dup | Maternal | 1 (M) | D/E/F | |
| 8 | 12940 | c.1377_1406dup30 | p.A461_470dup | De novo | Fetus (M) | G | |
| 9 | 14475 | c.1377_1406dup30 | p.A461_470dup | De novo | fetus (F) | H | |
| 10 | 14919 | c.1448 G>A | p.G483D | Maternal | Fetus (F) | ||
| 11 | 11643 | c.1508_1520del13 | p.G503AfsX48 | De novo | 1 (M) | I | |
| 12 | 9723 | Gene deletion | — | Unknown | 62 (F) | J | |
| 13 | SIX3 | 12248 | c.235 A>G | p.M79V | De novo | Fetus (F) | |
| 14 | 9823 | c.404_407dupGCGC | p.V137RfsX18 | De novo | 1 (F) | K | |
| 15 | 10482 | c.463_465delCAC | p.H155del | Maternal | 0† (F) | ||
| 16 | 8616 | c.736delA | p.T245fsX5 | De novo | 0† (F) | ||
| 17 | 8449 | c.769 C>G | p.R257G | Paternal | 16 (M) | ||
| 18 | 7583 | c.806 G>T | p.R269M | Maternal | 21 (M) | L/M | |
| 19 | 10536 | c.806 G>C | p.R269T | Paternal | 60 (M) | N/O | |
| 20 | 5726 | c.806 +1 G>T | Frameshift | De novo/germline | 2† (M) | ||
| 21 | 12519 | Gene deletion | — | De novo | Fetus (M) | P |
Abbreviations: M, male; F, female. †, deceased
PCR and sequence analysis
Blood samples (in duplo), cultured fibroblasts or amniotic fluid cells were used for DNA extraction following standard procedures. In addition, blood samples from both parents and additional family members were obtained when possible. Genetic analysis for the complete coding regions of genes SHH (HPE3, MIM no. 600725), SIX3 (HPE2, MIM no. 603714), ZIC2 (HPE5, MIM no. 603073) and TGIF (HPE4, MIM no. 602630) was performed by direct sequence analysis in both directions using the ABI Big Dye Terminator Cycle Sequencing Ready Reaction kit and the ABI3730 Genetic Analyzer (Applied Biosystems, Foster City, CA, USA).
Primer sets, as designed by Applied Biosystems (AB) for VariantSEQr SNP discovery or custom designed primers following the same standards were used. Full details of primer sequences, amplicon lengths and PCR conditions are presented in Supplementary Table 1. Sequences were aligned with known coding sequences (Genbank cDNA accession numbers: SHH; NM_000193.2, SIX3; NM_005413.2, ZIC2; NM_007129.2, TGIF; NM_003244.2) and mutations were confirmed on DNA extracted from the second blood sample if available. The frequency of novel mutations was determined in a control population of >100 healthy controls. Non-paternity was excluded in case of de novo mutations by multiplex VNTR analysis using the AmpFST Identifiler PCR Amplification kit (Applied Biosystems).
Multiplex ligation-dependent probe amplification (MLPA) analysis
A specific MLPA kit (SALSA MLPA kit P187, MRC-Holland bv, Amsterdam, The Netherlands) was developed to detect deletions/duplications within the coding regions of known HPE genes. This kit contains probes for all exons of genes SHH, SIX3, ZIC2 and TGIF. Details of the probes used for the MLPA procedure as well as standard protocols can be found at website http://www.mlpa.com. In case a deletion/duplication was detected with MLPA, they were confirmed with either FISH using a region/gene-specific probe and/or SNP array.
Affymetrix 250K Nsp I SNP array
The 250K Nsp1 SNP genotyping array analysis (Affymetrix Inc., Santa Clara, CA, USA) was performed according to the manufacturer's array-specific protocol. Briefly, 250 ng of DNA was digested with Nsp1, adaptors were ligated followed by PCR amplification. Subsequently the PCR product was fragmented, labeled and hybridized to the 250K Nsp1 SNP array. Genotype calls were obtained and analyzed using the Affymetric GTYPE analysis tool embedded in the GeneChip Operating Software (GCOS). For copy number estimation data were processed using the CNAT genotyping console (Affymetrix). The patients mapping data were analyzed against 50 reference data sets obtained on the same platform from unaffected individuals.
Results
In total, we detected 21 mutations in 86 patients (24%) (Table 1, Supplementary Table 2). The mutations included 2 gene deletions, 2 nonsense mutations, 5 frame shift mutations, 1 in-frame deletion, 1 repeat-expansion mutation (3 × ), 1 splice-site mutation and 7 missense mutations. Pathogenicity of mutations was based on several features including the de novo character of mutations, segregation with clinical features of HPE, exclusion of being a polymorphism (absence in control/HPE population), conservation and location of mutation. A overview of all detected mutations with genetic and clinical features is depicted in Tables 1 and 2. A summary of all currently known mutations in the four genes, including this study, is given in Supplementary Table 2. Some of our detected mutations were included in recent global studies to study number, type and location of mutations. No other information such as sex, clinical characteristics, inheritance, familial history, etc was included in these studies.14, 15, 16
Table 2. Typical HPE characteristics of index patients.
| Nr | Gene | Fam nr. | Mutation | Type of HPE | Microcephaly | Cleft lip/palate | Hypotelorism | Single nostril | Flat nose/face | Mental retardation |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | SHH | 10304 | p.N50X | (Semi)lobar | X | X | X | X | ||
| 2 | 10959 | p.C102R | Alobar | X | X | X | ||||
| 3 | 14317 | p.L382P | None | |||||||
| 4 | ZIC2 | 8774 | p.P159RfsX58 | Lobar | X | |||||
| 5 | 9004 | p.Q364X | Semilobar | X | X | |||||
| 6 | 14914 | p.S442SfsX112 | Lobar | |||||||
| 7 | 7430 | p.A461_470dup | Semilobar | X | X | |||||
| 8 | 12940 | p.A461_470dup | Semilobar | |||||||
| 9 | 14475 | p.A461_470dup | Semilobar | |||||||
| 10 | 14919 | p.G483D | Alobar | |||||||
| 11 | 11643 | p.G503AfsX48 | Semilobar | X | X | |||||
| 12 | 9723 | gene deletion | Semilobar | X | ||||||
| 13 | SIX3 | 12248 | p.M79V | Alobar | X | X | X | |||
| 14 | 9823 | p.V137RfsX18 | Semilobar | |||||||
| 15 | 10482 | p.H155del | n.s. | X | X | |||||
| 16 | 8616 | p.T245fsX5 | Alobar | X | X | X | X | |||
| 17 | 8449 | p.R257G | Lobar | X | X | |||||
| 18 | 7583 | p.R269M | Semilobar | X | X | |||||
| 19 | 10536 | p.R269T | None. obligate carrier (son) | (X) | (X) | (X) | (X) | (X) | ||
| 20 | 5726 | c.806 +1 G>T (fs) | Semilobar | X | X | |||||
| 21 | 12519 | gene deletion | Semilobar |
NS, not specified, in brackets () clinical characteristics of son of asymptomatic obligate carrier.
The distribution of the 21 mutations over the genes was three in SHH (14%), nine in ZIC2 (43%), nine in SIX3 (43%) and 0 in TGIF (0%). Eleven (52%) mutations occurred de novo and non-paternity was excluded in all of them.
SHH mutations
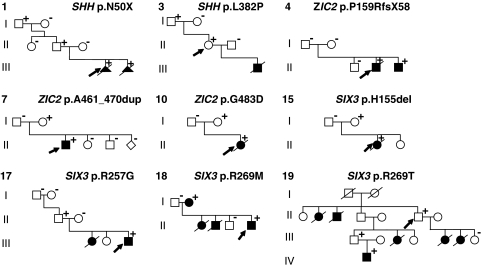
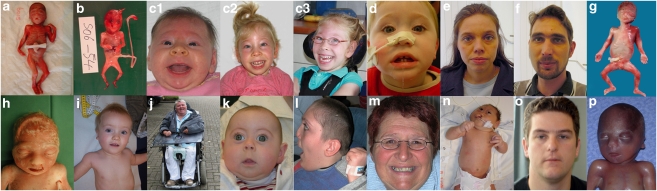
We detected three mutations in the SHH gene. Mutation c.148_166del19 (p.Asn50X) was detected in an aborted male fetus (21+4 weeks) with (semi)lobar HPE, median cleft lip/palate, hypotelorism, an underdeveloped nose and left isomerism of the lungs. The father showed microcephaly, hypoplastic alae nasi, thin upper lip and long uvula and carried the mutation as well as the paternal grandfather. Also a second pregnancy was terminated (14+3 weeks) due to HPE signs detected with ultrasound (cleft lip/palate, hypotelorism, underdeveloped nose) and detection of the mutation in amniotic cells (pedigree 1/Figure 1, picture A and B/Figure 2). This couple underwent pre-implantation genetic diagnosis (PGD) and delivered a healthy son in August 2008. Mutation c.304 T>C (p.Cys102Arg) was detected in an aborted female fetus (23 weeks) with alobar HPE and arose de novo. The autopsy report listed facial dysmorphism including hypotelorism and a flat nose. Mutation c.1145 T>C (p.Leu382Pro) was detected in a 46-year-old female showing no HPE clinical characteristics. She was tested because her first child died of HPE at the age of 6 months. This child was diagnosed with alobar HPE, cleft palate and epilepsy but no DNA was available for testing. The mother's asymptomatic father also carried the mutation (pedigree 3/Figure 1).
Figure 1.
Pedigrees of familial mutations. Family pedigrees according to numbering of Tables 1 and 2 are indicated at the top left corner of each pedigree. Specific mutation for the family is indicated in the right upper corner. +indicates mutation carriers; –indicates non-mutation carriers; squares indicates males, circles females; filled symbols are patients with HPE clinical characteristics; arrows indicate family index.
Figure 2.
Pictures of patients. Pictures of HPE patients or their relatives. Letters below the pictures are referred in the text as well as in Table 1. All photographs are published with informed consent of index patients or their parents.
ZIC2 mutations
We detected nine mutations in the ZIC2 gene. Mutation c.479delC (p.Pro159ArgfsX58) was detected in an 8-year-old boy with lobar HPE, deafness, mental retardation and spastic diplegia. Both parents were healthy and non-carriers but one older brother died of alobar HPE 3 weeks after birth. Cultured fibroblasts of this deceased older brother showed that he was a carrier of the same mutation (pedigree 4/Figure 1) suggesting germ line mosaicism. The mutation was not detected in semen of the father. Marker analysis indicated that the risk haplotype, shared by the two affected brothers and not by a healthy brother, was inherited from the mother (data not shown). Mutation c.1090 C>T (p.Gln364X) occurred de novo and was detected in a female first child with semilobar HPE including microcephaly, hypotelorism and a very slow general development (picture C1-C3/Figure 2). Mutation c.1325delC (p.S442SfsX112) occurred de novo and was detected in an aborted female fetus with lobar HPE and no further facial anomalies. The previously described alanine tract expansion mutation c.1377_1406dup30 (p.A461_470dup) was detected in three unrelated families: In the first family (7430) the mutation was detected in a male child with semilobar HPE, microcephaly, mental retardation, diabetes insipidus and tetraparesis. Now, the boy does not speak and does not walk at age 8 years. The asymptomatic mother with a normal brain MRI carried the expansion (pedigree 7/Figure 1, picture D, E, F/Figure 2). In the second family (12940) the alanine tract occurred de novo and was detected in an aborted male fetus (23+4 weeks) with semilobar HPE (picture G/Figure 2) and no apparent facial dysmorphism except for a broad mouth. In the third family (14475) the expansion also occurred de novo and was detected in an aborted female fetus (30+6 weeks) with semilobar HPE without further external malformations (picture H/Figure 2). Mutation c.1448 G>A (p.G483D) was found in an aborted female fetus (14 weeks) with alobar HPE in combination with an encephalocele (pedigree 10/Figure 1). The mother also carried the mutation but further clinical examination of the mother showed no microsigns of HPE. She refused an MRI or further familial genetic screening. Mutation c.1508_1520del13 (p.G503AfsX48) occurred de novo in a 1-year-old boy with semilobar HPE (picture I/Figure 2) hypotelorism and a very slow mental development. The ZIC2 deletion detected with MLPA analysis was found in an elderly female with mental retardation (picture J, Figure 2). The deletion was confirmed with FISH analysis and the 250k nspI Affymetrix SNP array. The deletion was too small to be detected by the default algorithms but with the exact location known could be determined at approximately 100 kb in size including the ZIC2 and ZIC5 genes. DNA of parents was not available.
SIX3 mutations
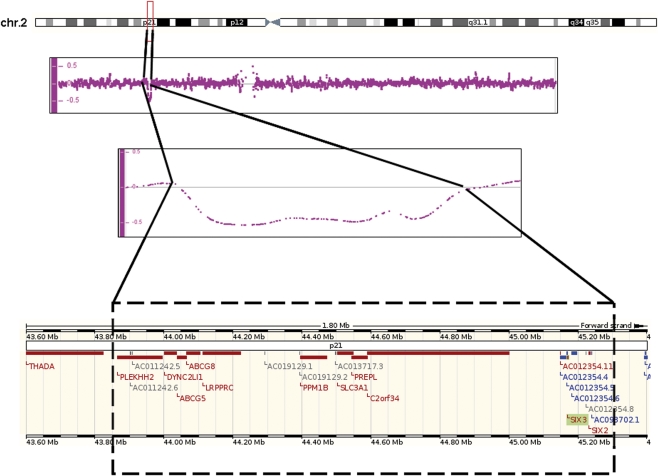
We detected nine mutations in the SIX3 gene. Mutation c.235 A>G (p.M79V) occurred de novo and was detected in an aborted female fetus (18+3 weeks) with alobar HPE and microcephaly, a single nostril, flat face and a malformed upper lip without cleft. De novo mutation c.404_407dupGCGC (p.V137RfsX18) was detected in a 10-month-old girl with semilobar HPE (partial hypoplasia of the corpus callosum)(picture K/Figure 2). Mutation c.463_465delCAC (p.H155del) was found in a baby girl who died shortly after birth due to HPE, bilateral cleft lip and palate and a single nostril. The mutation was also detected in the asymptomatic mother (pedigree 15/Figure 1). Mutation c.736delA (p.T245fsX5) occurred de novo in a baby girl that died 12 days after birth because of alobar HPE with a single nostril, flat nose, median cleft lip/palate and hypotelorism. Mutation c.769 C>G (p.R257G) was detected in a 16-year-old boy with lobar HPE, microcephaly and mental retardation. A younger sister died the second day after birth. The autopsy report on the sister did not indicate specific HPE characteristics as the forebrain was completely divided and the corpus callosum was present. There were, however, brain abnormalities including brain atrophy, absent nervi olphactorii and large ventricles. DNA of the sister was not available. The asymptomatic father carried the mutation, but his parents did not (pedigree 17/Figure 1). Mutation c.806 G>T (p.R269M) was detected in a 21-year-old man (picture L/Figure 2, photo taken at teenage years) with semilobar HPE cleft lip/palate, diabetes insipidus, epileptic fits and severe mental retardation (pedigree 18/Figure 1). Clinical diagnosis of HPE was made at age 4 years.17 Two younger children of this family died shortly after birth because of HPE but their DNA was not available.17 The mother has a solitary median maxillary incisor (SMMCI) and carries the mutation (picture M/Figure 2). Mutation c.806 G>C (p.R269T) was detected in an asymptomatic 60-year-old male who was screened because of multiple HPE-related deaths in his family (II-5, pedigree 19/Figure 1). A sister (II-2, bilateral cleft lip/palate, microcephaly), brother (II-3, cervical rachischisis, missing vertebrae), a niece (III-2, HPE, midface hypoplasia, high palatum), a nephew (IV-1, HPE, cleft lip/palate, absent nose, hypotelorism, picture N/Figure 2) and two daughters (III-4, III-5, HPE, bilateral cleft lip/palate, hydrocephalus) showed characteristics within the HPE spectrum. DNA was not available for testing. The nephew (IV-1, picture N/Figure 2) and asymptomatic father (III-1, picture O/Figure 2) of the index both carried the mutation (pedigree 19/Figure 1). Mutation c.806+1 G>T occurred de novo in an 8-month-old boy with semilobar HPE, microcephaly, epilepsy, psychomotor retardation and diabetes insipidus who died at age 2 years. The SIX3 gene deletion detected with MLPA occurred de novo in an aborted male fetus (23+4 weeks) with semilobar HPE and no facial dysmorphism (picture P/Figure 2). The deletion was confirmed with FISH analysis and the nspI 250k Affymetrix SNP array. The deletion was approximately 1.45 Mb in size (Nucleotide positions 43845329 to 45152135, 194 SNP's, hg. 19/NCBI Build 37), including the SIX3 and SIX2 genes at the centromeric end of the deletion (Figure 3).
Figure 3.
Characterization of SIX3 gene deletion. Family 12 519 with the SIX2/3 deletion originally detected with MLPA and confirmed with by 250K Affymetrix SNP array (194 SNP's =>1.45 Mb). The deleted area is highlighted and zoomed in with the SIX2/3 genes at the centromeric end of the deletion.
Discussion
Screening the four major HPE genes (SHH, SIX3, ZIC2 and TGIF) in 86 Dutch index patients and 54 family members led to the detection of 21 heterozygous mutations (24.4%). This mutation rate is comparable with previous reports for non-syndromic HPE.18 Only the alanine tract expansion in the ZIC2 gene and the c.404_407dupGCGC mutation in the SIX3 gene have been identified previously.9, 19, 20 Of the 31 family members tested for a familial mutation, nine were carriers of whom four showed major or minor HPE signs. Therefore around 55% of mutation carriers were completely asymptomatic.
We detected only 3 of 86 mutations in SHH (3.5%), which is significantly lower than the previously reported 121 of 1100 (10.7%) by Roessler et al.15 (P=0.047, χ2=3.94). Two of the mutations were familial and showed extremely wide phenotypic variability within the family as previously described.18 The p.N50X mutation truncates the SHH protein in the SHH-N domain, thereby eliminating the Hint motif necessary for autocatalytic processing and is considered a biologically null allele.15 Missense mutation p.C102R is also located in the important SHH-N region of the protein and is very highly conserved in other species up to c. elegans. Combined with the de novo character of this mutation, pathogenicity of this mutation is a likely explanation for the alobar HPE. Missense mutation p.L382P is located in the SRR region, in which several HPE mutations have been detected and which functions to bind cholesterol.15
We detected 9 of 86 mutations in ZIC2 (10.5%), a percentage comparable with a recent summarizing study from Roessler et al.16 Consistent with previous studies9, 16, 21 we found: (1) a high frequency (>55%) of frame shifts, nonsense mutations and gene deletions (Table 1), (2) >50% de novo mutations, (3) several frame shift mutations in the C-terminal end of the ZIC2 protein, leaving the zinc-finger domains theoretically intact, (4) the poly-alanine tract expansion as a prevalent mutation type. Inconsistencies with previous studies were also found: in two of three patients the alanine tract expansion was de novo, in the third patient maternally inherited, therefore not predominantly from the father as previously described. In addition, the mother was not a mosaic carrier of the mutation.19 The one missense mutation we detected, p.G483D, is located in the C-terminal end of the ZIC2 protein where so far only frame shifts and ins/del mutations have been detected. This mutation does not occur in a conserved region and the glycine amino-acid is not well conserved among species. However, as the mutation was not observed in a panel of 110 healthy controls and was never described in literature (>1000 HPE index patients) it is very unlikely to be a common polymorphism. The aborted fetus showed alobar HPE without facial anomalies and the asymptomatic mother also carried the mutation. As minor/absent facial dysmorphism combined with severe brain malformations and incomplete clinical penetrance are specifically consistent with ZIC2 mutations, we consider this variant most likely pathogenic.
Considering all 21 index patients with a mutation (Tables 1 and 2) shows that facial malformations in the ZIC2 patients are less apparent than in patients with mutations in the other genes. This milder facial phenotype with usually no clefts or single nostrils as seen in genes as SIX3 and SHH is consistent with literature.1, 19, 22 The specific facial features (bitemporal narrowing, upslanting palpebral fissures, flat nasal bridge, short nose, broad philtrum and large ears) as described recently by Solomon et al.22 could in part be fitted to our patients; specifically patient C, D and I fit several of these features. Distribution of HPE classifications (lobar, semilobar, alobar) were, however, mixed and not different from patients with other mutated genes. We detected 9 of 86 mutations in SIX3 (10.5%) a percentage significantly higher than published by Lacbawan et al.14 who detected 34 of 800 (4.25%) mutations in SIX3 (P=0.018, χ2=5.63). The more prevalent maternal inheritance and a 1.5:1 female/male ratio described in this study could not be confirmed in our population (Table 1), probably due to the smaller sample size. Our population, however, does seem to support the more severe phenotype in SIX3 index patients (Table 2)14, 23 and a high penetrance of clinical symptoms within familial mutation carriers. In a recent study Domené et al.20 developed a set of robust and sensitive assays in zebra fish to functionally test SIX3 mutations and characterized 46 mutations. The already known SIX3 mutation c.404_407dupGCGC (Table 1) was also characterized in this study and yielded <50% of activity in the overexpression assay (c.q. ability to induce dorsalized phenotypes), but retained activity in the rescue assay (c.q. ability to rescue the headless phenotype).20 Interestingly they characterized two different mutants in codon 257, namely c.769 C>T (p.R257W) and c.770 G>C (p.R257P) leading to different amino acid substitutions and completely opposite functional properties. As we detected yet another mutation in this codon c.769 C>G (p.R257G), and this codon therefore appears to be prone to mutation, it would be interesting to see what phenotype this mutation gives in the assay. A second nearby codon susceptible to mutation is amino acid p.R269. Domené et al.20 characterized c.807 G>C (p.R269S), which retained intermediate activity (50–90%). We detected two other mutations c.806 G>T (p.R269M) and c.806 G>C (p.R269T) in the same codon and a splice mutation c.806+1 G>T. These mutations would also be very interesting in functional assays and would maybe explain why these two arginine positions at the end of Homeo-domain and C-terminal motifs are so apparently prone to mutate.
We did not detect any mutation in the TGIF gene. This is not statistically different from other studies and our population may have been simply too small as the percentage of TGIF mutations is very low (1–2%). The added value of a technique to determine duplications or deletions such as MLPA for the four major HPE genes is evident as we detected two gene deletions, representing 12% of the total number of mutations detected. Earlier, Bendavid et al.24 already described several microdeletions in other genes (SHH and TGIF) using the subtelomeric MLPA kit as well as precisely sized duplications and deletions at HPE loci using array-CGH.25 It is therefore clear that complete gene deletions of all major HPE genes are possible and should be included in diagnostic testing.
Our future research will focus on increasing the mutation rate (currently 24%), as not all currently known HPE genes are as yet routinely tested and a large portion of the non-syndromic patients with a normal karyotype remain unexplained. We aim at including GLI2 (MIM no. 165230, HPE9), PTCH1 (MIM no. 601309, HPE7) and TDGF1 (MIM no. 187395) in the near future. Furthermore, recent studies24, 25 have indicated more candidate loci for HPE, which may also increase the current mutation rate. Obviously, functional in vitro/in vivo studies that may elucidate biological and molecular pathways and/or discriminate between the functional effects of unique mutations are imperative in further unraveling the HPE etiology. A major future challenge will be the search for additional genetic or environmental risk factors that modulate or even evoke the phenotypic variability within families.
Acknowledgments
We thank the patients and their families for participating in this study and all clinicians who referred cases.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies the paper on European Journal of Human Genetics website (http://www.nature.com/ejhg)
Supplementary Material
References
- Dubourg C, Bendavid C, Pasquier L, Henry C, Odent S, David V. Holoprosencephaly. Orphanet J Rare Dis. 2007;2:8. doi: 10.1186/1750-1172-2-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen MM., Jr Holoprosencephaly: clinical, anatomic, and molecular dimensions. Birth Defects Res A Clin Mol Teratol. 2006;76:658–673. doi: 10.1002/bdra.20295. [DOI] [PubMed] [Google Scholar]
- Demyer W, Zeman W. Alobar holoprosencephaly (arhinencephaly) with median cleft lip and palate: clinical, electroencephalographic and nosologic considerations. Confin Neurol. 1963;23:1–36. doi: 10.1159/000104278. [DOI] [PubMed] [Google Scholar]
- Simon EM, Hevner RF, Pinter JD, et al. The middle interhemispheric variant of holoprosencephaly. AJNR Am J Neuroradiol. 2002;23:151–156. [PMC free article] [PubMed] [Google Scholar]
- Demyer W, Zeman W, Palmer CG. The face predicts the brain: diagnostic significance of median facial anomalies for holoprosencephaly (arhinencephaly) Pediatrics. 1964;34:256–263. [PubMed] [Google Scholar]
- Hahn JS, Plawner LL. Evaluation and management of children with holoprosencephaly. Pediatr Neurol. 2004;31:79–88. doi: 10.1016/j.pediatrneurol.2004.03.006. [DOI] [PubMed] [Google Scholar]
- Roessler E, Belloni E, Gaudenz K, et al. Mutations in the human Sonic Hedgehog gene cause holoprosencephaly. Nat Genet. 1996;14:357–360. doi: 10.1038/ng1196-357. [DOI] [PubMed] [Google Scholar]
- Wallis DE, Roessler E, Hehr U, et al. Mutations in the homeodomain of the human SIX3 gene cause holoprosencephaly. Nat Genet. 1999;22:196–198. doi: 10.1038/9718. [DOI] [PubMed] [Google Scholar]
- Brown SA, Warburton D, Brown LY, et al. Holoprosencephaly due to mutations in ZIC2, a homologue of Drosophila odd-paired. Nat Genet. 1998;20:180–183. doi: 10.1038/2484. [DOI] [PubMed] [Google Scholar]
- Gripp KW, Wotton D, Edwards MC, et al. Mutations in TGIF cause holoprosencephaly and link NODAL signalling to human neural axis determination. Nat Genet. 2000;25:205–208. doi: 10.1038/76074. [DOI] [PubMed] [Google Scholar]
- Roessler E, Du YZ, Mullor JL, et al. Loss-of-function mutations in the human GLI2 gene are associated with pituitary anomalies and holoprosencephaly-like features. Proc Natl Acad Sci USA. 2003;100:13424–13429. doi: 10.1073/pnas.2235734100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ming JE, Kaupas ME, Roessler E, et al. Mutations in PATCHED-1, the receptor for SONIC HEDGEHOG, are associated with holoprosencephaly. Hum Genet. 2002;110:297–301. doi: 10.1007/s00439-002-0695-5. [DOI] [PubMed] [Google Scholar]
- Roessler E, Ma Y, Ouspenskaia MV, et al. Truncating loss-of-function mutations of DISP1 contribute to holoprosencephaly-like microform features in humans. Hum Genet. 2009;125:393–400. doi: 10.1007/s00439-009-0628-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacbawan F, Solomon BD, Roessler E, et al. Clinical spectrum of SIX3-associated mutations in holoprosencephaly: correlation between genotype, phenotype and function. J Med Genet. 2009;46:389–398. doi: 10.1136/jmg.2008.063818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roessler E, El-Jaick KB, Dubourg C, et al. The mutational spectrum of holoprosencephaly-associated changes within the SHH gene in humans predicts loss-of-function through either key structural alterations of the ligand or its altered synthesis. Hum Mutat. 2009;30:E921–E935. doi: 10.1002/humu.21090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roessler E, Lacbawan F, Dubourg C, et al. The full spectrum of holoprosencephaly-associated mutations within the ZIC2 gene in humans predicts loss-of-function as the predominant disease mechanism. Hum Mutat. 2009;30:E541–E554. doi: 10.1002/humu.20982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwetsloot CP, Brouwer OF, Maaswinkel-Mooy PD. Holoprosencephaly: variation of expression in face and brain in three sibs. J Med Genet. 1989;26:274–276. doi: 10.1136/jmg.26.4.274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazaro L, Dubourg C, Pasquier L, et al. Phenotypic and molecular variability of the holoprosencephalic spectrum. Am J Med Genet A. 2004;129A:21–24. doi: 10.1002/ajmg.a.30110. [DOI] [PubMed] [Google Scholar]
- Brown LY, Odent S, David V, et al. Holoprosencephaly due to mutations in ZIC2: alanine tract expansion mutations may be caused by parental somatic recombination. Hum Mol Genet. 2001;10:791–796. doi: 10.1093/hmg/10.8.791. [DOI] [PubMed] [Google Scholar]
- Domene S, Roessler E, El-Jaick KB, et al. Mutations in the human SIX3 gene in holoprosencephaly are loss of function. Hum Mol Genet. 2008;17:3919–3928. doi: 10.1093/hmg/ddn294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubourg C, Lazaro L, Pasquier L, et al. Molecular screening of SHH, ZIC2, SIX3, and TGIF genes in patients with features of holoprosencephaly spectrum: Mutation review and genotype-phenotype correlations. Hum Mutat. 2004;24:43–51. doi: 10.1002/humu.20056. [DOI] [PubMed] [Google Scholar]
- Solomon BD, Lacbawan F, Mercier S, et al. Mutations in ZIC2 in human holoprosencephaly: description of a novel ZIC2-specific phenotype and comprehensive analysis of 157 individuals J Med Genet 2009. e-pub ahead of print. [DOI] [PMC free article] [PubMed]
- Ribeiro LA, El-Jaick KB, Muenke M, Richieri-Costa A. SIX3 mutations with holoprosencephaly. Am J Med Genet A. 2006;140:2577–2583. doi: 10.1002/ajmg.a.31377. [DOI] [PubMed] [Google Scholar]
- Bendavid C, Dubourg C, Pasquier L, et al. MLPA screening reveals novel subtelomeric rearrangements in holoprosencephaly. Hum Mutat. 2007;28:1189–1197. doi: 10.1002/humu.20594. [DOI] [PubMed] [Google Scholar]
- Bendavid C, Rochard L, Dubourg C, et al. Array-CGH analysis indicates a high prevalence of genomic rearrangements in holoprosencephaly: an updated map of candidate loci. Hum Mutat. 2009;30:1175–1182. doi: 10.1002/humu.21016. [DOI] [PubMed] [Google Scholar]
- Nanni L, Ming JE, Bocian M, et al. The mutational spectrum of the sonic hedgehog gene in holoprosencephaly: SHH mutations cause a significant proportion of autosomal dominant holoprosencephaly. Hum Mol Genet. 1999;8:2479–2488. doi: 10.1093/hmg/8.13.2479. [DOI] [PubMed] [Google Scholar]
- Kato M, Nanba E, Akaboshi S, et al. Sonic hedgehog signal peptide mutation in a patient with holoprosencephaly. Ann Neurol. 2000;47:514–516. [PubMed] [Google Scholar]
- Goetz JA, Singh S, Suber LM, Kull FJ, Robbins DJ. A highly conserved amino-terminal region of sonic hedgehog is required for the formation of its freely diffusible multimeric form. J Biol Chem. 2006;281:4087–4093. doi: 10.1074/jbc.M511427200. [DOI] [PubMed] [Google Scholar]
- Hehr U, Gross C, Diebold U, et al. Wide phenotypic variability in families with holoprosencephaly and a sonic hedgehog mutation. Eur J Pediatr. 2004;163:347–352. doi: 10.1007/s00431-004-1459-0. [DOI] [PubMed] [Google Scholar]
- Muenke M, Gurrieri F, Bay C, et al. Linkage of a human brain malformation, familial holoprosencephaly, to chromosome 7 and evidence for genetic heterogeneity. Proc Natl Acad Sci USA. 1994;91:8102–8106. doi: 10.1073/pnas.91.17.8102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odent S, Atti-Bitach T, Blayau M, et al. Expression of the Sonic hedgehog (SHH ) gene during early human development and phenotypic expression of new mutations causing holoprosencephaly. Hum Mol Genet. 1999;8:1683–1689. doi: 10.1093/hmg/8.9.1683. [DOI] [PubMed] [Google Scholar]
- Bertolacini CD, Richieri-Costa A, Ribeiro-Bicudo LA.Sonic hedgehog (SHH) mutation in patients within the spectrum of holoprosencephaly Brain Dev 2009. e-pub ahead of print 26 April 2009. [DOI] [PubMed]
- Nanni L, Ming JE, Du Y, et al. SHH mutation is associated with solitary median maxillary central incisor: a study of 13 patients and review of the literature. Am J Med Genet. 2001;102:1–10. doi: 10.1002/1096-8628(20010722)102:1<1::aid-ajmg1336>3.0.co;2-u. [DOI] [PubMed] [Google Scholar]
- El-Jaick KB, Brunoni D, Castilla EE, Moreira MA, Orioli IM. SHH Ile111Asp in alobar holoprosencephaly in a proposita, whose mother had only a solitary median maxillary incisor. Am J Med Genet A. 2005;136:345. doi: 10.1002/ajmg.a.30624. [DOI] [PubMed] [Google Scholar]
- Marini M, Cusano R, De Biasio P, et al. Previously undescribed nonsense mutation in SHH caused autosomal dominant holoprosencephaly with wide intrafamilial variability. Am J Med Genet A. 2003;117:112–115. doi: 10.1002/ajmg.a.10163. [DOI] [PubMed] [Google Scholar]
- Orioli IM, Castilla EE, Ming JE, et al. Identification of novel mutations in SHH and ZIC2 in a South American (ECLAMC) population with holoprosencephaly. Hum Genet. 2001;109:1–6. doi: 10.1007/s004390100537. [DOI] [PubMed] [Google Scholar]
- Santiago G, Abramides DV, De-Vitto LP, Ribeiro LA, Meira SG., Jr Richieri-Costa A: Language skills and neuropsychological performance in patients with SHH mutations and a holoprosencephaly-like phenotype. Am J Med Genet A. 2006;140:2085–2090. doi: 10.1002/ajmg.a.31311. [DOI] [PubMed] [Google Scholar]
- Richieri-Costa A, Ribeiro LA. Holoprosencephaly-like phenotype: clinical and genetic perspectives. Am J Med Genet A. 2006;140:2587–2593. doi: 10.1002/ajmg.a.31378. [DOI] [PubMed] [Google Scholar]
- Roessler E, Belloni E, Gaudenz K, et al. Mutations in the C-terminal domain of Sonic Hedgehog cause holoprosencephaly. Hum Mol Genet. 1997;6:1847–1853. doi: 10.1093/hmg/6.11.1847. [DOI] [PubMed] [Google Scholar]
- Schimmenti LA, de la Cruz J, Lewis RA, et al. Novel mutation in sonic hedgehog in non-syndromic colobomatous microphthalmia. Am J Med Genet A. 2003;116A:215–221. doi: 10.1002/ajmg.a.10884. [DOI] [PubMed] [Google Scholar]
- Pasquier L, Dubourg C, Gonzales M, et al. First occurrence of aprosencephaly/atelencephaly and holoprosencephaly in a family with a SIX3 gene mutation and phenotype/genotype correlation in our series of SIX3 mutations. J Med Genet. 2005;42:e4. doi: 10.1136/jmg.2004.023416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasquier L, Dubourg C, Blayau M, et al. A new mutation in the six-domain of SIX3 gene causes holoprosencephaly. Eur J Hum Genet. 2000;8:797–800. doi: 10.1038/sj.ejhg.5200540. [DOI] [PubMed] [Google Scholar]
- El-Jaick KB, Fonseca RF, Moreira MA, et al. Single median maxillary central incisor: New data and mutation review. Birth Defects Res A Clin Mol Teratol. 2007;79:573–580. doi: 10.1002/bdra.20380. [DOI] [PubMed] [Google Scholar]
- Brown L, Paraso M, Arkell R, Brown S. In vitro analysis of partial loss-of-function ZIC2 mutations in holoprosencephaly: alanine tract expansion modulates DNA binding and transactivation. Hum Mol Genet. 2005;14:411–420. doi: 10.1093/hmg/ddi037. [DOI] [PubMed] [Google Scholar]
- Richieri-Costa A, Ribeiro LA. Variable phenotypic manifestations of a K44N mutation in the TGIF gene. Brain Dev. 2008;30:203–205. doi: 10.1016/j.braindev.2007.07.012. [DOI] [PubMed] [Google Scholar]
- El-Jaick KB, Powers SE, Bartholin L, et al. Functional analysis of mutations in TGIF associated with holoprosencephaly. Mol Genet Metab. 2007;90:97–111. doi: 10.1016/j.ymgme.2006.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CP, Chern SR, Du SH, Wang W. Molecular diagnosis of a novel heterozygous 268C-->T (R90C) mutation in TGIF gene in a fetus with holoprosencephaly and premaxillary agenesis. Prenat Diagn. 2002;22:5–7. doi: 10.1002/pd.202. [DOI] [PubMed] [Google Scholar]
- Chen M, Kuo SJ, Liu CS, et al. A novel heterozygous missense mutation 377T > C (V126A) of TGIF gene in a family segregated with holoprosencephaly and moyamoya disease. Prenat Diagn. 2006;26:226–230. doi: 10.1002/pd.1385. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.