Abstract
We evaluated 966 consecutive pediatric patients with various developmental disorders by high-resolution microarray-based comparative genomic hybridization and found 10 individuals with pathogenic copy number variants (CNVs) on the short arm of chromosome 8 (8p), representing approximately 1% of the patients analyzed. Two patients with 8p terminal deletion associated with interstitial inverted duplication (inv dup del(8p)) had different mechanisms leading to the formation of a dicentric intermediate during meiosis. Three probands carried an identical ∼5.0 Mb interstitial duplication of chromosome 8p23.1. Four possible hotspots within 8p were observed at nucleotide coordinates of ∼10.45, 24.32–24.82, 32.19–32.77, and 38.94–39.72 Mb involving the formation of recurrent genomic rearrangements. Other CNVs with deletion- or duplication-specific start or stop coordinates on the 8p provide useful information for exploring the basic mechanisms of complex structural rearrangements in the human genome.
Keywords: microarray-based comparative genomic hybridization, short arm of chromosome 8, copy number variant, genomic disorders
Introduction
The short arm of human chromosome 8 (8p) spans about 44 million base pairs containing 484 annotated genes (NCBI Build 36.3 of the human genome).1 Point mutations in more than 50 genes on the 8p are associated with various genetic disorders and diseases (http://www.ncbi.nlm.nih.gov/omim).
8p is especially prone to various genomic rearrangements mainly because of the existence of the two olfactory receptor gene clusters (REPD and REPP) flanking an ∼5 Mb region of 8p23.1.2, 3, 4, 5, 6 These REPD- and/or REPP-related 8p genomic rearrangements include (1) the 8p23.1 deletion or duplication between REPD and REPP,6, 7, 8, 9 (2) the 8p23.1 paracentric inversion between REPD and REPP,8, 10 (3) the pericentric inversion (inv(8)(p23.1q22.1)) and recombinant chromosome 8 (rec(8)dup(8q)inv(8)(p23.1q22.1)),11 (4) the 8p interstitial inverted duplication with associated terminal deletion (inv dup del(8p)),5, 6, 8, 10, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24 (5) the 8p translocations involving the 8p23.1,25, 26 and (6) different types of supernumerary chromosome 8 (SMC(8)) involving the breakpoints within 8p23.1.4, 27 In addition to these defined 8p genomic abnormalities, other pathogenic genomic changes have been identified,28, 29, 30 whereas numerous genomic imbalances on 8p are still described as copy number variants (CNVs) of unknown clinical significance or CNVs without apparent clinical significance (benign CNVs) (http://projects.tcag.ca/variation).
In this study, we describe a comprehensive CNV profile of 8p derived from the tests of a large number of pediatric patients with diverse clinical phenotypes using a high-resolution microarray-based comparative genomic hybridization (aCGH) platform, and discuss plausible mechanisms for the formation of these genomic rearrangements.
Materials and methods
Specimen acquisition
The DNA samples used for this research study came from 966 consecutive pediatric patients referred for genome-wide screen testing by aCGH in our laboratory. Each patient was evaluated because of one or more of the following categorical clinical findings: global developmental delay, autism, dysmorphism, seizures, or multiple congenital anomalies. The study protocol was approved by the institutional review board of Children's Mercy Hospitals and Clinics.
Testing using chromosome banding analysis (GTG banding) and fluorescence in situ hybridization technique
Standard GTG banding analyses were performed on the peripheral blood previously (patients 2, 3, 4, 6, 8, and 10) or concurrently (patients 1, 5 and 7, 8, 9), and on the cultured skin fibroblast cells (patient 10). Fluorescence in situ hybridization (FISH) tests were performed for patient 3 using an 8p subtelomeric probe (AFM 197XG5), and for patient 10 using a chromosome 8 centromeric probe (CEP 8) as recommended by the manufacturer (Abbott Molecular Inc., Des Plaines, IL, USA).
Testing using microarray-based comparative genomic hybridization
The aCGH platform used in this study is the Agilent Human Genome Microarray Kit 244K, a genome-wide screening platform, containing 10 960 distinct 60-mer oligonucleotide probes on chromosome 8, with average probe spacing of 13.3 kb (Agilent Technologies, Santa Clara, CA, USA). All aCGH tests were performed and analyzed following the protocols described previously.31
aCGH result verification and parental follow-up
The GTG banding analysis results for patients 1, 2, 3, 7, and 10 and FISH results for patients 2, 3, and 10 were used for verification of the abnormal aCGH findings on 8p (Table 1; Supplementary Figure S1). Quantitative real-time PCR (qPCR) were performed as previously described for verification of some abnormal aCGH findings on 8p using different test primers targeting genes within corresponding abnormal regions (RECQL4 for patient 1, GATA4 gene for patients 2-6, DLC1 gene for patient 3, NKX3 gene for patient 8, and MAK16 for patient 9 (Table 1; Supplementary Table S1)).32 GTG banding, FISH, or qPCR was also carried out for available corresponding parental follow-up studies in this study (Table 1).
Table 1. Pathogenic CNVs on the short arm of chromosome 8.
| Patient | Chromosome and FISH findings | aCGH findings | Start coordinate (bp) | Stop coordinate (bp) | Size (bp) | Verification for aCGH findings | Parents availability, and test findings |
|---|---|---|---|---|---|---|---|
| 1 | 46,XY,del(8)(p23.1) [C] | 8p23.3p23.1 × 1 | 1 | 10 453 863 | 10 453 862 | GTG banding | Both parents, normal findings by GTG banding |
| 2 | 46,XY,add(8)(p12).ish add(8)(P12)(wcp8+) [R] | 8p23.3p23.1 × 1 8p23.1p21.2 × 3 8q24.3 × 3 | 1 12 601 939 145 084 438 | 6 929 891 24 823 981 146 265 046 | 6 929 890 12 222 042 1 180 608 | GTG banding, FISH and qPCR | Both parents, normal findings by GTG banding |
| 3 | 46,XX,add(8)(p23.1).ish add(8)(p23.1)(8pter−) [R] | 8p23.3p23.1 × 1 8p23.1p21.2 × 3 | 1 7 358 840 | 6 913 476 25 707 713 | 6 913 475 18 348 873 | GTG banding, FISH and qPCR | Both parents, normal findings by GTG banding and qPCR |
| 4 | 46,XX [R] | 8p23.1 × 3 | 7 256 029 | 11 898 119 | 4 642 090 | qPCR | Inherited from mother; father NA |
| 5 | 46,XY,dup(8)(p23.1p23.2) [C] | 8p23.1 × 3 | 7 256 029 | 12 512 055 | 5 256 026 | qPCR | Both parents, normal findings by qPCR |
| 6 | 46,XY,var(8)(p23.1p23.2) [R] | 8p23.1 × 3 | 7 358 840 | 11 904 101 | 4 545 261 | qPCR | Mother normal by qPCR; father NA |
| 7 | 46,XY,del(8)(p12p22) [C] | 8p22p12 × 1 | 18 375 250 | 32 765 636 | 14 390 386 | GTG banding | Both parents, normal findings by GTG banding |
| 8 | 46,XX,t(8;18;14) (p21.3;q23.1; q24.1) [R] | 2q35q36.1 × 1 4q35.2 × 1 8p21.3p21.2 × 1 14q22.2 × 1 | 217 249 378 188 428 673 22 680 632 54 012 870 | 222 041 732 189 965 537 24 395 832 54 614 836 | 4 792 354 1 536 864 1 715 200 601 966 | qPCR | Both parents, normal findings by GTG banding and qPCR |
| 9 | 46,XY [C] | 8p12 × 1 | 32 189 595 | 35 280 851 | 3 091 256 | qPCR | Both parents, inherited from father |
| 10 | 47,XY,+mar.ish mar(8) (D8Z2+) from blood; 47,XY,+mar[20]/46,XY[7] from skin [R] | 8p11.23q11.1 × 3 | 38 944 542 | 47 575 850 | 8 631 308 | GTG banding and FISH | Both parents, normal findings by GTG banding |
Abbreviations: C, concurrent chromosome analysis; R, retrospective chromosome analysis; Dup/Del, duplication/deletion; NA, not available.
Results
Results from GTG banding and FISH testing
The GTG banding and FISH testing results are summarized in Table 1. Of the 10 patients with abnormal aCGH results, 2 showed normal karyotypes (patients 4 and 9) and the remaining patients appeared to have chromosomal rearrangements (Table 1). Images from GTG banding and/or FISH testing for patients 2, 3, and 10 are presented in Supplementary Figure S1.
Pathogenic CNVs on 8p identified by aCGH-244K
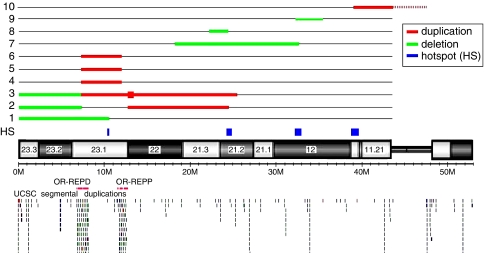
Pathogenic 8p CNVs were found in 10 individuals (patients 1–10) in our study, representing ∼1% of the patients analyzed (Figure 1; Table 1). There are no other pathogenic genomic abnormalities except in patients 2 and 8. Patient 1 has a 10.45 Mb terminal deletion with proximal breakpoint residing between the MSRA and RP1L1 genes within the REPD–REPP region of 8p23.1 (Figure 1; Supplementary Figure S2a). Two patients (patients 2 and 3) have inv dup del(8p) (Figure 1; Supplementary Figure S2a). Patient 2 has three genomic imbalances, a 6.93 Mb terminal deletion, a 12.22 Mb interstitial duplication of 8p, and a 1.18 Mb terminal duplication of 8q. Patient 3 has two genomic imbalances, a 6.91 Mb terminal deletion, and an 18.35 Mb interstitial duplication of 8p. Within the 18.35 Mb interstitial duplication, there is a 563 kb quintuplicated region, calculated by the base-2 logarithm ratio of 1.33 in an aCGH analysis. Both patients 2 and 3 have an ∼6.90 Mb terminal deletion of 8p with the breakpoints residing within the REPD region. The distal breakpoints of the 8p23.1p21.2 duplications reside within REPP in patient 2 and REPD in patient 3, respectively. Three patients (4, 5, and 6) have an ∼5.0 Mb interstitial duplication of 8p23.1 flanked by REPD and REPP (Figure 1; Supplementary Figure S2b). The proximal breakpoint of the 14.39 Mb deletion in patient 7, and the distal breakpoint of the 3.09 Mb deletion in patient 9 all reside within Neuregulin 1 (NRG1) gene within the 8p11.23 region (Figure 1; Supplementary Figure S2c). aCGH testing identified four cryptic de novo deletions in four chromosomes in patient 8 (Supplementary Figure S1c), including one of them with involvement of 8p21.3p21.2 (Figure 1; Supplementary Figure S2c). Patient 10 has a de novo mosaic pericentric SMC(8) (47,XY,+mar[20]/46,XY[7]) (Supplementary Figure S1d). aCGH testing in this patient identified an 8.63 Mb duplication of 8p11.23q11.1 (Figure 1; Supplementary Figure S2c).
Figure 1.
Locations of genomic abnormalities identified on chromosome 8p in this study. Numbers 1–10 represent the two patients with genomic abnormalities on chromosome 8p.
Verification of abnormal aCGH results in probands and parental follow-up studies
The abnormal aCGH findings were verified in patients 1, 7, and 10 or partially verified in patients 2 and 3 by GTG banding and FISH results (Supplementary Figure S1a–c). The remaining abnormal aCGH results were verified by qPCR (Table 1; Supplementary Table S1).
Eighteen parental samples (eight couples and two single parents) were available for determination of the inheritance pattern of the genomic abnormalities in their respective children (Table 1). The ∼4.6 Mb duplication of 8q23.1 in patient 4 was inherited from the mother; the ∼4.5 Mb duplication of 8q23.1 in patient 6 was negative in the mother, but the father was not available for testing; the ∼3.1 Mb deletion of 8p12 in patient 9 was inherited from the father. The remaining tested parental samples showed negative results (Table 1).
Benign CNVs on 8p identified by aCGH-244K
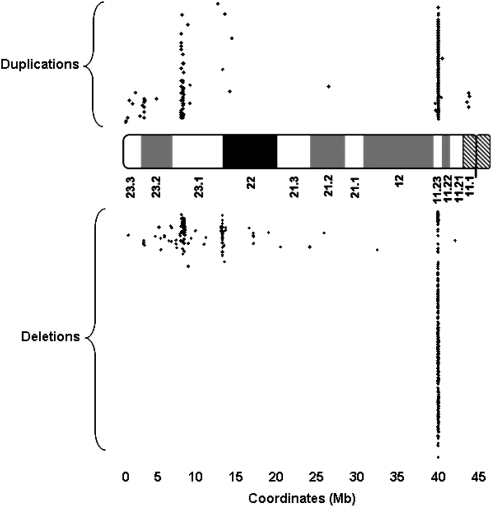
In addition to the pathogenic genomic abnormalities listed in Table 1, 915 benign CNVs were observed on 8p in this study, 482 deletions and 433 duplications (Figure 2; Supplementary Table S2). The majority of these benign CNVs (93%) reside within three regions: 149 CNVs (50 dup/99 del) in region 1 (6 706 776–8 349 519 of the REPD region), 31 CNVs (2 dup/41 del) in region 2 (11 903 701–12 630 984 of the REPP region), and 655 CNVs (349 dup/306 del) in region 3 (39 323 211–39 586 403) (Figure 2; Supplementary Table S3).
Figure 2.
Distribution of benign copy number variants (CNVs) across 8p. Each dot represents a single benign CNV.
The 915 benign CNVs derived from the combination of 81 start and 85 stop breakpoints (Supplementary Tables S4 and S5). Although most benign CNV regions showed both deletion and duplication, deletion- or duplication-specific benign CNVs were also observed in this study (Supplementary Table S2). Some start (Supplementary Table S4) or stop (Supplementary Table S5) breakpoints existed only in deleted CNVs and others occurred only in duplicated CNVs.
Clinical features in 10 patients with genomic abnormalities on 8p
Clinical justification for aCGH testing in these 10 patients with genomic abnormalities on 8p varied, but global development delay and dysmorphic features were the most common indications. Detailed description about the clinical findings in these patients is beyond the scope of this article, but is available in Supplementary Table S6.
Discussion
aCGH techniques have revolutionized the understanding of the human genome structure and are rapidly becoming new standard methods for clinical cytogenetics.33 These techniques have facilitated the identification of novel genomic disorders and precisely defined the breakpoint(s) of various genomic abnormalities. The findings in this study provide additional perspectives in viewing the underlying mechanisms causing genomic rearrangements on 8p.
REPD and REPP-mediated inv dup del(8p)
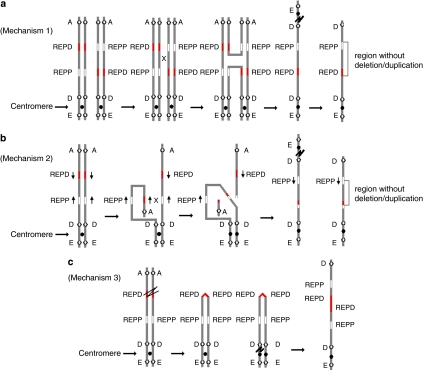
Patients 2 and 3 in this study are postulated to have inv dup del(8p). Three mechanisms may explain the origin of inv dup del(8p) (Figure 3) with detailed explanations referred to the recent reports.4, 8, 24, 34 In brief, all three mechanisms involve the formation of a dicentric chromosome 8 (dic(8)) in meiosis I followed by breakage of the dic(8) either during meiotic division or during early stages of embryonic development leading to the production of an inv dup del(8p). However, the events causing the formation of a dic(8) differ among the three mechanisms.
Figure 3.
(a) Mechanism 1 in which the chromosome carrying the paracentric inversion between REPD and REPP pairs with its homologue by forming an inversion loop. Crossing-over and recombination within the loop create an unstable dicentric chromosome and an acentric fragment. The dicentric chromosome breaks outside the inverted region lead to the formation of a monocentric chromosome with a terminal deletion and an inverted duplication with a single copy region between the duplication. (b) Mechanism 2 in which the inverted LCRs within REPD or REPP in the same short arm of chromosome 8. Pairing and recombination between the inverted repeats on sister chromatids results in the formation of a dicentric chromosome and an acentric fragment. Breakage of the dicentric outside the inverted repeats leads to a monocentric chromosome with a terminal deletion and an inverted duplication with a single copy region between the duplication, which will be flanked by the inverted repeats. (c) Mechanism 3, which involves an initial premeiotic double-strand break of the two sister chromatids. Fusion of the broken ends results in a symmetric U-type reunion between the sister chromatids leading to the formation of a dicentric chromosome. Breakage distal to the fusion site outside the fusion region results in a monocentric chromosome with a terminal deletion and an inverted duplication without a single copy region between the duplication.
Mechanism 1 involves a single parent (usually maternal) who carries a paracentric inversion of 8p23.1 between REPD and REPP. During meiosis I, the chromosome carrying the paracentric inversion pairs with its normal homologue by forming an inversion loop. Crossing-over and recombination within the loop create an unstable dic(8). Mechanism 2 involves the presence of inverted LCRs within the REPD or REPP of 8p. Partial folding of one homologue onto itself with a recombination event between the inverted repeats leads to the formation of a dic(8). Mechanism 3 involves an initial premeiotic double-strand break of the two sister chromatids of 8p. Fusion of the broken ends results in a symmetric U-type reunion between the sister chromatids leading to the formation of a dic(8). Both mechanisms 1 and 2 produce inv dup del(8p) with a single copy region between the duplicated regions on the derivative chromosome 8, whereas mechanism 3 produces inv dup del(8p) without a single copy region between the duplicated regions. The difference between mechanisms 1 and 2 is that the single copy region between the duplications in mechanism 2 will be flanked by the inverted repeats.
The inv dup del(8p) in patient 2 should be explained by mechanism 2 because of the possible existence of inverted LCRs within the REPD or REPP on 8p, whereas mechanism 3 should explain the formation of inv dup del(8p) in patient 3. The majority of the reported patients with inv dup del(8p) should be explained by mechanism 25, 6, 8, 10, 12, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23 with few exceptions (Table 2), which could be explained by mechanism 3.13, 24 This finding is in contrast to the observation that mechanism 3 is the most frequent mechanism for this type of genomic rearrangements in all other chromosome arms.24 Although the inv dup del(8p) in patients 3 and two previously reported patients (Table 2) was caused by mechanism 3,13 there are several differences among them. First, although there are no LCRs or other repeat sequences around the breakpoints of the 3.9 Mb terminal deletion in published case B and the 619 kb terminal deletion in published case C, the breakpoint of the 6.91 Mb terminal deletion in patient 3 resides within the 8p REPD region, indicating that an unstable REPD region could facilitate the initiation of a double-strand break of the two sister chromatids and the formation of a symmetric U-type reunion between the sister chromatids producing a dic(8). Second, both the distal and proximal breakpoint sites of the 8p-duplicated regions in these patients are different (Table 2). Third, the mechanisms to stabilize the broken chromosome ends are different, with telomere capture in reported case of patient B, but not in reported case of patient C and our patient 3 (Table 2).
Table 2. Comparison of findings in patients 2 and 3 in this study with published case reports.
| Patient/published case report | Findings | Start coordinate (Mb) | Stop coordinate (Mb) | Size (Mb) | Formation of dicentric chromosome | Telomere capture |
|---|---|---|---|---|---|---|
| Patient 2 in this study | 8pterp23.1 × 1 8p23.1p21.2 × 3 8q24.3qter | 0 12.60 145.08 | 6.93 24.82 146.26 | 6,93 12,22 1,18 | NAHR (mechanism 2) with involvement of REPD and REPP | Yes? |
| Patient 3 in this study | 8pterp23.1 × 1 8p23.1p21.2 × 3 | 0 7.36 | 6.91 25.71 | 6,91 18,35 | U-type exchange (mechanism 3) with involvement of REPD | No |
| Case A in Buysse et al13 | 8pterp23.1 × 1 8p23.1p21.2 × 3 8q24.13qter | 0 12.63 125.34 | 6.90 16.97 146.25 | 6,90 3.40 20.90 | NAHR (mechanism 2) with involvement of REPD and REPP | Yes |
| Case B in Buysse et al13 | 8pterp23.1 × 1 8p23.1p21.2 × 3 8q24.13qter | 0 4.48 117.93 | 3.90 24.31 146.25 | 3,90 19.83 28.30 | U-type exchange (mechanism 3) without involvement of REPD or REPP | Yes |
| Case C in Rowe et al24 | 8pterp23.1 × 1 8p23.1p21.2 × 3 | 0 0.62 | 0.61 37.79 | 0.61 18,35 | U-type exchange (mechanism 3) without involvement of REPD or REPP | No |
| Cases D–H in Shimokawa et al10 | 8pterp23.1 × 1 8p23.1p21.2 × 3 | 0 12.62 | 5.8–8.3 ∼39.72 | 5.8–8.3 26–31 | NAHR (mechanism 2) with involvement of REPD and REPP | No |
Abbreviation: NAHR, nonallelic homologous misalignment and unequal recombination.
For explanations of ‘mechanism 2' and ‘mechanism 3', see text.
Terminal deletions can result in severe genomic instability if not properly repaired.24 Thus, broken chromosome ends must acquire a new telomeric cap to remain structurally stable by ‘telomere healing,' in which telomeric sequences can be acquired de novo,24 or by ‘telomere capture,' in which broken chromosome obtains the telomeric end of another chromosome.35, 36, 37 Alternatively, stabilization can occur through circularization of the inv dup del chromosome, leading to the formation of a ring chromosome.38
In addition to the 6.93 Mb 8p terminal deletion associated with a 12.22 Mb interstitial duplication of 8p23.1p21.2, an additional 1.18 Mb terminal duplication of 8q was identified by aCGH in patient 2 (Figure 1; Table 1). It is possible that the broken chromosome end of 8p has been stabilized by telomere capture through an additional rearrangement with distal 8q material, similar to three previously reported patients with inv dup del(8p).13, 39 No additional tissue from our patient 2 was available for confirmation of this hypothesis using FISH methods.
The distal end of the 563 kb de novo quintuplicated region in patient 3 exists just at the proximal end of the LCR–REPP region. It is likely that the occurrence of this quintuplicated region may involve more complex genomic rearrangements than we postulate here. However, the mechanism causing this complexity is unknown.
8p23.1 duplication
Although more than 10 patients having isolated 8p23.1 duplication have been reported,7, 40, 41, 42 only 6 of them in four families have been characterized by FISH or aCGH.7, 42 Three probands (patients 4–6) in this study were observed to have the 8p23.1 duplication (Table 1; Figure 1; Supplementary Figure S1b). The 8p23.1 duplication in patient 4 was inherited from the affected mother. The 8p23.1 duplication in patient 5 occurred de novo. The inheritance pattern for the duplication in patient 6 could not be determined because of negative results in the mother and the unavailability of the father.
This ∼5.0 Mb duplication of 8p23.1 is flanked by REPD and REPP, which share a high level of identity of complex genomic repeats involving retroviral elements, olfactory repeat regions, and variable copy numbers of defensin genes.4, 7, 8, 42 The 8p23.1 duplication is likely caused by intrachromosomal recombination between REPD and REPP within the 8p23.1 region through LCR-mediated nonallelic homologous misalignment and unequal recombination (NAHR) during meiosis.6, 7, 8, 9, 43, 44
The copy numbers of different types of defensin and olfactory receptor genes in the REPD and REPP regions are highly variable, leading to cytogenetically visible euchromatic variants if the copies are sufficiently high.4, 45, 46 However, these euchromatic variants are indistinguishable from the genuine 8p23.1 duplication under a light microscope. For example, two of the three patients with 8p23.1 duplication in this study were initially reported as euchromatic variants to the referring physicians. These results show the power of aCGH with high resolution to reveal unexpected imbalances in affected patients without specific clinical findings as well as the sensitivity to discriminate genuine 8p23.1 duplication from euchromatic variants caused by high copies of defensin and olfactory receptor genes. On the basis of the finding of three probands with 8p23.1 duplication in this study, or a 0.31% detection rate, we suspect that the 8p23.1 duplication may be more common than previously suspected.7
Rare genomic abnormalities reveal the possible existence of multiple hotspots leading to variable genomic rearrangements on 8p
The specific genomic abnormalities of patients 1 and 7–10 in this study have not been previously reported. However, these rare genomic abnormalities reveal the occurrence of several unstable regions leading to recurrent genomic rearrangements on 8p. The proximal breakpoint of the 10.45 Mb deletion within the REPD–REPP region of 8p23.1 in patient 1 was recently reported in a transmitted 8p23.1p23.2 duplication in an individual with autism.47
Patient 8 in this study was found to have an apparently balanced complex translocation (46,XX,t(8;18;14)(p21.3;q23.1;q24.1)), but aCGH testing identified four cryptic de novo deletions of possible clinical significance (Table 1; Supplementary Figure S2c). The two deletions of 2q35q36.1 and 4q35.2 were not apparently associated with the complex translocation. Either the two deletions occur independently without involvements of the complex translocation, or more complex chromosomal rearrangements with involvements of the two deletions were missed by chromosome analysis. However, there is no available specimen from this patient to further verify the complexity of the chromosomal rearrangements. Such discoveries support the hypothesis that some ‘balanced' rearrangements include cryptic deletions or more complex rearrangements,48 further emphasizing the benefit of high-resolution aCGH analysis. The proximal breakpoint of the 1.72 Mb deletion of 8p21.3p21.2 in this patient is flanked by repeat-rich sequences, indicating that this region could be an unstable region for genomic arrangements. This hypothesis is supported by the observation that the proximal breakpoints of the 12.82 Mb duplication in patient 2 and the 19.83 Mb duplication in reported case 2 reside within the same gene desert region (Table 2). In addition, the proximal breakpoint of the duplication in reported case 4 appeared to map to this region (Table 2).5
The NRG1 gene was disrupted in two of the ten abnormal patients (patients 7 and 9) in this study. Breakpoints of genomic rearrangements occurring in the NRG1 gene were observed in other reported cases with 8p genomic abnormalities,49, 50 indicating that unknown genomic structural features within the NRG1 gene may be susceptible to these various genomic rearrangements. Alternatively, patients with disruption of this gene may be more likely to be investigated because of the impact of the NRG1 gene on cardiac and neural development.
More than 60 cases with SMC(8) have been reported (http://www.med.uni-jena.de/fish/sSMC/08.htm). Ten published cases with SMC(8) include genomic material similar to patient 10 in this study.27, 51 Although no annotated repeat sequences occur around the breakpoints of this SMC(8) in patient 10, the 8p breakpoint (38.94 Mb) of this SMC(8) is adjacent to the proximal breakpoint (∼39.72 Mb) of the inverted duplicated regions in five reported cases with inv dup del(8p).10 This region appears to be extremely unstable because 72% (847 of 915) of the benign CNVs in this study carry proximal and/or distal breakpoints within the region (Figure 2; Table 2). On the basis of these rare genomic abnormalities on 8p, we may conclude that the regions with nucleotide coordinates at ∼10.45, 24.32–24.82, 32.19–32.77, and 38.94–39.72 Mb on 8p are particularly susceptible to genomic rearrangements.
Mechanisms leading to the formation of benign CNVs on 8p
Ninety-three percent of the benign CNVs on 8p identified in this study reside within three regions: the REPD and REPP regions of 8p23.1, and the 8p11.23 region (Figure 2; Supplementary Table S3). These results are concordant to the data documented in the Database of Genomic Variants (http://projects.tcag.ca/variation/), reflecting the extreme instability of these three regions. Variable copy numbers of defensin and olfactory receptor genes within the REPD and REPP regions,4 and the large numbers of simple tandem repeats within the 8p11.23 region should be the structural basis leading to the formation of these benign copy variations.
Different underlying mechanisms were proposed to explain the formation of CNVs based on the structural features of the human genome.52, 53, 54, 55, 56 However, none of the proposed mechanisms can fully explain the occurrence of some deletion- or duplication-specific start or stop breakpoints of CNVs and some deletion- or duplication-specific CNVs observed in this study (Figure 2; Supplementary Tables S2–S5). The actual mechanisms leading to this phenomenon remain unknown, and it is possible that yet-to-be discovered mechanisms prohibit the formation of certain deletions or duplications.
In summary, we report an integrated high-resolution CNV profile of human chromosome 8p derived from the analysis of 966 consecutive individuals using the aCGH-244K platform, and present plausible mechanisms for the formation of inv dup del(8p) and 8p23.1 duplication. Several regions within 8p are proposed to be hotspots leading to the formation of recurrent genomic rearrangements. CNVs with deletion- or duplication-specific start or stop breakpoints provide useful information for exploring underlying mechanisms leading to the formation of complex structural rearrangements in the human genome.
Acknowledgments
We thank Dr Holly H Ardinger for the critical review and comments on the article.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies the paper on European Journal of Human Genetics website (http://www.nature.com/ejhg)
Supplementary Material
References
- Nusbaum C, Mikkelsen TS, Zody MC, et al. DNA sequence and analysis of human chromosome 8. Nature. 2006;439:331–335. doi: 10.1038/nature04406. [DOI] [PubMed] [Google Scholar]
- Vermeesch JR, Thoelen R, Salden I, Raes M, Matthijs G, Fryns JP. Mosaicism del(8p)/inv dup(8p) in a dysmorphic female infant: a mosaic formed by a meiotic error at the 8p OR gene and an independent terminal deletion event. J Med Genet. 2003;40:e93. doi: 10.1136/jmg.40.8.e93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giorda R, Ciccone R, Gimelli G, et al. Two classes of low-copy repeats comediate a new recurrent rearrangement consisting of duplication at 8p23.1 and triplication at 8p23.2. Hum Mutat. 2007;28:459–468. doi: 10.1002/humu.20465. [DOI] [PubMed] [Google Scholar]
- Hollox EJ, Barber JC, Brookes AJ, Armour JA. Defensins and the dynamic genome: what we can learn from structural variation at human chromosome band 8p23.1. Genome Res. 2008;18:1686–1697. doi: 10.1101/gr.080945.108. [DOI] [PubMed] [Google Scholar]
- Cooke SL, Northup JK, Champaige NL, et al. Molecular cytogenetic characterization of a unique and complex de novo 8p rearrangement. Am J Med Genet A. 2008;146A:1166–1172. doi: 10.1002/ajmg.a.32248. [DOI] [PubMed] [Google Scholar]
- Shimokawa O, Miyake N, Yoshimura T, et al. Molecular characterization of del(8)(p23.1p23.1) in a case of congenital diaphragmatic hernia. Am J Med Genet A. 2005;136:49–51. doi: 10.1002/ajmg.a.30778. [DOI] [PubMed] [Google Scholar]
- Barber JC, Maloney VK, Huang S, et al. 8p23.1 duplication syndrome; a novel genomic condition with unexpected complexity revealed by array CGH. Eur J Hum Genet. 2008;16:18–27. doi: 10.1038/sj.ejhg.5201932. [DOI] [PubMed] [Google Scholar]
- Giglio S, Broman KW, Matsumoto N, et al. Olfactory receptor-gene clusters, genomic-inversion polymorphisms, and common chromosome rearrangements. Am J Hum Genet. 2001;68:874–883. doi: 10.1086/319506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devriendt K, Matthijs G, Van Dael R, et al. Delineation of the critical deletion region for congenital heart defects, on chromosome 8p23.1. Am J Hum Genet. 1999;64:1119–1126. doi: 10.1086/302330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimokawa O, Kurosawa K, Ida T, et al. Molecular characterization of inv dup del(8p): analysis of five cases. Am J Med Genet A. 2004;128A:133–137. doi: 10.1002/ajmg.a.30063. [DOI] [PubMed] [Google Scholar]
- Graw SL, Sample T, Bleskan J, Sujansky E, Patterson D. Cloning, sequencing, and analysis of inv8 chromosome breakpoints associated with recombinant 8 syndrome. Am J Hum Genet. 2000;66:1138–1144. doi: 10.1086/302821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weleber RG, Verma RS, Kimberling WJ, Fieger HG, Jr, lubs HA. Duplication-deficiency of the short arm of chromosome 8 following artificial insemination. Ann Genet. 1976;19:241–247. [PubMed] [Google Scholar]
- Buysse K, Antonacci F, Callewaert B, et al. Unusual 8p inverted duplication deletion with telomere capture from 8q. Eur J Med Genet. 2009;52:31–36. doi: 10.1016/j.ejmg.2008.10.007. [DOI] [PubMed] [Google Scholar]
- Floridia G, Piantanida M, Minelli A, et al. The same molecular mechanism at the maternal meiosis I produces mono- and dicentric 8p duplications. Am J Hum Genet. 1996;58:785–796. [PMC free article] [PubMed] [Google Scholar]
- Pabst B, Arslan-Kirchner M, Schmidtke J, Miller K. The application of region-specific probes for the resolution of duplication 8p: a case report and a review of the literature. Cytogenet Genome Res. 2003;103:3–7. doi: 10.1159/000076280. [DOI] [PubMed] [Google Scholar]
- Dill FJ, Schertzer M, Sandercock J, Tischler B, Wood S. Inverted tandem duplication generates a duplication deficiency of chromosome 8p. Clin Genet. 1987;32:109–113. doi: 10.1111/j.1399-0004.1987.tb03335.x. [DOI] [PubMed] [Google Scholar]
- Barber JC, James RS, Patch C, Temple IK. Protelomeric sequences are deleted in cases of short arm inverted duplication of chromosome 8. Am J Med Genet. 1994;50:296–299. doi: 10.1002/ajmg.1320500315. [DOI] [PubMed] [Google Scholar]
- Minelli A, Floridia G, Rossi E, et al. D8S7 is consistently deleted in inverted duplications of the short arm of chromosome 8 (inv dup 8p) Hum Genet. 1993;92:391–396. doi: 10.1007/BF01247342. [DOI] [PubMed] [Google Scholar]
- Guo WJ, Callif-Daley F, Zapata MC, Miller ME. Clinical and cytogenetic findings in seven cases of inverted duplication of 8p with evidence of a telomeric deletion using fluorescence in situ hybridization. Am J Med Genet. 1995;58:230–236. doi: 10.1002/ajmg.1320580307. [DOI] [PubMed] [Google Scholar]
- de Die-Smulders CE, Engelen JJ, Schrander-Stumpel CT, et al. Inversion duplication of the short arm of chromosome 8: clinical data on seven patients and review of the literature. Am J Med Genet. 1995;59:369–374. doi: 10.1002/ajmg.1320590318. [DOI] [PubMed] [Google Scholar]
- Macmillin MD, Suri V, Lytle C, Krauss CM. Prenatal diagnosis of inverted duplicated 8p. Am J Med Genet. 2000;93:94–98. doi: 10.1002/1096-8628(20000717)93:2<94::aid-ajmg3>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]
- Felbor U, Knotgen N, Schams G, Buwe A, Steinlein C, Schmid M. Mosaicism for an ectopic NOR at 8pter and a complex rearrangement of chromosome 8 in a patient with severe psychomotor retardation. Cytogenet Genome Res. 2004;106:55–60. doi: 10.1159/000078561. [DOI] [PubMed] [Google Scholar]
- Zuffardi O, Bonaglia M, Ciccone R, Giorda R. Inverted duplications deletions: underdiagnosed rearrangements. Clin Genet. 2009;75:505–513. doi: 10.1111/j.1399-0004.2009.01187.x. [DOI] [PubMed] [Google Scholar]
- Rowe LR, Lee JY, Rector L, et al. U-type exchange is the most frequent mechanism for inverted duplication with terminal deletion rearrangements. J Med Genet. 2009;46:694–702. doi: 10.1136/jmg.2008.065052. [DOI] [PubMed] [Google Scholar]
- Zollino M, Murdolo M, Marangi G, et al. On the nosology and pathogenesis of Wolf–Hirschhorn syndrome: genotype–phenotype correlation analysis of 80 patients and literature review. Am J Med Genet C Semin Med Genet. 2008;148C:257–269. doi: 10.1002/ajmg.c.30190. [DOI] [PubMed] [Google Scholar]
- Ozkinay F, Kanit H, Onay H, et al. Prenatal diagnosis of de novo unbalanced translocation 8p;21q using subtelomeric probes. Genet Couns. 2006;17:315–320. [PubMed] [Google Scholar]
- Liehr T, Mrasek K, Weise A, et al. Small supernumerary marker chromosomes – progress towards a genotype–phenotype correlation. Cytogenet Genome Res. 2006;112:23–34. doi: 10.1159/000087510. [DOI] [PubMed] [Google Scholar]
- Fellermann K, Stange DE, Schaeffeler E, et al. A chromosome 8 gene-cluster polymorphism with low human beta-defensin 2 gene copy number predisposes to Crohn disease of the colon. Am J Hum Genet. 2006;79:439–448. doi: 10.1086/505915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho SC, Yim SH, Yoo HK, et al. Copy number variations associated with idiopathic autism identified by whole-genome microarray-based comparative genomic hybridization. Psychiatr Genet. 2009;19:177–185. doi: 10.1097/YPG.0b013e32832bdafa. [DOI] [PubMed] [Google Scholar]
- Hollox EJ, Huffmeier U, Zeeuwen PL, et al. Psoriasis is associated with increased beta-defensin genomic copy number. Nat Genet. 2008;40:23–25. doi: 10.1038/ng.2007.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu S, Bittel DC, Kibiryeva N, Zwick DL, Cooley LD. Validation of the Agilent 244K oligonucleotide array-based comparative genomic hybridization platform for clinical cytogenetic diagnosis. Am J Clin Pathol. 2009;132:349–360. doi: 10.1309/AJCP1BOUTWF6ERYS. [DOI] [PubMed] [Google Scholar]
- Yu S, Kielt M, Stegner AL, Kibiryeva N, Bittel DC, Cooley LD. Quantitative real-time polymerase chain reaction for the verification of genomic imbalances detected by microarray-based comparative genomic hybridization. Genet Test Mol Biomarkers. 2009;13:751–760. doi: 10.1089/gtmb.2009.0056. [DOI] [PubMed] [Google Scholar]
- Shinawi M, Cheung SW. The array CGH and its clinical applications. Drug Discov Today. 2008;13:760–770. doi: 10.1016/j.drudis.2008.06.007. [DOI] [PubMed] [Google Scholar]
- Ciccone R, Mattina T, Giorda R, et al. Inversion polymorphisms and non-contiguous terminal deletions: the cause and the (unpredicted) effect of our genome architecture. J Med Genet. 2006;43:e19. doi: 10.1136/jmg.2005.037671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballif BC, Gajecka M, Shaffer LG. Monosomy 1p36 breakpoints indicate repetitive DNA sequence elements may be involved in generating and/or stabilizing some terminal deletions. Chromosome Res. 2004;12:133–141. doi: 10.1023/b:chro.0000013165.88969.10. [DOI] [PubMed] [Google Scholar]
- Ballif BC, Wakui K, Gajecka M, Shaffer LG. Translocation breakpoint mapping and sequence analysis in three monosomy 1p36 subjects with der(1)t(1;1)(p36;q44) suggest mechanisms for telomere capture in stabilizing de novo terminal rearrangements. Hum Genet. 2004;114:198–206. doi: 10.1007/s00439-003-1029-y. [DOI] [PubMed] [Google Scholar]
- Chabchoub E, Rodriguez L, Galan E, et al. Molecular characterisation of a mosaicism with a complex chromosome rearrangement: evidence for coincident chromosome healing by telomere capture and neo-telomere formation. J Med Genet. 2007;44:250–256. doi: 10.1136/jmg.2006.045476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knijnenburg J, van Haeringen A, Hansson KB, et al. Ring chromosome formation as a novel escape mechanism in patients with inverted duplication and terminal deletion. Eur J Hum Genet. 2007;15:548–555. doi: 10.1038/sj.ejhg.5201807. [DOI] [PubMed] [Google Scholar]
- Kostiner DR, Nguyen H, Cox VA, Cotter PD. Stabilization of a terminal inversion duplication of 8p by telomere capture from 18q. Cytogenet Genome Res. 2002;98:9–12. doi: 10.1159/000068536. [DOI] [PubMed] [Google Scholar]
- Kennedy SJ, Teebi AS, Adatia I, Teshima I. Inherited duplication, dup (8) (p23.1p23.1) pat, in a father and daughter with congenital heart defects. Am J Med Genet. 2001;104:79–80. doi: 10.1002/1096-8628(20011115)104:1<79::aid-ajmg1598>3.0.co;2-t. [DOI] [PubMed] [Google Scholar]
- Tsai CH, Graw SL, McGavran L. 8p23 duplication reconsidered: is it a true euchromatic variant with no clinical manifestation. J Med Genet. 2002;39:769–774. doi: 10.1136/jmg.39.10.769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barber JC, Maloney V, Hollox EJ, et al. Duplications and copy number variants of 8p23.1 are cytogenetically indistinguishable but distinct at the molecular level. Eur J Hum Genet. 2005;13:1131–1136. doi: 10.1038/sj.ejhg.5201475. [DOI] [PubMed] [Google Scholar]
- Slavotinek A, Lee SS, Davis R, et al. Fryns syndrome phenotype caused by chromosome microdeletions at 15q26.2 and 8p23.1. J Med Genet. 2005;42:730–736. doi: 10.1136/jmg.2004.028787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devriendt K, De Mars K, De Cock P, Gewillig M, Fryns JP. Terminal deletion in chromosome region 8p23.1-8pter in a child with features of velo-cardio-facial syndrome. Ann Genet. 1995;38:228–230. [PubMed] [Google Scholar]
- Barber JC, Joyce CA, Collinson MN, et al. Duplication of 8p23.1: a cytogenetic anomaly with no established clinical significance. J Med Genet. 1998;35:491–496. doi: 10.1136/jmg.35.6.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelen JJ, Moog U, Evers JL, Dassen H, Albrechts JC, Hamers AJ. Duplication of chromosome region 8p23.1 → p23.3: a benign variant. Am J Med Genet. 2000;91:18–21. [PubMed] [Google Scholar]
- Glancy M, Barnicoat A, Vijeratnam R, et al. Transmitted duplication of 8p23.1–8p23.2 associated with speech delay, autism and learning difficulties. Eur J Hum Genet. 2009;17:37–43. doi: 10.1038/ejhg.2008.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins AW, Alkuraya FS, Bosco AF, et al. Characterization of apparently balanced chromosomal rearrangements from the developmental genome anatomy project. Am J Hum Genet. 2008;82:712–722. doi: 10.1016/j.ajhg.2008.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willemsen MH, de Leeuw N, Pfundt R, de Vries BB, Kleefstra T. Clinical and molecular characterization of two patients with a 6.75 Mb overlapping deletion in 8p12p21 with two candidate loci for congenital heart defects. Eur J Med Genet. 2009;52:134–139. doi: 10.1016/j.ejmg.2009.03.003. [DOI] [PubMed] [Google Scholar]
- Klopocki E, Fiebig B, Robinson P, et al. A novel 8 Mb interstitial deletion of chromosome 8p12–p21.2. Am J Med Genet A. 2006;140:873–877. doi: 10.1002/ajmg.a.31163. [DOI] [PubMed] [Google Scholar]
- Bettio D, Baldwin EL, Carrozzo R, et al. Molecular cytogenetic and clinical findings in a patient with a small supernumerary r(8) mosaicism. Am J Med Genet A. 2008;146A:247–250. doi: 10.1002/ajmg.a.32083. [DOI] [PubMed] [Google Scholar]
- Makoff AJ, Flomen RH. Detailed analysis of 15q11–q14 sequence corrects errors and gaps in the public access sequence to fully reveal large segmental duplications at breakpoints for Prader–Willi, Angelman, and inv dup(15) syndromes. Genome Biol. 2007;8:R114. doi: 10.1186/gb-2007-8-6-r114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupski JR. Structural variation in the human genome. N Engl J Med. 2007;356:1169–1171. doi: 10.1056/NEJMcibr067658. [DOI] [PubMed] [Google Scholar]
- Gu W, Zhang F, Lupski JR. Mechanisms for human genomic rearrangements. Pathogenetics. 2008;1:4. doi: 10.1186/1755-8417-1-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Smith AJ, Walters RG, Coin LJ, et al. Small deletion variants have stable breakpoints commonly associated with alu elements. PLoS One. 2008;3:e3104. doi: 10.1371/journal.pone.0003104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hastings PJ, Ira G, Lupski JR. A microhomology-mediated break-induced replication model for the origin of human copy number variation. PLoS Genet. 2009;5:e1000327. doi: 10.1371/journal.pgen.1000327. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.