Abstract
Asthma is characterized by an oxidant/antioxidant imbalance in the lungs leading to activation of redox-sensitive transcription factors, nuclear factor κB (NF-κB), and activator protein-1 (AP-1). To develop therapeutic strategies for asthma, we used a chemogenomics approach to screen for small molecule inhibitor(s) of AP-1 transcription. We developed a β-strand mimetic template that acts as a reversible inhibitor (pseudosubstrate) of redox proteins. This template incorporates an enedione moiety to trap reactive cysteine nucleophiles in the active sites of redox proteins. Specificity for individual redox factors was achieved through variations in X and Y functionality by using a combinatorial library approach. A limited array (2 × 6) was constructed where X was either NHCH3 or NHCH2 Ph and Y was methyl, phenyl, m-cyanophenyl, m-nitrophenyl, m-acetylaniline, or m-methylbenzoate. These analogs were evaluated for their ability to inhibit transcription in transiently transfected human lung epithelial A549 cells from either an AP-1 or NF-κB reporter. A small-molecule inhibitor, PNRI-299, was identified that selectively inhibited AP-1 transcription (IC50 of 20 μM) without affecting NF-κB transcription (up to 200 μM) or thioredoxin (up to 200 μM). The molecular target of PNRI-299 was determined to be the oxidoreductase, redox effector factor-1 by an affinity chromatography approach. The selective redox effector factor-1 inhibitor, PNRI-299, significantly reduced airway eosinophil infiltration, mucus hypersecretion, edema, and IL-4 levels in a mouse asthma model. These data validate AP-1 as an important therapeutic target in allergic airway inflammation.
Asthma has reached epidemic proportions with ≈200 million individuals affected worldwide (1). This complex lung inflammatory disease is characterized by airway infiltration by CD4+ T cells with a T helper (Th)2 phenotype, eosinophils, and other inflammatory cells, mucus hypersecretion, and edema. The eosinophils and other leukocytes undergo respiratory burst activation with release of reactive oxygen species (ROS), including superoxide and its dismutation product, hydrogen peroxide (H2O2) (2). Increased airway levels of ROS (3, 4), coupled with decreased levels of antioxidants such as glutathione (5), may lead to an oxidant/antioxidant imbalance in the lungs of patients with asthma and activation of redox-sensitive transcription factors activator protein-1 (AP-1) and nuclear factor κB (NF-κB). These transcription factors, in collaboration with Th2-specific transcription factors (i.e., c-maf, GATA-3, Stat6), control expression of Th2 cytokines (e.g., IL-4, IL-5, IL-13), the molecular hallmarks of asthma (6). Corticosteroids, which inhibit both NF-κB and AP-1 transcription, are the most effective treatment for asthma but have severe adverse effects when administered systemically (7). A subset of asthmatics, which responds poorly to corticosteroids and is at risk for respiratory failure, has increased activity of AP-1 in peripheral blood mononuclear cells (8).
There is increasing evidence that redox regulation of AP-1 and NF-κB (9–11) transcription is important. AP-1 transcriptional activity is redox-sensitive with increased AP-1 expression induced by reactive oxygen species (e.g., H2O2) (11–13). Paradoxically, AP-1 DNA binding is decreased by oxidation of critical cysteine residues (e.g., cysteine 252 in cJun) and increased when these residues undergo reduction (or mutation to serine in vJun) (11, 14). Peptides derived from NF-κB bind in an extended strand conformation to the active site cleft of their cognate redox regulator, thioredoxin (TRX) (15). TRX is a member of the oxidoreductase superfamily, which augments the production of IL-2Rα in human T-lymphotrophic virus-1-infected T cells by reduction of the S-NO form of Cys-62 within the DNA binding domain of Rel family members (16–18).
To capture the therapeutic value of the data derived from the recent sequencing of the human genome and translate this information into pharmaceutical agents for treatment of various disease states, the information must be converted into small-molecule chemistry to “pharmacologically validate” a new molecular target. Toward this goal, we have developed a unique set of chemistries that can be performed in a highly automated combinatorial fashion to very rapidly synthesize nonpeptide, biologically stable, secondary structure mimics (19–21). These unique “privileged template” secondary structure mimics have enormous potential for the development of “pharmacological knockouts” (and eventually therapeutics), because of the manner in which proteins have evolved. Through the utilization of 20-aa side chains displayed on a limited array of topological motifs (i.e., the common secondary structures, reverse turns, extended strands, and α-helices), nature has provided the elements required to delineate a multitude of ligand–receptor, enzyme–substrate interactions, and other signal transduction processes. We have used an extended-strand templated library to develop a small-molecule inhibitor of TRX and NF-κB transcription (22, 23). Here, we use a chemogenomic approach to develop a small-molecule inhibitor of redox-regulated AP-1 transcription and validate it as a therapeutic target for asthma.
Methods
A549 Cell Cytosolic and Nuclear Extracts.
A549 cells were grown in normal media until confluent. After media removal, the cells were washed with PBS and scraped from the plates in cell suspension buffer. After transferring the cells to a clean Eppendorf tube and centrifugation (at 200 × g), the supernatant was discarded and the cells were resuspended in suspension buffer containing 0.1% NP-40 (to lyse the cell membrane). The cells were centrifuged (at 12,000 × g), and the cytosolic extracts were saved. The nuclear pellet was resuspended in minimal high-salt buffer to lyse the nuclear membrane. After centrifugation (at 12,000 × g), the supernatant containing the soluble nuclear protein was collected.
Affinity Chromatography.
A549 cell-soluble cytosolic and nuclear extracts were incubated with biotinylated affinity reagents 162-150 and 162-149. After treatment with the analogs, streptavidin beads were added to isolate biotinylated compounds and their bound targets. After centrifugation (at 1,000 × g), the beads and supernatant were separated. Reduced sample buffer was added to the supernatant (containing unbound proteins) and boiled for 5 min before SDS/PAGE analysis. The streptavidin beads were washed extensively with PBS to remove unbound proteins, resuspended in a minimal amount of reduced sample buffer, and boiled for 5 min to release bound proteins before SDS/PAGE analysis.
Mouse Asthma Model Studies.
All animal use procedures were approved by the University of Washington Animal Care Committee. Female BALB/c mice (age 6–8 wk; The Jackson Laboratory) received an i.p. injection of 100 μg of ovalbumin (OVA) complexed with alum on days 0 and 14. Mice received an intranasal (i.n.) dose of 50 μg of OVA on days 14, 25, 26, and 27. The control group received normal saline with alum i.p. on days 0 and 14 and saline without alum i.n. on days 14, 25, 26, and 27. A group of OVA-treated mice was administered PNRI-299 i.n. at a dose of 0.75 or 2.0 mg/kg, 30 min before OVA on days 25–27. The mice were anesthetized before the administration of PNRI-299 with xylazine (0.44 mg/ml)/ketamine (6.5 mg/ml). The volume of administration was 50 μl, vehicle = 0.16% DMSO, and there was a vehicle-only group (0.16% DMSO in saline), which had no effect on the OVA-induced airway inflammatory response. On day 28, 24 h after the last i.n. administration of either normal saline or OVA, each mouse was exsanguinated by cardiac puncture, bronchoalveolar lavage was performed on the right lung, and left lung tissue was obtained for morphology (24). For light microscopy, the lung sections were stained with hematoxylin and eosin to assess the inflammatory cell infiltrate and mucus secretion (24, 25). Airway edema was assessed in hematoxylin- and eosin-stained lung sections by morphometry (0–2 scale). Morphometry was performed by individuals blinded to the protocol design (24). A minimum of 10 fields throughout the upper and lower left lung tissue was randomly examined for the morphologic analyses. The data are shown as the mean ± SE. Total RNA was isolated from the right lung of each mouse, and mRNA levels for IL-4, IL-5, IL-13, eotaxin, and CCR3 were determined by RT-PCR (26).
Results
Development of Small-Molecule Inhibitor of AP-1 Transcription by Using a Combinatorial Library Approach.
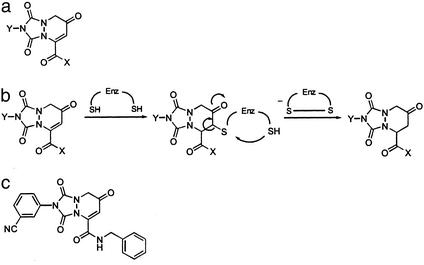
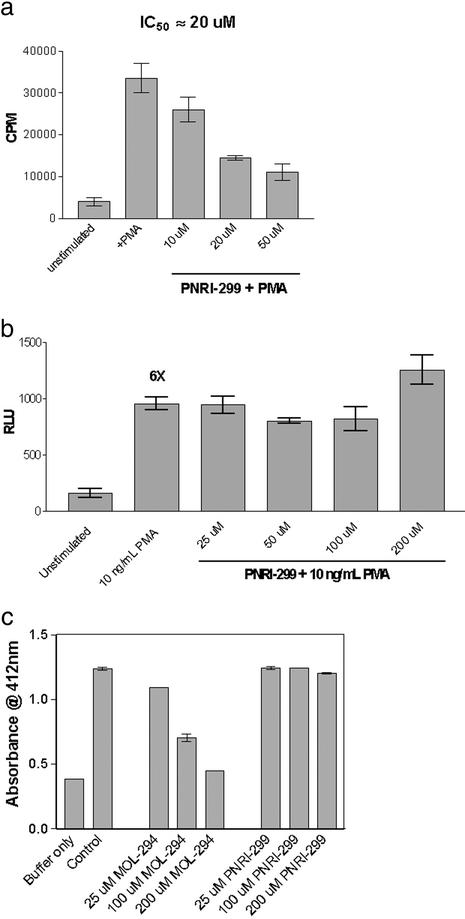
We developed a β-strand mimetic template (Fig. 1a) designed to act as a reversible inhibitor (pseudosubstrate) of redox proteins. This template is a modified version of our β-strand template (21), which incorporates an enedione moiety to trap reactive cysteine nucleophiles in the active sites of redox proteins (Fig. 1b). By using a combinatorial library approach, it was anticipated that, through variations in X and Y functionality, specificity for individual redox factors could be achieved. We constructed a limited array (2 × 6) where X was either NHCH3 or NHBn and Y was methyl, phenyl, m-cyanophenyl, m-nitrophenyl, m-acetylaniline, or m-methylbenzoate. These analogs were evaluated for their ability to inhibit transcription in transiently transfected human lung epithelial A549 cells from either an AP-1 or NF-κB reporter (22). One compound, PNRI-299 (Fig. 1c), selectively inhibited AP-1 transcription with an IC50 of 20 μM (Fig. 2a) without affecting NF-κB transcription (up to 200 μM; Fig. 2b). This analog was also nonreactive with TRX (up to 200 μM; Fig. 2c).
Figure 1.
Development of small-molecule inhibitor of Ref-1. (a) Pseudosubstrate oxidoreductase template. (b) Modified β-strand template serves as an oxidoreductase pseudosubstrate. (c) Molecular structure of AP-1 inhibitor, PNRI-299.
Figure 2.
Selective inhibition of AP-1 transcription by PNRI-299. (a) Inhibition in A549 cells transiently transfected with an AP-1 luc reporter gene construct stimulated with phorbol 12-myristate 13-acetate (PMA). (b) Inhibition in A549 cells transiently transfected with an NF-κB luc reporter gene construct stimulated with PMA. (c) Inhibition of TRX activity by assessing reduction of Ellman's reagent by TRX reductase-activated TRX (22).
Identification of Redox Effector Factor-1 (Ref-1) as Molecular Target of AP-1 Inhibitor PNRI-299 by Using an Affinity Chromatography Approach.
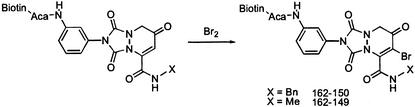
We used an affinity chromatography approach to determine the molecular target(s) of the analog PNRI-299 (27). The initial screen indicated that an acylaniline analog had decreased activity only slightly (by a factor of 2). To increase the likelihood of affinity-purifying the molecular target(s), the enedione portion was modified by addition of bromine to provide analog 162-150 (Fig. 3). The affinity reagent 162-150 incorporated an aminocaproic acid linker (Aca) to provide a sufficient distance between the affinity probe and the biotin moiety that was used to bind to the agarose-streptavidin beads. As a negative control, we prepared analog 162-149 (Fig. 3), which was based on an inactive analog.
Figure 3.
Affinity probe synthesis. Generation of affinity reagents 162-150 and 162-149.
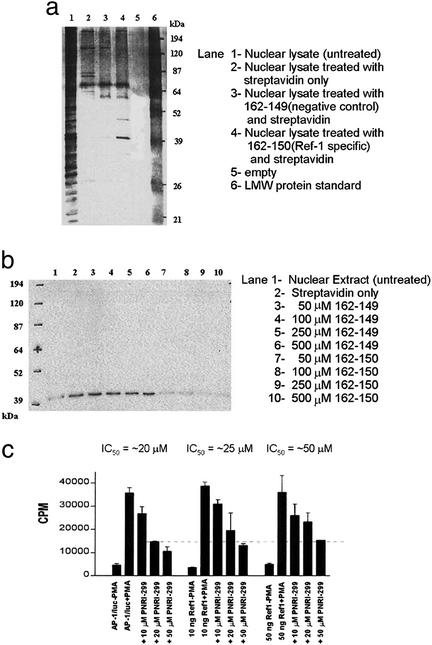
Soluble cytosolic and nuclear extracts were treated with biotinylated compounds 162-150 or 162-149, and streptavidin beads were then added to pull out the biotinylated compounds and their bound protein targets. Proteins retained by affinity probe 162-150 vs. affinity probe 162-149 from A549 epithelial cell nuclear extracts are shown in Fig. 4a. The major band that was selectively retained by affinity probe 162-150 (Fig. 4a, lane 4) ran with an apparent molecular mass of ≈39 kDa. Mass spectral analysis identified this band as Ref-1. Western blotting with an anti-Ref-1 antibody (Santa Cruz Biotechnology) showed depletion of Ref-1 in the nuclear extracts treated with 162-150 but not 162-149 (Fig. 4b). Transfection of a Ref-1 expression vector in A549 cells transfected with an AP-1 luc reporter gene construct was demonstrated to increase the IC50 for PNRI-299 (from 20 to 50 μM) (Fig. 4c). These data establish that PNRI-299 specifically reacts with Ref-1, inhibits AP-1 transcription, and overexpression of the molecular target. Ref-1 attenuates PNRI-299 inhibition of AP-1 transcription.
Figure 4.
Identification of Ref-1 as molecular target of AP-1 inhibitor PNRI-299. (a) Affinity purification of proteins using biotinylated compound (162-150) targeting Ref-1 protein, in A549 nuclear lysates. (b) Western blot of Ref-1 in supernatant of A549 nuclear extracts treated with affinity probe 162-150 (50–500 μm) but not 162-149 (50–500 μm) is shown. (c) Cotransfection of Ref-1 expression vector in A549 cells transfected with an AP-1 luc reporter gene construct.
Molecular Interaction of PNRI-299 with Ref-1.
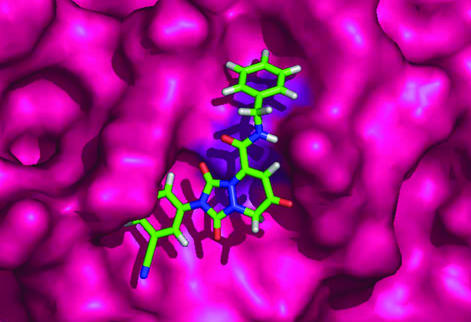
We next investigated the potential molecular interaction of PNRI-299 with Ref-1. AP-1 is composed of either a homodimer of Jun or a heterodimer of Jun and Fos. The mechanism by which Ref-1 activates AP-1 transcription is through reduction of the S-nitroso group of Jun Cys-252 (14) located within the DNA binding region of the transcription factor. Ref-1 has been shown to stimulate AP-1 DNA binding in vitro (28). Experiments in BA/F3 cells demonstrated that AP-1 transcriptional activity was enhanced by overexpression of Ref-1 and attenuated by the introduction of antisense Ref-1 (29). Independent of its characterization as a transcriptional regulator, Ref-1 was shown to act together with several other DNA repair proteins and possess apurinic/apyrimidinic endonuclease (APE) activity (30). The redox and DNA-repair activities of Ref-1 are encoded by nonoverlapping domains (31). Based on site-directed mutagenesis experiments (32) and the x-ray crystal structure of Ref-1 (33), we have modeled (Fig. 5) PNRI-299 interacting with the redox nucleophile Cys-65, to aid in the interpretation of structure activity relationships (SARs). In the Ref-1 structure, the side chain of Cys-65 is pointing into a hydrophobic pocket where the inhibitor is bound. This pocket includes Trp-67, Trp-75, Trp-83, Leu-92, and Pro-311, consistent with the limited SAR data obtained from the initial 12-member library.
Figure 5.
Docking model of inhibitor PNRI-299 bound to Ref-1. The molecular modeling was carried out by using the FirstDiscovery drug design suite component GLIDE (Schrödinger, Portland, OR).
Inhibition of IL-4-Driven Airway Inflammation in a Mouse Asthma Model by Ref-1 Inhibitor PNRI-299.
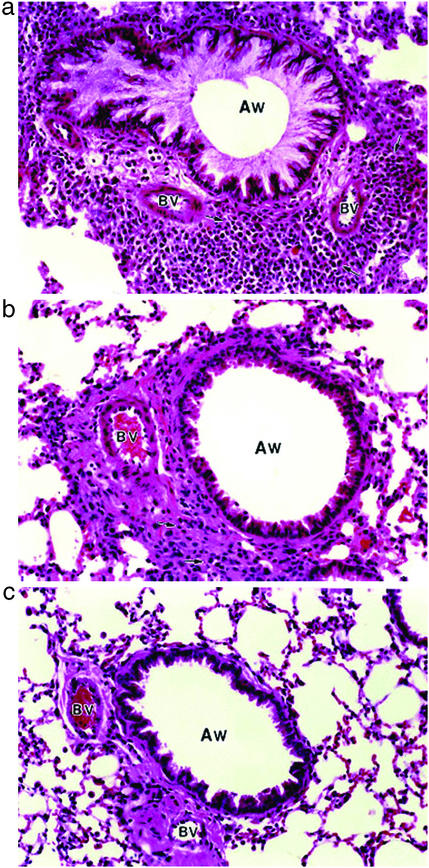
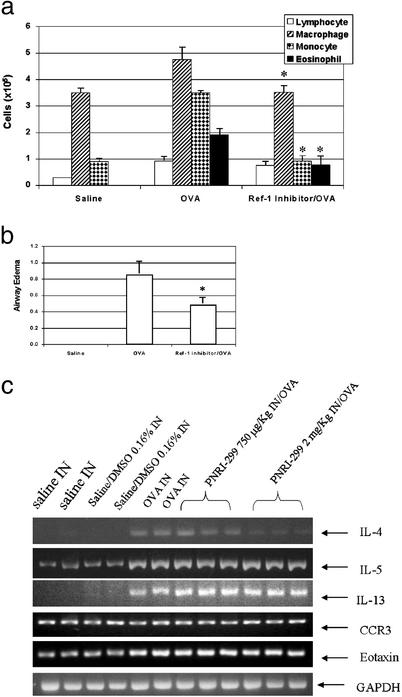
We examined the effect of Ref-1 inhibitor PNRI-299 on allergic airway inflammation in a mouse model that reproduced key features of human asthma (25). The OVA-sensitized/challenged mice developed a striking infiltration of the airways by eosinophils and other inflammatory cells and mucus hypersecretion (Fig. 6a) that was not observed in saline-treated controls (Fig. 6c). Treatment with PNRI-299 significantly decreased the influx of eosinophils, monocytes, and macrophages (no significant reduction of lymphocytes was observed) into the lung interstitium (Fig. 6b) and bronchoalveolar lavage fluid (Fig. 7a), and airway mucus (Fig. 6b) and edema (Fig. 7b) observed in OVA-treated mice. We conclude that PNRI-299 reduces the pathophysiological responses seen in this murine model of allergic asthma.
Figure 6.
Ref-1 inhibitor PNRI-299 reduces allergy airway inflammation. (a) OVA-treated mice have a dense inflammatory cell infiltrate (arrows) surrounding the airways (AW) and blood vessels (BV) and mucus hypersecretion not seen in saline controls (c). (b) PNRI-299 (0.75 mg/kg) reduced the airway inflammatory cell infiltration (arrows) and mucus release in OVA-treated mice.
Figure 7.
Reduction of asthmatic response. Bronchoalveolar lavage fluid eosinophils, monocytes, macrophages, and lymphocytes (a) and airway edema (b) were determined in controls (Saline) and OVA-treated mice in the absence (OVA) or presence (Ref-1 inhibitor/OVA) of PNRI-299. *, P < 0.05 vs. OVA by Student's two-tailed t test. (c) RT-PCR assay of IL-4, IL-5, IL-13, eotaxin, and CCR3 pulmonary gene expression.
Th2 cytokine and chemokine expression are molecular hallmarks of asthma. IL-4 gene expression is transcriptionally regulated through complex interactions of multiple factors that bind to the proximal promoter region. Cooperative and coordinated recruitment of both nuclear factor of activated T cells and AP-1 is required for inducible transcription of IL-4 during T-cell activation (34, 35). Although the genes for IL-4, IL-5, and IL-13 are closely located on human chromosome 5q31 (mouse chromosome 11) (36), there is evidence for the unique transcriptional regulation of IL-5 and IL-13 that is independent of AP-1 binding (37, 38). Based on this information, one would predict that PNRI-299 should decrease IL-4 but not IL-5 or IL-13 expression. We confirmed by RT-PCR analysis that gene expression of IL-4, IL-5, and IL-13 was markedly increased, eotaxin was minimally increased, and CCR3 was unchanged in whole lung tissue of the OVA-treated mice compared with saline-treated controls (Fig. 7c). The increased gene expression of IL-4 in the OVA-treated mice was decreased in a dose-dependent manner (0.75–2.0 mg/kg i.n. administration) by PNRI-299 (Fig. 7c). The increased expression of IL-5, IL-13, and eotaxin in the OVA-treated mice was unaffected by either dose of PNRI-299 (Fig. 7c). The selective reduction of IL-4 message by PNRI-299 is consistent with its inhibition of Ref-1 and thereby AP-1 transcription and its ameliorating effect on the asthmatic response in the OVA-treated mice.
Discussion
In summary, using a powerful chemogenomics approach, we found a selective inhibitor of AP-1 transcription and identified its molecular target as Ref-1. We demonstrated that this selective small-molecule inhibitor, PNRI-299, decreases airway eosinophilia, mucus hypersecretion, and edema, and inhibits the expression of IL-4 in a mouse asthma model. The critical role of IL-4 in the development and maintenance of the Th2 phenotype has been highlighted recently by the creation of triple-knockout (i.e., IL-5, IL-9, IL-13) mice that potently retain the major Th2 effector functions (i.e., eosinophilia, mastocytosis, and goblet cell hyperplasia) (39). Granulocyte/macrophage colony-stimulating factor and IL-13 cytokines, which are present in asthmatic lungs and during the initiation of fibrosis, enhance AP-1 DNA binding and Ref-1 production in normal alveolar macrophages (40). Inhibitors of Ref-1 may prove beneficial for the treatment of asthma and airway fibrosis.
Furthermore, the covalent reversible modification of cysteine residues in proteins by nitric oxide (NO) or related oxidative processes can regulate a variety of cellular responses, including gene expression (17). The oxidoreductase superfamily is extremely large with estimates of nearly 1,000 members in the human genome. Recently, it has become evident that some of the members of this superfamily of oxidoreductases are involved in regulating inflammatory processes. Other members of the oxidoreductase superfamily that may play important roles in inflammatory processes include nucleoredoxin (41), which significantly enhances tumor necrosis factor-α-induced NF-κB reporter activity (42); PICOT, a 37.5-kDa PKCθ-interacting protein that regulates AP-1 and NF-κB transcription (43); and TRP32, a 32-kDa protein that interacts with the catalytic domain of the protein kinase MST (44). The chemogenomics approach we used to develop the selective Ref-1 inhibitor PNRI-299 can be used to validate these oxidoreductase superfamily members as important targets for the design of agents to treat asthma and other inflammatory disorders.
Acknowledgments
We thank Dr. Tom Curran (St. Jude Children's Research Hospital, Memphis, TN) for providing the Ref-1 expression vector; Gertrude Chiang, Falaah Jones, Peter Ong Lim, and Ying-tzang Tien for excellent technical assistance; and Rachel Norris for typing this manuscript. This work was supported by National Institutes of Health Grant AI42989 and the Choong Wae Pharmaceutical Corporation, Seoul, Korea.
Abbreviations
- AP-1
activator protein-1
- NF-κB
nuclear factor κB
- Ref-1
redox effector factor-1
- Th
T helper
- TRX
thioredoxin
- OVA
ovalbumin
References
- 1.Holgate S T. Nature. 1999;402:B2–B4. doi: 10.1038/35037000. [DOI] [PubMed] [Google Scholar]
- 2.Nordberg J, Arner E S J. Free Radical Biol Med. 2001;31:1287–1312. doi: 10.1016/s0891-5849(01)00724-9. [DOI] [PubMed] [Google Scholar]
- 3.Saleh D, Ernst P, Lim S, Barnes P J, Giaid A. FASEB J. 1998;12:929–937. [PubMed] [Google Scholar]
- 4.MacPherson J C, Comhair S A, Erzurum S, Klein D F, Lipscomb M F, Kavuru M S, Samosznk M K, Hazen S L. J Immunol. 2001;166:5763–5772. doi: 10.4049/jimmunol.166.9.5763. [DOI] [PubMed] [Google Scholar]
- 5.Rahman I, MacNee W. Eur Respir J. 2000;16:534–554. doi: 10.1034/j.1399-3003.2000.016003534.x. [DOI] [PubMed] [Google Scholar]
- 6.Brightling C E, Symon F A, Birring S S, Bradding P, Pavord I D, Wardla A J. J Allergy Clin Immunol. 2002;110:899–905. doi: 10.1067/mai.2002.129698. [DOI] [PubMed] [Google Scholar]
- 7.Barnes P J. Respir Med. 2002;96, Suppl. A:S9–S15. [PubMed] [Google Scholar]
- 8.Adcock I M, Lane S J, Brown C R, Lee T H, Barnes P J. J Exp Med. 1995;192:1951–1958. doi: 10.1084/jem.182.6.1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Barnes P J, Adcock I M. Eur Respir J. 1998;12:221–234. doi: 10.1183/09031936.98.12010221. [DOI] [PubMed] [Google Scholar]
- 10.Sasada T, Sono H, Yodoi J. J Toxicol Sci. 1996;21:285–287. doi: 10.2131/jts.21.5_285. [DOI] [PubMed] [Google Scholar]
- 11.Schenk H, Klein M, Erdbrugger W, Droge W, Schulze-Osthoff K. Proc Natl Acad Sci USA. 1994;91:1672–1676. doi: 10.1073/pnas.91.5.1672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Timblin C R, Janssen Y M, Goldberg J L, Mossman B T. Free Radical Biol Med. 1998;24:632–642. doi: 10.1016/s0891-5849(97)00325-0. [DOI] [PubMed] [Google Scholar]
- 13.Singh N, Aggarwal S. Int J Cancer. 1995;62:107–114. doi: 10.1002/ijc.2910620120. [DOI] [PubMed] [Google Scholar]
- 14.Abate C, Patel L, Rauscher F J, III, Curran T. Science. 1990;249:1157–1161. doi: 10.1126/science.2118682. [DOI] [PubMed] [Google Scholar]
- 15.Qin J, Clore G M, Kennedy W M, Huth J R, Gronenborn A M. Structure (London) 1995;3:289–297. doi: 10.1016/s0969-2126(01)00159-9. [DOI] [PubMed] [Google Scholar]
- 16.Yodoi J, Okada M, Tagaya Y, Taniguchi Y, Teshigawara K, Kasahara T, Dinarello C A, Matsushima K, Honko T, Uchiyama T, et al. Adv Exp Med Biol. 1987;213:139–148. doi: 10.1007/978-1-4684-5323-2_14. [DOI] [PubMed] [Google Scholar]
- 17.Marshall H E, Stamler J S. Biochemistry. 2001;40:1688–1693. doi: 10.1021/bi002239y. [DOI] [PubMed] [Google Scholar]
- 18.Marshall H E, Merchant K, Stamler J S. FASEB J. 2000;14:1889–1900. doi: 10.1096/fj.00.011rev. [DOI] [PubMed] [Google Scholar]
- 19.Ogbu C O, Qabar M N, Boatman P D, Urban J, Meara J P, Ferguson M D, Tulinsky J, Lum C, Babu S, Blaskovich M A, et al. Bioorg Med Chem Lett. 1998;8:2321–2326. doi: 10.1016/s0960-894x(98)00420-x. [DOI] [PubMed] [Google Scholar]
- 20.Eguchi M, Lee M, Nakanishi H, Stasiak M, Lovell S, Kahn M. J Am Chem Soc. 1999;121:12204–12205. [Google Scholar]
- 21.Boatman P D, Ogbu C O, Eguchi M, Kim H O, Nakanishi H, Cao B, Shea J P, Kahn M. J Med Chem. 1999;42:1367–1375. doi: 10.1021/jm980354p. [DOI] [PubMed] [Google Scholar]
- 22.Misra-Press A, McMillan M, Cudaback E, Khoury D, Kozu G, Thompson P, Qabar M, Ruan F, Nguyen M, Mathew J, et al. Curr Med Chem Antiinflamm Antiallergy Agents. 2002;1:29–39. [Google Scholar]
- 23.Lazebnik Y A, Kaufmann S H, Desnoyers S, Poirier G G, Earnshaw W C. Nature. 1994;371:346–347. doi: 10.1038/371346a0. [DOI] [PubMed] [Google Scholar]
- 24.Henderson W R, Jr, Lewis D B, Albert R K, Zhang Y, Lamm W J, Chiang G K, Jones F, Eriksen P, Tien Y T, Jonas M, Chi E Y. J Exp Med. 1996;184:1483–1494. doi: 10.1084/jem.184.4.1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Oh S-W, Pae C I, Lee D-K, Jones F, Chiang G K, Kim H O, Moon S-H, Cao B, Ogbu C, Jeong K-W, et al. J Immunol. 2002;168:1992–2000. doi: 10.4049/jimmunol.168.4.1992. [DOI] [PubMed] [Google Scholar]
- 26.Henderson W R, Jr, Chi E Y, Albert R K, Chu S J, Lamm W J, Rochon Y, Jonas M, Christie P E, Harlan J M. J Clin Invest. 1997;100:3083–3092. doi: 10.1172/JCI119863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rosania G R, Chang Y T, Perez O, Sutherlin D, Dong H, Lockhart D J, Schultz P G. Nat Biotechnol. 2000;18:304–308. doi: 10.1038/73753. [DOI] [PubMed] [Google Scholar]
- 28.Xanthoudakis S, Miao G, Wang F, Pan Y C, Curran T. EMBO J. 1992;11:3323–3335. doi: 10.1002/j.1460-2075.1992.tb05411.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yan M, Xu W, Lu L, Sun L, Liu X, Zheng Z. Biochem Biophys Res Commun. 2000;278:462–469. doi: 10.1006/bbrc.2000.3826. [DOI] [PubMed] [Google Scholar]
- 30.Xu Y J, Kim E Y, Demple B. J Biol Chem. 1998;273:28837–28844. doi: 10.1074/jbc.273.44.28837. [DOI] [PubMed] [Google Scholar]
- 31.Xanthoudakis S, Miao G G, Curran T. Proc Natl Acad Sci USA. 1994;91:23–27. doi: 10.1073/pnas.91.1.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Walker L J, Robson C N, Black E, Gillespie D, Hickson I D. Mol Cell Biol. 1993;13:5370–5376. doi: 10.1128/mcb.13.9.5370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gorman M A, Morera S, Rothwell D G, de La Fortelle E, Mol C D, Tainer J A, Hickson I D, Freemont P S. EMBO J. 1997;16:6548–6558. doi: 10.1093/emboj/16.21.6548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rooney J W, Sun Y L, Glimcher L H, Hoey T. Mol Cell Biol. 1995;15:6299–6310. doi: 10.1128/mcb.15.11.6299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Burke T F, Casolaro V, Georas S N. Biochem Biophys Res Commun. 2000;270:1016–1023. doi: 10.1006/bbrc.2000.2508. [DOI] [PubMed] [Google Scholar]
- 36.Loots G G, Locksley R M, Blankespoor C M, Wang Z E, Miller W, Rubin E M, Frazer K A. Science. 2000;288:136–140. doi: 10.1126/science.288.5463.136. [DOI] [PubMed] [Google Scholar]
- 37.Mori A, Kaminuma O, Mikami T, Inoue S, Okumura Y, Akiyama K, Okudaira H. J Allergy Clin Immunol. 1999;103:S429–S436. doi: 10.1016/s0091-6749(99)70158-2. [DOI] [PubMed] [Google Scholar]
- 38.Maclan F, Garcia-Rodriguez C, Rao A. EMBO J. 2000;19:4783–4785. doi: 10.1093/emboj/19.17.4783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fallon P G, Jolin H E, Smith P, Emson C L, Townsend M J, Fallon R, McKenzie A N. Immunity. 2002;17:7–17. doi: 10.1016/s1074-7613(02)00332-1. [DOI] [PubMed] [Google Scholar]
- 40.Flaherty D M, Monick M M, Carter A B, Peterson M W, Hunninghake G W. Am J Respir Cell Mol Biol. 2001;25:254–259. doi: 10.1165/ajrcmb.25.2.4446. [DOI] [PubMed] [Google Scholar]
- 41.Kurooka H, Kato K, Minoguchi S, Takahashi Y, Ikeda J, Habu S, Osawa N, Buchberg A M, Moriwaki K, Shisa H, Honjo T. Genomics. 1997;39:331–339. doi: 10.1006/geno.1996.4493. [DOI] [PubMed] [Google Scholar]
- 42.Hirota K, Matsui M, Murata M, Takashima Y, Cheng F S, Itoh T, Fukuda K, Junji Y. Biochem Biophys Res Commun. 2000;274:177–182. doi: 10.1006/bbrc.2000.3106. [DOI] [PubMed] [Google Scholar]
- 43.Witte S, Villalba M, Bi K, Liu Y, Isakov N, Altman A. J Biol Chem. 2000;275:1902–1909. doi: 10.1074/jbc.275.3.1902. [DOI] [PubMed] [Google Scholar]
- 44.Lee K K, Murakawa M, Takahashi S, Tsubuki S, Kawashima S, Sakamaki K, Yonehara S. J Biol Chem. 1998;273:19160–19166. doi: 10.1074/jbc.273.30.19160. [DOI] [PubMed] [Google Scholar]