Abstract
DNA delivery of IL-12 has shown promise in reducing the toxic side effects associated with administration of recombinant human (h)IL-12 protein while maintaining the ability to inhibit tumor growth and abolish tumor metastases in animal models. We have developed a more potent version of IL-12 by using DNA shuffling and screening to improve its expression in human cells and specific activity on human T cells. The most improved evolved IL-12 (EvIL-12) derived from seven mammalian genes encoding both the p35 and p40 subunits of IL-12 showed a 128-fold improvement in human T cell proliferation compared with native hIL-12 during the initial screening of supernatants from transected cells. When purified hIL-12 and EvIL-12 proteins were compared in vitro in human T cell proliferation and Th1 differentiation assays, it was demonstrated that EvIL-12 exhibited a concomitant 10-fold increase in the specific activity of the protein compared with hIL-12. Furthermore, DNA shuffling improved the level of expression and homogeneity of the heterodimer synthesized by 293 human embryonic kidney cells transfected with EvIL-12 by at least 10-fold. Molecular analysis of the variant revealed strategic placement of amino acid substitutions that potentially may facilitate heterodimer formation and product expression. The enhanced expression and biological activity of EvIL-12 may improve the effectiveness of IL-12 gene-based vaccines and therapeutics without the toxic side effects sometimes associated with hIL-12 protein administration.
IL-12 is a heterodimeric cytokine involved in the regulation of both innate and adaptive immunity (1, 2). It has pleiotropic effects on T cells, B cells, and natural killer (NK) cells and is a key regulator of Th1 cell differentiation. IL-12 primes T cells to differentiate into Th1 cells producing high levels of IL-2 and IFN-γ, resulting in improved innate defense mechanisms through enhanced activation of NK and antigen-presenting cells. The adjuvant properties of IL-12 have been demonstrated in a number of infectious disease models, including bacterial (3–5), fungal (6, 7), and parasitic diseases (8, 9). Deficiencies in either IL-12 or IL-12 receptor in humans have been associated with impaired immunity against intracellular bacteria (10–14). Moreover, administration of IL-12 as a component of cancer vaccines enhances the activity of CD8+ cytotoxic T cells, resulting in protection and tumor regression in vivo (15, 16). Thus, IL-12 has a crucial role in the regulation of cellular immune responses to infectious disease and cancer antigens.
The biological activities of IL-12 depend on the formation of a high-affinity ligand-receptor complex through binding of the disulfide-linked p35-p40 heterodimer to receptors, IL-12Rβ1, and IL-12Rβ2 (17). The p35 subunit is a member of the four-helix bundle cytokines, IL-6, and granulocyte–colony-stimulating factor, whereas p40 resembles the soluble extracellular domains of the IL-6, growth hormone, and ciliary neurotrophic factor receptors (18–20). The p35 subunit is also capable of binding to a soluble cytokine receptor, EBI3, but no biological activity has been found for this complex (21). In contrast, the p40 subunit has been shown to form homodimers and is a natural antagonist of IL-12 (22). In addition, the p40 subunit has been shown to associate with a novel protein derived from dendritic cells to form IL-23, which has similar but distinct biological properties to IL-12 (23).
Several clinical trials have been conducted to evaluate the therapeutic potential of IL-12 in humans (24, 25). Dose-limiting toxicity has been observed in patients treated with soluble recombinant hIL-12, but it has been proposed that gene-based approaches may overcome these limitations (25–27). However, two hurdles for using IL-12 in gene-based applications are its relatively low expression and activity in vivo (28, 29), which depend on the proper formation of the p70 heterodimer. An attractive approach for optimizing expression and activity of IL-12 is directed molecular evolution by DNA shuffling and screening. DNA shuffling followed by screening typically comprises a genetic-combinatorial method that rapidly breeds functional sequence diversity through reiterative mutation and recombination and allows sampling of sequence space not accessible by conventional methods of mutagenesis (30). These technologies can be used for a wide range of applications (31–32) and have been successful, for example, in evolving subtilisin, IFN-α, costimulatory molecules, and viruses (33–36).
In this report, IL-12 was evolved for use as a genetic adjuvant/vaccine after two rounds of DNA shuffling and screening. The most active evolved IL-12 (EvIL-12) produced ≈128-fold improved activity from culture supernatants of transfected COS-7 cells compared with human IL-12 (hIL-12). In vitro analysis comparing affinity purified EvIL-12 and hIL-12 revealed the following significant improvements: (i) enhanced specific activity as measured by T cell proliferation and Th1 differentiation assays, and (ii) improved expression in mammalian cells resulting from the formation of a more homogenous p70 heterodimer attributed to amino acid substitutions that reposition critical regions of both subunits required for heterodimer formation.
Materials and Methods
DNA Shuffling.
DNA sequences encoding IL-12 subunits, p35 and p40, were obtained by RT-PCR (Stratagene) of human, rhesus monkey, cow, pig, goat, dog, and cat total or poly (A+)RNA from peripheral blood mononuclear cells (PBMC) activated with 1 ng/ml phorbol 12-myristate 13-acetate and 500 ng/ml ionomycin (Sigma). Shuffled p35 (sh-p35) and p40 subunit libraries were generated from partial or full-length cDNA clones by using DNA shuffling as described by Crameri et al. (37). The resultant shuffled sequences were ligated into pCDNA3.1+ (Invitrogen) and transformed into Escherichia coli 294.
Transfection.
DNA from individual clones was made by using the Qiaprep Turbo procedure for the Qiagen BioRobot 9600 (Qiagen, Valencia, CA). Single clones from each subunit library were cotransfected in a 96-well format with their corresponding human or shuffled subunit partner into either COS-7 cells for screening or 293 human embryonic kidney (HEK) cells for production using Superfect Reagent (Qiagen) to reconstitute the biologically active p70 heterodimer. In addition, plasmids encoding GFP or hIL-12 were transfected in triplicate as negative and positive controls, respectively.
Isolation of Human PBMC and T Cells.
Human PBMC were isolated from buffy coats by density-gradient centrifugation by using Histopaque (Sigma) (38). The isolated PBMC were cultured in T75 flasks at 2 × 106/ml for 4 days in the presence of 5 μg/ml phytohemagglutinin (PHA) (Sigma). Purified T cells were isolated by negative selection by using immunomagnetic beads (Dynal, Oslo) and mAbs specific for CD14, CD16, CD19, and CD56 (PharMingen). Labeled PBMC were incubated with the beads for 30 min at 4°C with gentle rotation, and positive cells were removed with a magnet. Subsequently, the T cells were washed with PBS containing 2% FBS and resuspended in culture medium (39). The purity of the cells was determined by analyzing the percentage of CD3+ cells by flow cytometry (FACS Calibur and cell quest software, Becton Dickinson) and was consistently found to be >98% (data not shown).
T Cell Proliferation Assay.
T cell proliferation assay was performed as described (40). Briefly, PHA-activated T cells were incubated for 72 h with either crude culture supernatants or purified protein from individual clones, and 3H-thymidine incorporation was measured during the last 16 h. Culture supernatants containing human/shuffled p70 heterodimers initially were screened in duplicate at a 1- to 200-fold dilution with 4 × 104 PHA-activated human T lymphocytes. Culture supernatants exhibiting activity greater than that of hIL-12 were subsequently reassayed by using serial 1:1 dilutions in triplicate.
Th1 Cell Differentiation Assay.
Differentiation of purified T cells into Th1 cells producing IFN-γ was analyzed essentially as described (38). Briefly, purified T cells were cultured in 24-well plates (Costar) at 1 × 106 cells/well for 5 days in medium containing serial dilutions of purified hIL-12 or EvIL-12. The cells were then collected and stimulated with 5 μg/ml of anti-CD3 mAbs and 5 μg/ml anti-CD28 mAbs for 48 h. Finally, the concentration of IFN-γ from the culture supernatants was measured by using an ELISA kit (R & D Systems).
Protein Purification.
The DNA sequence encoding each of the human and sh-p35 subunits was modified at the C terminus with an E-tag (Pharmacia-Amersham Pharmacia) to facilitate purification and quantitation of human and EvIL-12 from culture supernatants of scale-up transfections by using an anti-E-tag affinity column (Pharmacia-Amersham Pharmacia). The purified protein was buffered exchanged into PBS and characterization was determined by OD280 and SDS/PAGE (4–12% NuPAGE gel; NOVEX, San Diego) by using either silver staining (NOVEX) or chemiluminescent detection with either a 1:2,000 goat anti-hIL-12 p35 specific mAb (PharMingen) or a 1:1,500 dilution of mouse anti-His tag mAb (Accurate Scientific, Westbury, NY) followed by a 1:2,000 dilution of goat anti-mouse IgG conjugated to horseradish peroxidase (Pharmacia-Amersham Pharmacia). The isoelectric point of purified proteins was calculated by using vector nti suite software (InforMax, North Bethesda, MD).
Results and Discussion
DNA Shuffling.
DNA shuffling of cDNAs encoding human, rhesus monkey, cow, pig, goat, dog, and cat p35 and p40 subunits was used to construct two separate subunit libraries (Lib1-p35 and Lib1-p40) (Fig. 1). DNA sequencing indicated that all 10 randomly selected clones were recombined, and 80% contained multiple blocks of sequences derived from all of the parents (data not shown). Furthermore, a prescreen of supernatants from 100 first-round sh-p35 library clones combined with human p40 (h-p40) revealed a ≥95% frequency of functional heterodimers capable of inducing proliferation of human T cells. The frequency of biologically active clones was somewhat lower (43%) for the sh-p40 library, which is consistent with the notion that high-affinity binding to IL-12 receptor is mediated by multiple interactions with p40 (17, 22, 41), suggesting that p40 may be more sensitive to deleterious mutations than p35.
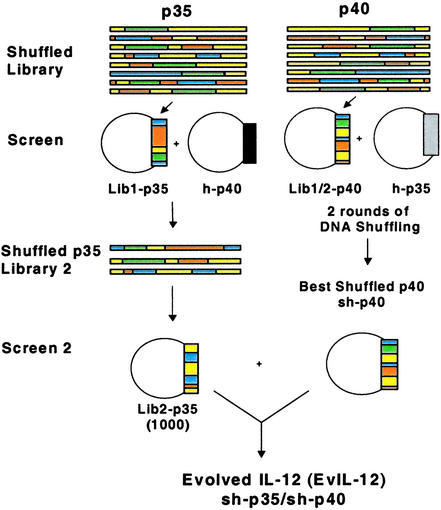
Figure 1.
Library and screening. sh-p35 and sh-p40 libraries for the first round of shuffling (Lib1-p35 and Lib1-p40, respectively) were constructed by using the corresponding cDNA sequences from seven mammalian species. COS-7 cells were cotransfected with a single plasmid from Lib1-p40 and a plasmid encoding h-p35. After 48 h, individual culture supernatants containing either human or chimeric IL-12 were screened for activity in a human T cell proliferation assay. The best six clones were reshuffled to create shuffled Lib2-p40 and the screening repeated with h-p35. The best second round sh-p40 clones were isolated and large-scale DNA preps made for screening with round II sh-p35 clones. Round I shuffled Lib1-p35 was screened in the same manner except COS-7 cells were cotransfected with a plasmid encoding h-p40. Three clones were identified with improved activity after the first round and used to create Lib2-p35. One thousand individual clones from Lib2-p35 were cotransfected along with plasmid DNA encoding sh-p40 and screened in the proliferation assay to yield a representative EvIL-12 molecule.
Identification of Clones with Biological Activity Greater than hIL-12.
In the first round of shuffling/screening, 2,200 clones from each library, Lib1-p35 and Lib1-p40, were screened as illustrated in Fig. 1. Single clones were cotransfected into COS-7 cells together with their matching human subunit, and cell culture supernatants from individual transfections were screened for their capacity to induce proliferation of human PHA-activated T-cells (Fig. 1). Supernatants were tested in a serial dilution format, and several shuffled clones were shown to possess a higher degree of activity than hIL-12. Three chimeras containing a sh-p35 clone were 2-fold more potent, whereas 11 chimeras containing a sh-p40 clone exhibited 4-fold more activity (data not shown). These clones were selected for the next cycle of shuffling/screening to further improve their biological activity.
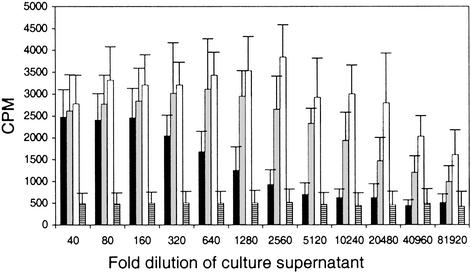
The 11 best sh-p40 clones from Lib1-p40 were used to generate a second library, Lib2-p40, of heterodimers with shuffled subunits. The frequency of active clones was ≈85%. The most active 18 clones were selected, rescreened, and six clones were further characterized. In two independent experiments, using different blood donors and separate transfections, culture supernatant from COS-7 cells cotransfected with the most potent sh-p40 clone (sh-p40) and human p35 (h-p35) showed a dramatic 64-fold increase in the levels of T cell proliferation compared with supernatants derived from COS-7 cells transfected with h-p35 and h-p40 plasmids (Fig. 2). The negative control, culture medium from cells transfected with pCDNA3.1 encoding GFP alone, did not induce proliferation (Fig. 2).
Figure 2.
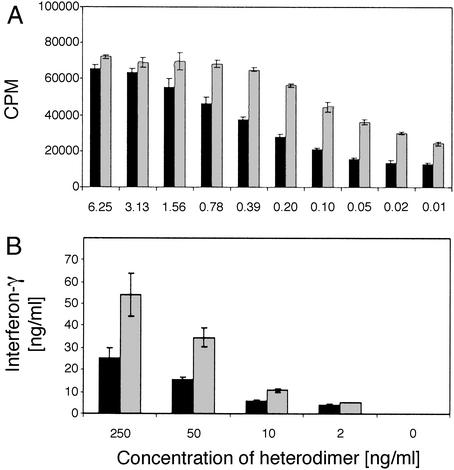
Human T cell proliferation induced by sh-p40 after two rounds of shuffling and screening. Human PHA-activated T cells cultured with serial dilutions of COS-7 supernatants containing hIL-12 (black bar), IL-12 chimera h-p35/sh-p40 (gray bar), Evolved (Ev)IL-12 (sh-p35/sh-p40, white bar), or pCDNA-GFP (hatched bar). 3H-thymidine incorporation (cpm) was measured during the last 16 h of a 72-h culture. Data were derived from two separate transfections per construct and analyzed by using T cells derived from two separate donors to generate a total of 24 data points per dilution.
Additional improvements in activity were achieved by shuffling three improved first-round sh-p35 clones and cotransfecting them with various sh-p40s to create a library of 1,000 fully shuffled heterodimers (Fig. 1). Activities of the best-shuffled heterodimers (sh-p35/sh-p40) were up to 128-fold improved over hIL-12 (Fig. 2).
Expression and Glycosylation of Purified EvIL-12 Protein.
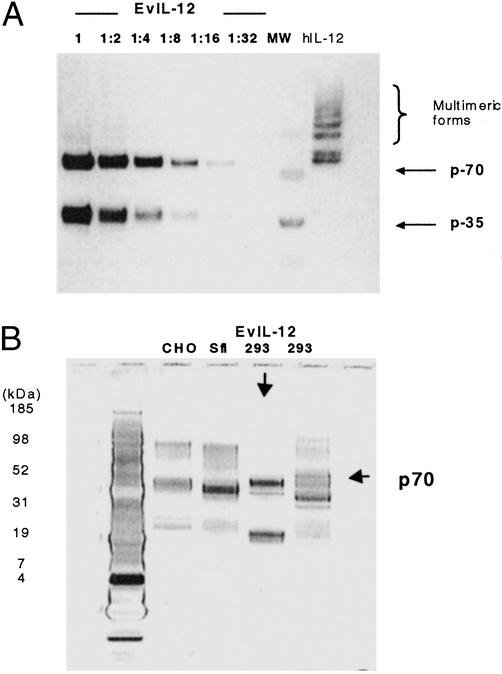
Purified hIL-12 and fully shuffled EvIL-12 (shp40.1/shp35) proteins were used to determine the factor(s) that contributed to the 128-fold improvement observed with crude conditioned medium from transfected cells. This was accomplished by attaching an E-epitope tag (Pharmacia Biotech) to the C terminus of sh-p35 and h-p35 subunits to facilitate purification of the heterodimers by FPLC by using a HiTrap anti-E epitope affinity column (Pharmacia Biotech). Physical characterization of purified hIL-12 and EvIL-12 by Western blot showed their mobility patterns were distinct, with the latter exhibiting a more homogenous product, as illustrated by the reduction of higher molecular weight species (Fig. 3A). More specific interactions between the subunits may be directed by the difference in the putative pI of sh-40 (8.24) compared with h-p40 (5.43), thereby creating a larger overall charge differential between the shuffled subunits (pI-6.5 sh-p35), which might produce a stronger and more specific interaction compared with h-p35 (pI-6.1), and h-p40 subunits. Alternatively, this may reflect alterations in glycosylation resulting from either the loss of a NXS/T (asparagine-X-serine/threonine) recognition site or sequence preference (42). In fact, two mutations present in sh-p40 corresponding to positions S283 and T105 in hIL-12 eliminated a putative glycosylation and preferred NXT recognition site, respectively (Fig. 5). The loss of glycosylation at S283K in sh-p40 may play an important role in altering the conformation of the heterodimer; however, the absence of this putative glycosylation site and its relevance to immunogenicity of EvIL-12 are unknown. Furthermore, although the type of host cell and culture conditions can influence glycosylation profiles (43), the comparison of purified EvIL-12 derived from 293HEK with commercially purified forms of hIL-12 showed EvIL-12 produces a more specific heterodimer regardless of whether hIL-12 was derived from 293HEK, Chinese hamster ovary, or Sfi expression systems (Fig. 3B). Also, the migration of 293-derived EvIL-12 was slightly faster than that of hIL-12. This translates into a molecular weight difference that is greater than the predicted 77 daltons calculated from the theoretical values of hIL-12 and EvIL-12. Nevertheless, future studies using analytical methods such as peptide mapping, MALDI-TOF, and oligosaccharide mapping of EvIL-12 would be useful to determine the relationship of glycosylation with product homogeneity and immunogenicity.
Figure 3.
Homogeneity of p70 heterodimers. Purified EvIL-12 and hIL-12 derived from transfected 293 cells were fractionated on a 4–12% NuPAGE gel, blotted, and detected with an anti-e-tag mAb. (A) Serial dilutions of the EvIL-12 were conducted to demonstrate the homogeneity of the p70 product compared with hIL-12. (B) Silver-stained gel comparing 293 HEK-derived EvIL-12 (indicated with a down arrow) with CHO-, Sfi-, and 293 HEK-derived hIL-12.
Figure 5.
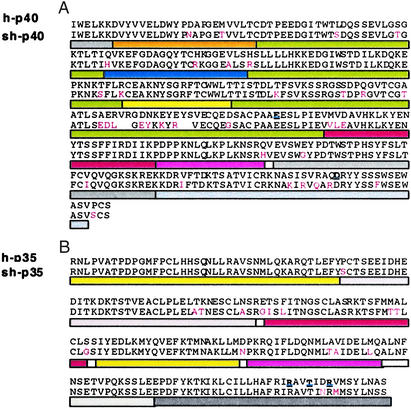
Alignment of representative round 2 sh-p35 and sh-p40 amino acid sequences with parental protein sequences. Sequence alignment of the most improved sh-p40 (A) and sh-p35 (B) with hIL-12. Numbering starts at the first amino acid residue of the mature form of the human subunit proteins. Colored bars identify predicted parental relationships to shuffled parents. Accession nos. of p35/p40: yellow, human AF180526/AF180563; gray, rhesus U19842/U19841; blue, bovine U14416/U11815; green, porcine L35765/U08317; orange, capra AF003542/AF007576; rose, canine U49085/U49100; and red, feline U83185/U83184. Regions showing homology to multiple species have been indicated with colored dots as follows: bovine–capra–canine–feline (blue dots); canine–feline (red dots); porcine–rhesus–human–feline (green dots); human–rhesus–feline (black dots); no homology (white). Amino acids highlighted in red indicate amino acids different from hIL-12; predicted contact residues for heterodimer formation (44) are highlighted in bold black.
Proliferation and Th1 Differentiation of Purified EvIL-12 Protein.
The C-terminal tag attached to the p35 subunits of purified EvIL-12 and hIL-12 heterodimers did not affect the biological activity of these proteins compared with nontagged EvIL-12 or hIL-12 (data not shown). Therefore, the affinity-purified C-terminal tagged heterodimers were used to further investigate their biological activities in cultures of PHA-activated human T cells. In three independent experiments, ≈10-fold less purified EvIL-12 than hIL-12 was required to induce half-maximal T cell proliferation (0.02 ng/ml vs. 0.2 ng/ml), indicating that the specific activity of the shuffled heterodimer was higher compared with hIL-12 (Fig. 4A).
Figure 4.
Functional analysis of purified EvIL-12 and hIL-12. (A) T cell proliferation. Shown is a representative experiment of a series of three assays comparing proliferation of human PHA-activated T cells by affinity purified e-tag EvIL-12 (gray bars) and hIL-12 (black bars), derived from transiently transfected human 293 cells. Samples were assayed in triplicate for each dilution and mean ± SD is shown. (B) Induction of human Th1 cell differentiation. Purified human T cells were cultured in the presence of varying concentrations of purified EvIL-12 (gray bars) or hIL-12 (black bars) followed by activation with anti-CD3 and anti-CD28 mAbs. A representative of three experiments is shown.
When purified human T cells were incubated with EvIL-12 for 5 days followed by activation with anti-CD3/anti-CD28 mAbs, a dose-dependent increase in IFN-γ induction by human T cells was observed, and ≈5- to 10-fold less EvIL-12 was required to obtain the same level of IFN-γ production when compared with cultures activated with hIL-12 (Fig. 4B). These data support the conclusion that EvIL-12 exhibits a higher specific activity than hIL-12 for inducing Th1 cell differentiation. In addition, quantitative experiments with purified hIL-12 and EvIL-12 using dot blot analysis, in conjunction with scanning densitometry, have shown that EvIL-12 expression is at least 10 times higher than hIL-12 made by transient transfection of human 293 HEK cells (data not shown). Collectively, the experiments with purified protein corroborate our findings with crude supernatants: the ≈128-fold improvement in activity of EvIL-12 is associated with enhanced expression and specific activity.
Structural Implications.
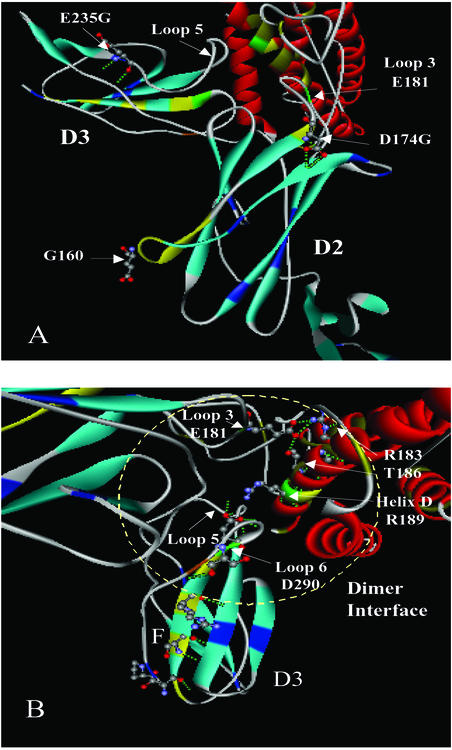
Sequence analysis determined that the improved sh-p40s contain sequence from all of the parental sequences (Fig. 5). The two most improved sh-p40s were 88% identical, and the best clone contained a shorter porcine sequence in place of the corresponding sequence (QGVTCGAATLSAERVRGDNKEYEYS) found in domain 2 (D2) of h-p40. Theoretically, this substitution may result in a shortening of β strand E and alter charge interactions between amino acid residues due to the numerous charge reversals (see Fig. 7 and Supporting Text, which are published as supporting information on the PNAS web site, www.pnas.org). It has been proposed that domain 2 of h-p40 contains the important amino acid E181 on loop 3, which is believed to be crucial for heterodimer formation in hIL-12 (Fig. 6A) (44). Notably, the improved sh-p40s retained E181. Furthermore, all of the sh-p40 subunits differed from h-p40 by a D174G. This substitution at the end of β strand E may facilitate E181 to make more favorable contacts with R183 and T186 on p35 by virtue of increased flexibility at β strand E and loop 3. All of the clones containing non-human sequence in this region produced higher levels of p70 heterodimer than hIL-12, but not all shuffled clones containing the porcine sequence were associated with the specific activity observed in sh-p40.1, suggesting that other substitutions may be important in ligand–receptor interactions and signaling. For instance, E235G at the end of β strand B in domain 3 (D3) of p40 may also influence the formation of the hydrophobic interaction of loop 5 with p35 (Fig. 6A). A number of amino acid substitutions along strand F (S283K, S285R, R287Q, and Q289R) and strand G (S296F) of domain 3 may also influence the orientation of amino acids on loop 6 (Fig. 6B). Interestingly, serine, which comprises a small side chain, has been replaced in each of these positions with an amino acid containing a bulky side chain, such as lysine, arginine, and phenylalanine. Such replacement could effect β-sheet packing of domain 3, creating more efficient binding to p35 or IL-12 receptors.
Figure 6.
Influence on heterodimer formation and receptor binding by amino acid substitutions. The location of selected amino acid substitutions derived from DNA shuffling is shown by using a molecular model of hIL-12 (PDB1F45). (A) Positioning of glycine residues at the end of β strands. In domain 2 (D2) of p40, two glycine substitutions (G160 and D174G) are shown on β strand E connecting a heterodimer contact residue E181 on loop 3. The exact location of glycine, which corresponds to position 160 in hIL-12, is uncertain due to low resolution of atoms between residues 156 and 164 in the crystal structure and the deletion of four amino acids in strand E of the sh-p40 subunit. Another substitution, E235G, at the end of β strand B in domain 3 (D3) of p40 is shown that may influence the hydrophobic interaction of loop5 with p35. (B) Replacement of bulky side-chain R groups in domain 3 of p40. Substitutions S283K, S285R, R287Q, and Q289R are highlighted in yellow on β strand F along with the dominant contact residue, D290, binding to p35.
Although significant recombination had occurred in the sh-p35 subunits, the two most improved clones were very similar (95% identity) (Fig. 5). Sequence analysis of these two clones revealed that both sh-p35 clones included the substitution, D188N, which may influence the orientation of R189 by eliminating charge interactions or modifying hydrogen bonding with Y292 on p40 (Fig. 6B). One of the sh-p35s used as a parent for the second round of DNA shuffling contained this substitution, which originated from the feline parental p35 sequence. Previously, Yoon et al. had mutated D188 to alanine and found a 2-fold increase in p70 formation (44). Our results corroborate their findings, because we also observed an additional 2-fold increase in p70 formation when sh-40.1 was paired with either of the two sh-p35s. Three novel amino acid substitutions observed in the most improved sh-p35s (E79G in loop 1, S103G and D126N in helix B) were also discovered in regions distal to the dimer interface and may have contributed to the improvements.
Strategies to Reduce Potential Immunogenicity.
The protein sequences of the subunits of EvIL-12 were 89.1% (sh-p35) and 86.6% (sh-p40) identical to their human homologs. Previous studies have shown that rhesus and squirrel monkeys treated with hIL-12 develop high titer neutralizing antibodies to the recombinant human protein (45–47). The sequence identity of rhesus IL-12 to hIL-12 is ≈95%, and therefore it is possible that EvIL-12 (88/86% identity to human/rhesus) may also be immunogenic in non-human primates and humans. On the other hand, a small percentage of patients receiving recombinant hIL-12 at doses considered efficacious and safe also developed antibodies but without adverse clinical effect (48–50). Antibody formation to other therapeutic human recombinant proteins such as IFN-α, IFN-β, IL2, GM-CSF, and GH have also been detected in patients without adverse consequences, but the clinical significance of the presence of non-neutralizing antibodies has not been fully established (51, 52). Immunogenicity is also likely to depend on the route, dose, and frequency of administration. Strategies for thwarting potential immunogenicity of EvIL-12 include backcrossing with hIL-12 to remove unwanted immunoreactive epitopes while maintaining the enhanced activity of the molecule, and site-specific PEGylation (53) or glycosylation (54) to shield such epitopes.
Studies in mice using genetic delivery of hIL-12 have shown it to be as efficacious as systemically delivered rhIL-12 but without the toxic side effects of IL-12 protein therapy (15, 27, 55). A genetic dose of 1/400 to 1/4,000 of the dose of systemically administered hIL-12 protein was efficacious in eradicating or suppressing tumors in mice (28), and the lack of toxicity was attributed to a lower serum level of IFN-γ induced by genetic vs. protein delivery (27). Furthermore, IL-12 gene therapy in a human phase I study has been demonstrated to be safe (24). The enhanced specific activity and expression of EvIL-12 in human cells may strengthen natural killer/Th1-mediated immunity while avoiding the toxic adverse effects of IL-12 protein delivery. Moreover, when administered as an adjuvant, only a few low doses of DNA may be required for efficacy. It is anticipated that an efficacious dose of EvIL-12 with an acceptable safety and immunogenicity profile can be determined through the modulation of dose and number of treatments given to a patient. However, clinical studies and coordinated long-term monitoring of patients will provide the ultimate determination of a protein's safety and immunogenicity.
Conclusion and Future Directions.
We have shown that various desirable properties of a therapeutically relevant protein can be improved by directed molecular evolution: IL-12 was engineered for enhanced expression and biological activity using the functional sequence diversity from seven mammalian genes. One of the advantages of evolutionary approaches is that such approaches typically yield improvement by multiple unrelated mechanisms and often in unexpected ways, whereas rational design generally aims to improve a single physical property or mechanism. For example, the revelation of improved homogeneity of EvIL-12 demonstrates that DNA shuffling can solve problems associated with protein folding and expression while maintaining desired biological activities, thus eliminating the need for the development of costly refolding and separation processes used in the current manufacture of soluble protein human therapeutic, and possibly increasing the potency of genetically delivered cytokines produced by the target cells of the patient. In vivo studies comparing hIL-12 and EvIL-12 as a genetic adjuvant or treatment vaccine will be important to ascertain the safety and efficacy in non-human primates and humans. Last, optimization of structure through DNA shuffling to produce a more stable homogenous product with improved specific activity should facilitate more alternative choices of drug delivery, lower therapeutic dose, improved efficacy and safety, and reduced development cost.
Supplementary Material
Acknowledgments
We thank Emily Mundorff for the bioinformatics analyses of EvIL-12. This study was supported by a grant from Defense Advanced Research Projects Agency.
Abbreviations
- hIL-12
human IL-12
- EvIL-12
evolved IL-12
- PHA
phytohemagglutinin
- PBMC
peripheral blood mononuclear cells
- HEK
human embryonic kidney
- h-p40
human p40
- sh-p40
shuffled p40
Footnotes
References
- 1.Gately M K, Renzetti L M, Magram J, Stern A S, Adorin I L, Gubler U, Presky D H. Annu Rev Immunol. 1998;16:495–521. doi: 10.1146/annurev.immunol.16.1.495. [DOI] [PubMed] [Google Scholar]
- 2.Trinchieri G, Scott P. Curr Top Microbiol Immunol. 1999;238:57–78. doi: 10.1007/978-3-662-09709-0_4. [DOI] [PubMed] [Google Scholar]
- 3.Greenberger M J, Kunkel S, Strieter R, Lukacs N, Bramson J, Gauldie J, Graham F, Hitt M, Danforth J, Standiford T. J Immunol. 1966;157:3006–3012. [PubMed] [Google Scholar]
- 4.Silva R, Pais T, Appelberg R. J Immunol. 1998;161:5578–5585. [PubMed] [Google Scholar]
- 5.Miller M, Skeen M, Ziegler H. Cell Immunol. 1998;184:92–104. doi: 10.1006/cimm.1998.1270. [DOI] [PubMed] [Google Scholar]
- 6.Romani L, Bistoni F, Puccetti P. Chem Immunol. 1997;68:110–135. doi: 10.1159/000058688. [DOI] [PubMed] [Google Scholar]
- 7.Kawakami K, Tohyama M, Xie Q, Saito A. Clin Exp Immunol. 1996;104:208–214. doi: 10.1046/j.1365-2249.1996.14723.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mountford A, Anderson S, Wilson R. J Immunol. 1996;156:4739–4745. [PubMed] [Google Scholar]
- 9.Kenney R, Sacks D, Sypek J, Vilela L, Gam A, Evans-Davis K. J Immunol. 1999;163:4481–4488. [PubMed] [Google Scholar]
- 10.Altare F, Durandy A, Lammas D, Emile J F, Lamhamedi S, Le Deist F, Drysdale P, Jouanguy E, Doffinger R, Bernaudin F, et al. Science. 1998;280:1432–1435. doi: 10.1126/science.280.5368.1432. [DOI] [PubMed] [Google Scholar]
- 11.Altare F, Lammas D, Revy P, Jouanguy E, Doffinger R, Lamhamedi S, Drysdale P, Scheel-Toellner D, Girdlestone J, Darbyshire P, et al. J Clin Invest. 1998;102:2035–2040. doi: 10.1172/JCI4950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.de Jong R, Altare F, Haagen I, Elferink D, Boer T, van Breda Vriesman P, Kabel P, Draaisma J, can Dissel J, et al. Science. 1998;280:1435–1438. doi: 10.1126/science.280.5368.1435. [DOI] [PubMed] [Google Scholar]
- 13.Hensler T, Heidecke C, Hecker H, Heeg K, Bartels H, Zantl N, Wagner H, Siewert J, Holmann B. J Immunol. 1998;161:2655–2659. [PubMed] [Google Scholar]
- 14.Haraguchi S, Day N, Nelson R J, Emmanuel P, Duplantier J, Christodoulou C, Good R. Proc Natl Acad Sci USA. 1998;95:13125–13129. doi: 10.1073/pnas.95.22.13125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lode H, Dreier T, Xiang R, Varki N, Kang A, Reisfeld R. Proc Natl Acad Sci USA. 1998;95:2475–2480. doi: 10.1073/pnas.95.5.2475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rook A, Wood G, Yoo E, Elenitsas R, Kao D, Sherman M, Witmer W, Rockwell K, Shane R, Lessin S, Vonderheid E. Blood. 1998;94:902–908. [PubMed] [Google Scholar]
- 17.Presky D, Minetti L, Gillessen S, Wilkinson V, Wu C, Gubler U, Chizzonite R, Gately M. J Immunol. 1998;160:2174–2179. [PubMed] [Google Scholar]
- 18.Merberg D, Wolf S, Clark S. Immunol Today. 1992;2:77–78. doi: 10.1016/0167-5699(92)90140-3. [DOI] [PubMed] [Google Scholar]
- 19.Gearing D, Cosman D. Cell. 1991;66:9–10. doi: 10.1016/0092-8674(91)90131-h. [DOI] [PubMed] [Google Scholar]
- 20.Dwyer D. J Mol Graphics. 1996;3:148–157. doi: 10.1016/s0263-7855(96)00044-6. [DOI] [PubMed] [Google Scholar]
- 21.Devergnes O, Birkenbach M, Kieff E. Proc Natl Acad Sci USA. 1997;94:12041–12046. doi: 10.1073/pnas.94.22.12041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ling P, Gately M, Gubler U, Stern A, Lin P, Hollfelder K, Su C, Pan Y-C, Hakimi J. J Immunol. 1995;154:116–127. [PubMed] [Google Scholar]
- 23.Oppmann B, Lesley R, Blom B, Timas J, Xu Y, Hunte B, Vega F, Yu N, Wang J, Singh K, et al. Immunity. 2000;5:715–725. doi: 10.1016/s1074-7613(00)00070-4. [DOI] [PubMed] [Google Scholar]
- 24.Kang W K, Park C, Yoon H L, Kim W S, Yoon S S, Lee M H, Park K, Kim K, Jeong H S, Kim J A, et al. Hum Gene Ther. 2001;12:671–684. doi: 10.1089/104303401300057388. [DOI] [PubMed] [Google Scholar]
- 25.Robertson M J, Cameron C, Atkins M B, Gordon M S, Lotze M T, Sherman M L, Ritz J. Clin Cancer Res. 1999;5:9–17. [PubMed] [Google Scholar]
- 26.Sun Y, Jurgovsky K, Moller P, Alijagic S, Dorbic T, Georgieva J, Wittig B, Schadendorf D. Gene Ther. 1998;5:481–490. doi: 10.1038/sj.gt.3300619. [DOI] [PubMed] [Google Scholar]
- 27.Rakhmilevich A, Timmins J, Janssen K, Pohlmann E, Sheehy M, Yang N. J Immunother. 1999;22:135–144. doi: 10.1097/00002371-199903000-00005. [DOI] [PubMed] [Google Scholar]
- 28.Rakhmilevich A, Turner J, Ford M, McCabe D, Sun W, Sondel P, Grota K, Yang N-S. Proc Natl Acad Sci USA. 1996;93:6291–6296. doi: 10.1073/pnas.93.13.6291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chaplin P, Camon E, Villarreal-Ramos B, Flint M, Ryan M, Collins R. J Interferon Cytokine Res. 1999;19:235–241. doi: 10.1089/107999099314162. [DOI] [PubMed] [Google Scholar]
- 30.Stemmer W. Biotechnology. 1995;13:549–553. [Google Scholar]
- 31.Ness J, Del Cardayre S, Minshull J, Stemmer W. Adv Protein Chem. 2000;55:261–292. doi: 10.1016/s0065-3233(01)55006-8. [DOI] [PubMed] [Google Scholar]
- 32.Whalen R G, Kaiwar R, Soong N-W, Punnonen J. Curr Opin Mol Ther. 2001;3:31–36. [PubMed] [Google Scholar]
- 33.Powell S, Kaloss M, Pinkstaff A, McKee R, Burimski I, Pensiero M, Otto E, Stemmer W, Soong N-W. Nat Biotechnol. 2000;18:1279–1282. doi: 10.1038/82391. [DOI] [PubMed] [Google Scholar]
- 34.Ness J E, Welch M, Giver L, Bueno M, Cherry J R, Borchert T V, Stemmer W P, Minshull J. Nat Biotechnol. 1999;17:893–896. doi: 10.1038/12884. [DOI] [PubMed] [Google Scholar]
- 35.Lazetic S, Leong S R, Chang C-C J, Ong R, Dawes G, Punnonen J. J Biol Chem. 2002;277:38660–38668. doi: 10.1074/jbc.M205808200. [DOI] [PubMed] [Google Scholar]
- 36.Chang C-C, Chen T, Cox B, Dawes G, Stemmer W, Punnonen J, Patten P. Nat Biotechnol. 1999;17:793–797. doi: 10.1038/11737. [DOI] [PubMed] [Google Scholar]
- 37.Crameri A, Railard S, Bermudez E, Stemmer W. Nature. 1998;391:288–291. doi: 10.1038/34663. [DOI] [PubMed] [Google Scholar]
- 38.Chang C-C, Wright A, Punnonen J. J Immunol. 2000;165:3584–3591. doi: 10.4049/jimmunol.165.7.3584. [DOI] [PubMed] [Google Scholar]
- 39.Yssel H, de Vries J, Koken M, Van Blitterswijk W, Spits H. J Immunol Methods. 1994;72:219–227. doi: 10.1016/0022-1759(84)90450-2. [DOI] [PubMed] [Google Scholar]
- 40.Punnonen J, de Vries J. J Immunol. 1994;152:1094–1102. [PubMed] [Google Scholar]
- 41.Schoenhaut D, Chua A, Wolitzky A, Quinn P, Dwyer C, McComas W, Familletti P, Gately M, Gubler U. J Immunol. 1992;148:3433–3440. [PubMed] [Google Scholar]
- 42.Reddy A, Gibbs B, Lui Y, Coward J, Changchien L, Maley F. Glycobiology. 1999;9:547–555. doi: 10.1093/glycob/9.6.547. [DOI] [PubMed] [Google Scholar]
- 43.Werner R, Noe W, Kopp K, Schluter M. Arzneim-Forsch. 1998;48:870–880. [PubMed] [Google Scholar]
- 44.Yoon C, Johnston S, Tang J, Stahl M, Tobin J, Somers W. EMBO J. 2000;19:3530–3541. doi: 10.1093/emboj/19.14.3530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nadeau R, Ostrowski C, Ni-Wu G, Liberato D. J Pharmacol Exp Ther. 1995;274:78–83. [PubMed] [Google Scholar]
- 46.Sarmiento U, Riley J, Knaack P, Lipman J, Becker J, Gately M, Chizzonite R, Anderson T. Lab Invest. 1994;71:862–873. [PubMed] [Google Scholar]
- 47.Watanabe N, Sypek J, Mittler S, Reimann K, Flores-Villanueva P, Voss G, Lord C, Letvin N. AIDS Res Retroviruses. 1998;14:393–399. doi: 10.1089/aid.1998.14.393. [DOI] [PubMed] [Google Scholar]
- 48.Atkins M, Robertson M, Gordon M S, Lotze M, DeCoste M, DuBois J, Ritz J, Sandler A, Edington H, Garzone P, et al. Clin Cancer Res. 1997;3:409–417. [PubMed] [Google Scholar]
- 49.Carreno V, Zuezem S, Hopf U, Marcellin P, Cooksley W, Fevery J, Diago M, Reddy R, Peters M, Rittweger K, et al. J Hepatol. 2000;32:317–324. doi: 10.1016/s0168-8278(00)80078-1. [DOI] [PubMed] [Google Scholar]
- 50.Zuezem S, Hopf U, Carreno V, Diago M, Shiffman M, Grune S, Dudley F, Rakhit A, Rittweger K, Yap S, Koff R, Thomas H. Hepatology. 1999;29:1280–1287. doi: 10.1002/hep.510290429. [DOI] [PubMed] [Google Scholar]
- 51.Zeisel H, Lutz A, von Petrykowski W. Horm Res. 1992;37:47–55. doi: 10.1159/000182380. [DOI] [PubMed] [Google Scholar]
- 52.Vial T, Descotes J. Toxiciology. 1995;105:31–57. doi: 10.1016/0300-483x(95)03124-x. [DOI] [PubMed] [Google Scholar]
- 53.Leong S, DeForge L, Presta L, Gonzalez T, Fan A, Reichert M, Chuntharapai A, Kim K, Tumas D, Lee W, et al. Cytokine. 2001;16:106–119. doi: 10.1006/cyto.2001.0936. [DOI] [PubMed] [Google Scholar]
- 54.Cheng-Mayer C, Brown A, Harhouse J, Luciw P, Mayer A. J Virol. 1999;73:5294–5300. doi: 10.1128/jvi.73.7.5294-5300.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kim J, Ayyavoo V, Bagarazzi M, Chattergoon M, Dang K, Wang B, Boyer J, Weiner D. J Immunol. 1997;158:816–826. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.