Abstract
Eosinophils are implicated prominently in allergic diseases and the host response to parasitic infections. Eosinophils may be activated in vitro by diverse classes of agonists such as immunoglobulins, lipid mediators, and cytokines. The leukocyte Ig-like receptors (LIRs) comprise a family of inhibitory and activating cell-surface receptors. Inhibitory LIRs down-regulate cellular responses through cytoplasmic immunoreceptor tyrosine-based inhibitory motifs. There are limited data on the action of the activating LIRs, which are thought to signal through the Fc receptor γ chain, which contains an immunoreceptor tyrosine-based activation motif. We now demonstrate the expression of LIR1 (inhibitory), LIR2 (inhibitory), LIR3 (inhibitory), and LIR7 (activating) on eosinophils from 4, 4, 12, and 11, respectively, of 12 healthy donors. Cross-linking of LIR7 with plate-bound antibody elicited the dose- and time-dependent release of eosinophil-derived neurotoxin and leukotriene C4. Eosinophils activated with antibodies to LIR7 embedded in gel-phase EliCell preparations showed leukotriene C4 generation at the nuclear envelope and the release of IL-12 but not IL-4 by vesicular transport. Thus, LIR7 is an activating receptor for eosinophils that elicited the release of cytotoxic granule proteins, de novo lipid mediator generation, and cytokine release through vesicular transport.
Eosinophils are important in host responses to parasitic infections and in allergic diseases (1). Eosinophils elicit their effects through the generation of a range of mediators, both preformed and newly generated after exposure to appropriate stimuli. Their specific granules contain toxic products such as major basic protein, eosinophil peroxidase, eosinophil cationic protein, and eosinophil-derived neurotoxin (EDN) (2). Specific eosinophil granules have also been reported to contain a wide variety of preformed cytokines, growth factors, and chemokines (3). Furthermore, in response to appropriate stimuli, phospholipase A2 is activated to release lysophospholipids and arachidonic acid (4), leading to the generation of leukotriene (LT)C4 (5), which is implicated prominently in the pathobiology of asthma (6). Eosinophils may be activated by a diverse array of molecules that act through G protein-coupled receptors (RANTES, eotaxin, and fMLP), cytokine receptors (IL-5, IFNγ, and IL-16), Ig superfamily receptors (IgG, IgA, and CD28), and tetraspanins (CD9) (1) to elicit secretory granule mediator release, cytokine production through either de novo protein synthesis or vesicular transport from preformed stores, and LTC4 generation at either the perinuclear membrane (7) or newly formed lipid bodies (8).
A complex network of inhibitory and activating signals likely regulates the immunological and/or inflammatory responses of eosinophils (9). The leukocyte Ig-like receptors (LIRs), also termed Ig-like transcripts, comprise a family of inhibitory and activating cell-surface receptors with extracellular Ig-like domains (10–13). The inhibitory LIRs (LIRs 1–3, 5, and 8) have long cytoplasmic domains with two to four immunoreceptor tyrosine-based inhibitory motifs. These receptors mediate inhibition of cell activation by recruiting the src homology 2 domain-containing phosphatase 1 to the phosphorylated immunoreceptor tyrosine-based inhibitory motif to inhibit or terminate signaling through nonreceptor tyrosine kinase cascades (14). The activating LIRs (LIRs 6a, 6b, and 7 and Ig-like transcripts 7, 8, and 11) are characterized by a short cytoplasmic domain and a positively charged arginine residue within the transmembrane domain that mediates association with the immunoreceptor tyrosine-based activation motif-containing Fc receptor γ chain (Fcγ) (15). A third type of LIR (LIR4) is a soluble molecule with no transmembrane domain (12). Although LIR1 and LIR2 are known to recognize a broad range of classical MHC class I molecules and the nonclassical HLA-G (16–18), the ligands for other LIRs are unknown. It has been proposed that LIRs regulate the threshold and amplitude of cellular activation (9).
The expression of LIRs is well documented on cells of monocytic lineage, T cells, B cells, and natural killer cells (10–19). The function of the inhibitory LIRs is also well documented in transfected cells, cells of monocytic lineage, and T cells (20). The only data on LIR expression in granulocytes have been the recognition of LIR7 (also termed Ig-like transcript 1) on polymorphonuclear leukocytes (15). Furthermore, the only data on the events elicited through activating LIRs are confined to the demonstration of Ca2+ flux in monocytes and transfected rat basophilic leukemia cells and the release of histamine from the latter in response to cross-linking of LIR7 (15). We now show that human eosinophils and neutrophils have a restricted pattern of cell-surface LIR expression with LIR3 and LIR7 being expressed in almost all donors, with occasional expression of LIR1 and/or LIR2. Eosinophils may be activated through LIR7 for release of EDN, perinuclear synthesis of LTC4, and secretion of preformed IL-12 but not IL-4 through vesicular transport.
Materials and Methods
Antibodies.
The generation of mouse IgG1 mAbs against the inhibitory LIRs 1–3, 5, and 8 and the activating LIRs 6 and 7 has been described (10, 21). These mAbs are designated m402, m421, m431, m451, m481, m467, and m471, respectively. Other antibodies used are nonimmune mouse IgG1 (BioSource International, Camarillo, CA); mouse IgG1 anti-CD9 (clone ALB 6), mouse mAb pair anti-CD45-FITC/anti-CD14-phycoerythrin, and irrelevant IgG1-FITC/IgG2a-phycoerythrin (Beckman Coulter); mouse IgG1 anti-MHC class I and FITC-conjugated mouse IgG1 anti-CD16 (PharMingen); normal goat serum and FITC- or Cy3-conjugated goat F(ab′)2 anti-mouse IgG [F(ab′)2-specific] with minimum crossreactivity to human, rat, and bovine serum (Jackson ImmunoResearch).
Eosinophil Isolation.
Venous blood from normal volunteers was anticoagulated with acid citrate. After dextran sedimentation of red cells, the buffy coat was layered over Ficoll/Hypaque (Amersham Pharmacia) and centrifuged at 500 × g for 30 min at room temperature. The supernatant and the mononuclear cells at the interface were discarded. Eosinophils were isolated by negative selection using magnetic affinity cell sorting and anti-CD16-conjugated magnetic beads to remove neutrophils (Miltenyi Biotec, Auburn, CA). The purity of eosinophils, assessed by Wright and Giemsa stain, was >95% (range 96–100%). The contaminating cells were neutrophils with occasional mononuclear cells. Viability, assessed by exclusion of trypan blue (Sigma), was ≥90% (range 90–100%). Purified eosinophils were used immediately for experiments.
Flow-Cytometric Analysis.
Isolated eosinophils, unfractionated granulocytes, and mononuclear cells were washed with cold PBS containing 0.05% NaN3 and 1% BSA (PAB buffer) and suspended in the same buffer at 2 × 106 cells per ml. Human serum (final concentration of 10%) was added to the cell suspension. Fifty-microliter portions of the cell suspension were incubated for 30 min at room temperature with mAbs to LIRs (5 μg/ml) or CD9 (2.5 μg/ml), control mouse IgG1, or saturating amounts of directly conjugated mAb to CD16, anti-CD45/CD14, or control dual-labeled antibodies. After two washes with cold PAB buffer, 500 μl of 1% paraformaldehyde in PBS was added to cells stained with directly conjugated antibodies and stored at 4°C in the dark. Cells stained with unconjugated antibodies were incubated on ice for 45 min with 10 μl (10 μg/ml) of FITC- or Cy3-conjugated F(ab′)2 goat anti-mouse IgG [F(ab′)2-specific], which was preabsorbed in an equal volume of normal goat serum. Cells were washed twice with PAB buffer, fixed with 1% paraformaldehyde in PBS, and analyzed by using a FACScan flow cytometer (Becton Dickinson). A unimodal shift in fluorescence intensity of cells stained with specific antibodies compared with cells stained with the isotype-matched negative control antibody was considered positive.
Stimulation of Eosinophils for EDN Release and LTC4 Production with Plate-Bound mAbs.
Eosinophils were stimulated by cross-linking cell-surface LIR7 or CD9 by mouse mAbs to LIR7 or CD9, immobilized onto the wells of tissue-culture plates as described (22). Each well in 96-well flat-bottom tissue-culture plates (Costar 3596) was coated overnight at 4°C with 100 μl of F(ab′)2 goat anti-mouse IgG, Fc-specific (Jackson ImmunoResearch), in PBS at 50 μg/ml. After aspiration of the solution, 50 μl of mAb to LIR7 or CD9, diluted to the desired concentrations in PBS containing 2.5% human serum albumin, was added to each well. Irrelevant mouse IgG1 and mAb to MHC class I were used as controls. After incubation for 2 h at 37°C and 5% CO2, wells were washed twice with 0.9% NaCl before use. Purified eosinophils were washed once with RPMI medium 1640 supplemented with 10 mM Hepes and 0.1% human serum albumin and resuspended in the same medium at 5 × 106 per ml. Twenty-microliter portions of eosinophils were added to 180 μl of medium in each well. After incubation for the desired time at 37°C and 5% CO2, cell-free supernatants were collected and frozen at −80°C for the assay of EDN by RIA (Amersham Pharmacia) and of LTC4 by enzyme-linked immunoassay (Cayman Chemical, Ann Arbor, MI).
Stimulation of Agarose-Embedded Eosinophils.
As described (23, 24), viable eosinophils were embedded in an agarose matrix enabling their morphology and generated/released products to be microscopically localized intracellularly (LTC4) or extracellularly (IL-4 or IL-12). Agarose-embedded eosinophils then were stimulated for 15 min to 3 h with the recombinant CC chemokine eotaxin (6–12 nM, R & D Systems), the calcium ionophore A23187 (0.1–0.5 μM, Sigma), mAb to CD9 (2.5–10 μg/ml), mAb to LIR7 (10 μg/ml), mAb to MHC class I (2.5–10 μg/ml, clone W6/32, Sigma), or isotype-matched mouse IgG1 control (10 μg/ml, R & D Systems).
For inhibition studies, eosinophils still in suspension were pretreated for 30 min with the 5-lipoxygenase (5-LO) inhibitor AA861 (10 μM), the 5-LO-activating protein inhibitor MK886 (10 μM, Biomol, Plymouth Meeting, PA), brefeldin A (0.1–10 μg/ml), cycloheximide (10 μM) or their vehicles (final concentration of DMSO was <0.01%). Cell stimulation was stopped by either fixing EliCell preparations with 2% paraformaldehyde for 5 min or simultaneously fixing/permeabilizing cells for 30 min with 0.5% water-soluble 1-ethyl-3-(3-dimethylamino-propyl) carbodiimide (Sigma) for LTC4 intracellular immunolocalization.
EliCell Assay for Detection of Released IL-4 and IL-12.
The EliCell assay, a gel-phase dual-antibody capture and detection assay based on microscopic observations of individual viable eosinophils embedded in an agarose matrix, was performed as described (24, 25) to enumerate the proportions of eosinophils releasing IL-4 and IL-12. Biotinylated goat polyclonal antibodies against IL-4 and IL-12 (20 μg/ml, R & D Systems) were used as capturing antibodies and paired with Alexa 546-labeled anti-IL-4 and anti-IL-12 mAb (400 μl of 10 μg/ml, R & D Systems) to detect released IL-4 and IL-12, respectively. Slides were viewed with a ×100 objective by both phase-contrast and fluorescence microscopy. Two hundred eosinophils were scored, and the percentages of those exhibiting fluorescent staining for extracellular cytokine release then were calculated. The amounts of immunoreactive IL-4 and IL-12 released by individual eosinophils were evaluated by quantitating fluorescence intensity around each cell by using the software program IPLAB 3.2.4 (Scanalytics, Fairfax, VA). Fifty consecutive eosinophils per experimental condition were evaluated. Fluorescence intensity at each pixel was quantitated in arbitrary units ranging from 0 to 255. The cumulative fluorescence intensities in excess of the background threshold were summed for all pixels overlying each analyzed eosinophil.
Staining of Lipid Bodies and Newly Formed Intracellular LTC4.
Lipid body formation and intracellular production of LTC4 within eosinophils embedded in an agarose matrix were evaluated as described (23, 26). Briefly, to stain lipid bodies, slides containing agarose-embedded eosinophils were fixed with 2% paraformaldehyde in Hanks' balanced salt solution lacking calcium and magnesium, rinsed in 0.1 M cacodylate buffer (pH 7.4), stained in 1.5% OsO4 for 30 min, rinsed in distilled water, immersed in 1% thiocarbohydrazide for 5 min, rinsed with 0.1 M cacodylate buffer, restained with 1.5% OsO4 for 3 min, dried, and mounted. Lipid bodies were enumerated by light microscopy with a ×100 objective lens in 25 consecutively scanned cells.
To immunolocalize LTC4 at its formation sites within agarose-embedded viable eosinophils, 1-ethyl-3-(3-dimethylamino-propyl) carbodiimide was used to cross-link eicosanoid carboxyl groups to amines in adjacent proteins, fixing newly formed LTC4 before its immunofluorescent localization with an Alexa 488-labeled rat anti-LTC4/LTD4/LTE4 mAb (clone 6E7, Sigma). The percentages of those eosinophils exhibiting green staining for intracellular immunoreactive LTC4 were calculated by analyzing 100 consecutive scanned cells by both phase-contrast and fluorescence microscopy. To document cell morphology and fluorescent immunolocalization of intracellular LTC4, cells were imaged by using a spot-cooled color digital camera (model 1.3.0, Diagnostic Instruments, Sterling Heights, MI) and Adobe PHOTOSHOP 5.5 (Adobe Systems, Mountain View, CA).
Statistical Analysis.
Data were expressed as means ± SD except where otherwise stated. For EliCell assays, percentage inhibition was calculated in comparison to stimulated increases in LTC4 production or IL-12 release above baselines. Statistical comparisons were done by ANOVA followed by Newman–Keuls Student's test. Differences were considered significant when P < 0.05.
Results and Discussion
LIR Expression on Human Peripheral Blood Eosinophils.
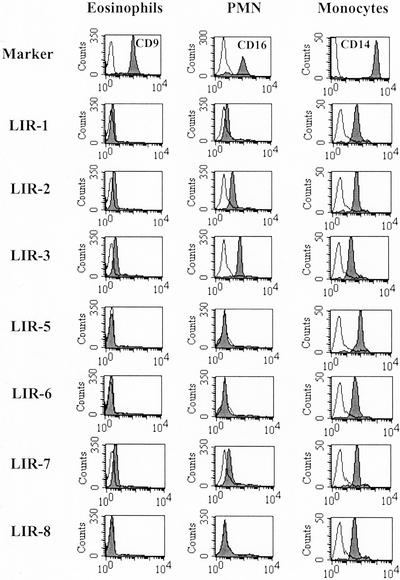
We used a panel of mouse mAbs to examine cell-surface expression of LIRs on peripheral blood eosinophils and, for comparison, unfractionated granulocytes (>90% CD16+ neutrophils). C14+ monocytes, gated by forward and right-angle light scatter of peripheral blood mononuclear cells, provided a positive control. Representative flow-cytometry histograms are shown in Fig. 1. The presence or absence of LIRs on cells from all donors is indicated in Table 1. Consistent with published data, monocytes expressed multiple LIRs. LIR3 (inhibitory) and LIR7 (activating) were expressed on eosinophils obtained from 12 of 12 donors and 11 of 12 donors, respectively (Table 1). Eosinophils from four donors expressed LIR2. Eosinophils from four donors expressed LIR1. The levels of expression of LIR1, LIR2, LIR3, and LIR7 on eosinophils were lower than on monocytes (Fig. 1). There was no detectable expression of LIR5, LIR6, or LIR8 on eosinophils isolated from any of the 12 donors. The pattern of LIR expression on polymorphonuclear leukocytes was similar to that observed on eosinophils (Table 1; Fig. 1).
Figure 1.
Flow-cytometric analysis of LIR expression. Cell-surface expression of LIRs on eosinophils, neutrophils (polymorphonuclear leukocytes, PMN), and monocytes was analyzed as described in Materials and Methods. mAbs directed to CD9, CD16, and CD14 were used as markers for eosinophils, polymorphonuclear leukocytes, and monocytes, respectively. Representative histograms are shown.
Table 1.
Cell-surface expression of LIRs on eosinophils, neutrophils, and monocytes of all donors studied
| LIR | Eosinophils (n = 12) | Neutrophils (n = 5) | Monocytes (n = 5) |
|---|---|---|---|
| 1 | 4 (33) | 4 (80) | 5 (100) |
| 2 | 4 (33) | 4 (80) | 5 (100) |
| 3 | 12 (100) | 5 (100) | 5 (100) |
| 5 | 0* | 0 | 5 (100) |
| 6 | 0 | 0 | 3 (60) |
| 7 | 11 (92) | 5 (100) | 5 (100) |
| 8 | 0 | 0 | 1 (20) |
Data are expressed as the number (percentage) of individuals with cells staining positive for each LIR.
n = 8.
The suggestion that LIRs and related receptors may determine the threshold and amplitude of an activation response in leukocytes (9) is supported by observations in mice with disruption of gp49, an immunoreceptor tyrosine-based inhibitory motif-containing inhibitory receptor homologous to the LIRs, that is expressed on mast cells and monocytes. Mice that lack expression of gp49 demonstrate augmented passive cutaneous anaphylaxis and augmented active cutaneous and systemic anaphylactic responses (27). The expression of both activating and inhibitory LIRs on eosinophils suggests that their responses after recruitment to a site of inflammation may be regulated by exposure to appropriate ligands for the LIRs, although these presently are unknown for LIR3 and LIR7.
EDN Release.
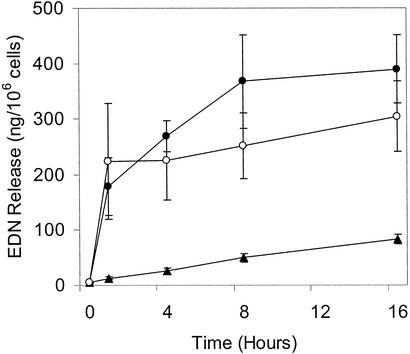
Although there is a wealth of data on the function of the inhibitory LIRs, there is little known about the events elicited by the activating LIRs. We therefore investigated the activation of eosinophils through LIR7 using immobilized mAb to LIR7. As described for stimulation through CD9 (22), we first coated 96-well plates with F(ab′)2 fragments of goat anti-mouse IgG (Fc-specific) to provide uniform orientation of the mAbs and improve the efficiency of cross-linking. Anti-CD9 was used as a positive control. Exposure of eosinophils to immobilized mAb to LIR7 induced EDN release in a dose- and time-dependent manner. EDN release was detected within 15 min (data not shown), and net release reached a plateau at 4–8 h (Fig. 2) in response to 5–10 μg/ml mAb to LIR7. Immobilized mouse IgG1 of irrelevant specificity and mAb to MHC class I elicited minimal EDN release (Fig. 2 and data not shown), indicating that EDN release was not elicited through Fc receptors for IgG nor merely as a result of eosinophil adherence to an antibody-coated plate. Irrelevant IgG1 and mAbs to MHC class I, CD9, and LIR7 elicited EDN release of 120 ± 95 (n = 7), 35 ± 18 (n = 4), 532 ± 290 (n = 10; P < 0.002 compared with IgG1 and mAb to MHC class I), and 419 ± 142 ng/106 cells (n = 10; P < 0.001 compared with IgG1 and mAb to MHC class I), respectively, 4 h after treatment with 5 μg/ml immobilized antibodies. For stimulation through LIR7, this represented 18% and 19% release of total cellular EDN content in two independent experiments. Eosinophil viability, analyzed 1 and 4 h after stimulation by uptake of calcein acetoxymethyl ester or ethidium homodimer 1 (Molecular Probes) was no different between eosinophils incubated with mAb to LIR7 and eosinophils incubated with negative control antibodies (data not shown). Thus, EDN release by eosinophils was not the result of cell death.
Figure 2.
Time course of release of EDN. Purified eosinophils were stimulated with plate-bound mAb (5 μg/ml) to LIR7 (filled circles), to CD9 (open circles), or to MHC class I (filled triangles) as described in Materials and Methods (n = 3). Results are means ± SE.
EDN is one of several preformed toxic granule products that, with eosinophil peroxidase, major basic protein, and eosinophil cationic protein, is stored in the specific granules of eosinophils (2). EDN release from eosinophils has been reported in response to a range of immobilized ligands including IgA, particularly secretory IgA, and IgG bound to Sepharose beads (28). The Fc portion of Ig was required for eosinophil activation, indicating signaling through FcαR and one of the activating FcγRs, likely FcγRII, expressed on human eosinophils (28). LIR7, in common with these receptors, signals through the immunoreceptor tyrosine-based activation motif-containing Fcγ (15). Consistent with this, cross-linking of LIR7 on human eosinophils elicited release of EDN (Fig. 2) in quantities and with kinetics similar to those observed in response to immobilized immunoglobulins (28).
LTC4 Generation.
Although EDN is preformed and released from secretory granules, LTC4, the major eicosanoid produced by human eosinophils, is generated de novo from arachidonic acid released from cell membrane phospholipids by the action of phospholipase A2. Engagement of LIR7 by immobilized mAbs elicited the dose- and time-dependent release of LTC4 that was maximal 1 h after the addition of eosinophils to plates coated with 5–10 μg/ml mAb to LIR7 (n = 2; data not shown). mAbs to MHC class I, CD9, and LIR7 elicited LTC4 release of 0.4 ± 0.5 (n = 6), 0.8 ± 1.2 (n = 6), and 2.8 ± 1.7 ng per 106 cells (n = 6; P < 0.01 compared with IgG1 anti-MHC class I), respectively. LTC4 generation and release in response to cross-linking of CD9 was variable, not observed in all donors, was less than that observed in response to cross-linking of LIR7, and did not reach statistical significance.
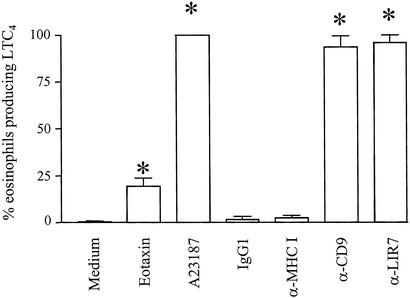
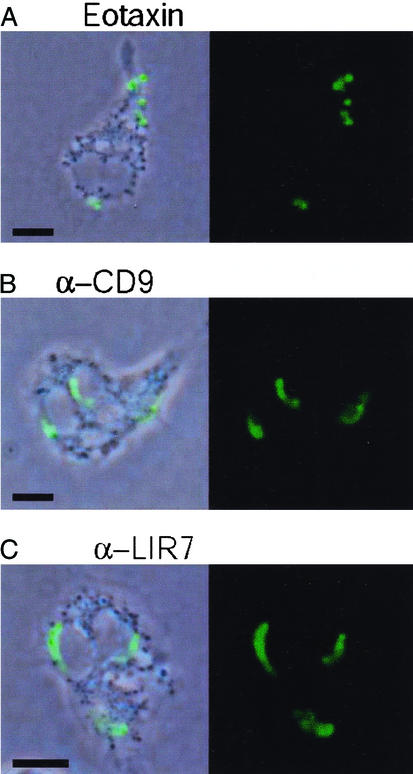
LTC4 generation in human eosinophils has been reported to occur in a perinuclear location (7) or in lipid bodies (8, 23). We used a new strategy for direct in situ immunolocalization of intracellular LTC4 (23) to ascertain the intracellular site of cysteinyl LT biosynthesis after cross-linking of membrane-expressed LIR7 (Figs. 3 and 4). Eosinophils were stimulated for 15 min to 3 h in agarose containing mAbs to CD9 (2.5 μg/ml), LIR7 (10 μg/ml), or MHC class I (10 μg/ml) or isotype-matched mouse IgG1 control (10 μg/ml). As positive controls, eosinophils were treated with eotaxin, a classical physiological stimulus for eosinophils (12 nM) or the calcium ionophore A23187 (0.1 μM), a cytolytic/exocytotic nonphysiological stimulus. Nonstimulated agarose-embedded eosinophils exhibited no immunofluorescence staining for LTC4 (Fig. 3). Stimulation with eotaxin elicited LTC4 synthesis in cytoplasmic, newly formed lipid bodies in ≈20% of eosinophils (Figs. 3 and 4A; ref. 23). In clear contrast, the activation of eosinophils through LIR7 did not induce lipid body formation (data not shown). Rather, cross-linking of LIR7 triggered LTC4 synthesis in nearly 100% of eosinophils (Fig. 3) with immunostaining in a predominantly perinuclear location (Fig. 4C), similar to the pattern observed in A23187-stimulated eosinophils (ref. 23 and data not shown). In eosinophils that released detectable LTC4 in response to cross-linking of CD9, perinuclear LTC4 synthesis was observed (Fig. 4B). The specificity of immunostaining for LTC4 was supported by the absence of staining for LTC4 in A23187-stimulated eosinophils when either (i) an Alexa 488-labeled isotype control antibody replaced the anti-LTC4/D4/E4 antibody or (ii) the cells were pretreated with inhibitors of the 5-LO pathway, AA861 and MK886 (data not shown).
Figure 3.
LTC4 generation by eosinophils. Agarose-embedded eosinophils were stimulated with recombinant eotaxin (12 nM), with A23187 (0.1 μM), with mAbs to CD9 (2.5 μg/ml), LIR7 (10 μg/ml), or MHC class I (10 μg/ml), or with irrelevant mouse IgG1 control (10 μg/ml) for 1 h. Cells were fixed and stained with Alexa 488-labeled anti-cysteinyl LT mAb. The data are expressed as the percentages of cells containing immunodetectable LTC4. *, P < 0.05 compared with medium alone or control IgG1 (n = 3).
Figure 4.
Intracellular localization of LTC4 biosynthesis. Agarose-embedded eosinophils were stimulated with 12 nM eotaxin (A), 2.5 μg/ml mAb to CD9 (B), or 10 μg/ml mAb to LIR7 (C) for 1 h. Cells then were fixed and stained with Alexa 488-labeled anti-cysteinyl LT mAb. To facilitate intracellular localization, anti-LTC4 immunoreactive sites (green staining) were overlaid on phase-contrast images. (Bar, 5 μm.) The figure is illustrative of three independent experiments.
The demonstration of LTC4 generation in response to cross-linking of LIR7 provides evidence of lipid mediator release in response to signaling through an activating LIR. Cytosolic phospholipase A2 and 5-LO exist in the cytosol and translocate to the perinuclear membrane in response to calcium flux (29, 30). 5-LO-activating protein and LTC4 synthase are present at the perinuclear membrane in both resting and stimulated cells (31, 32). Subsequent work implicated lipid bodies as a site of LT biosynthesis (26). Induction of lipid bodies containing cytosolic phospholipase A2, 5-LO, and LTC4 synthase (8, 33) primes eosinophils for enhanced LT biosynthesis (8, 26, 34). An adaptation of the EliCell assay demonstrated the presence of LTC4 in lipid bodies in response to eotaxin (23). The generation of LTC4 at a perinuclear site in response to cross-linking of LIR7 suggests a Ca2+-dependent process, consistent with previous data demonstrating Ca2+flux after cross-linking of LIR7 in monocytes and RBL cells and with signaling through the immunoreceptor tyrosine-based activation motif-containing Fcγ (15). Cross-linking of LIR7 did not elicit lipid body formation. This contrasts with the observation that stimulation of eosinophils with plate-bound IgG for 1 h elicited the formation of lipid bodies and LTC4 generation, both of which depended on platelet-activating factor biosynthesis (35). Thus, although FcγRII and LIR7 both signal through Fcγ, they may elicit different downstream signaling events. The local generation of LTC4 at distinct intracellular sites within eosinophils may be important for the roles of this mediator, both as a paracrine agent in allergic inflammation and as an intracrine signal-transducing mediator that may regulate cellular responses.
Cytokine Release.
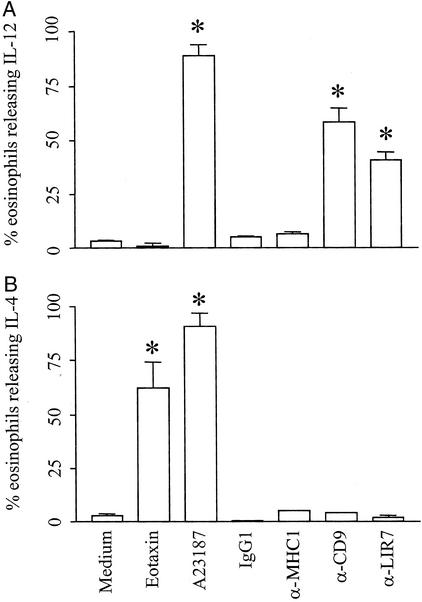
Human eosinophils have the potential to release >24 cytokines including prototypical T helper (Th)1 (e.g., IL-12) and Th2 (e.g., IL-4) cytokines, likely through vesicular transport (3). By using the EliCell assay (24), we confirmed that the calcium ionophore A23187 (0.5 μM) stimulated the release of both IL-12 and IL-4, detectable extracellularly around eosinophils (ref. 23; Fig. 5). Consistent with previous data, eotaxin elicited IL-4 release (ref. 25; Fig. 5B). Interestingly, eotaxin failed to induce secretion of IL-12 (Fig. 5). In contrast, stimulation of eosinophils through CD9 or LIR7 triggered release of IL-12 but not IL-4 (Fig. 5). Incubation of eosinophils with irrelevant mouse IgG1 or mAb to MHC class I did not elicit IL-12 or IL-4 secretion (Fig. 5). No extracellular staining for IL-4 or IL-12 was detectable around nonstimulated eosinophils (Fig. 5) or when the Alexa 546-labeled anti-IL-12 or anti-IL-4 detection antibodies were replaced by an Alexa 546-labeled isotype-matched control antibody (data not shown). Moreover, IL-12 and IL-4 were not detectable when the biotinylated anti-IL-12 and anti-IL-4 capture antibodies were substituted with a biotinylated irrelevant control antibody (not shown). The latter condition assured that neither intracellular nor membrane-bound cytokines were being detected in the nonpermeabilized eosinophils.
Figure 5.
IL-12 and IL-4 release by eosinophils. Agarose-embedded eosinophils were stimulated with eotaxin (6 nM), A23187 (0.5 μM), mAb to CD9 (2.5 μg/ml), mAb to LIR7 (10 μg/ml), mAb to MHC class I (10 μg/ml), or mouse IgG1 control (10 μg/ml). IL-12 and IL-4 release was determined by using the EliCell assay. The data are reported as the percentage of cells with detectable cytokine release. Shown is release of IL-12 (A) and IL-4 (B) in response to each stimulus at 1 h. The data are expressed as percentage of cells releasing each cytokine. *, P < 0.05 compared with medium alone or control IgG1 (n = 3–5).
Stimulation through CD9 or LIR7 was detectable by 15 and 60 min, respectively. The rapid and selective release of IL-12 in response to engagement of LIR7 or CD9 suggested the release of preformed cytokines by a process of vesicular transport. Consistent with this mechanism, pretreatment of eosinophils with 10 μM cycloheximide did not inhibit IL-12 release in response to any stimulus tested (Table 2), indicating that IL-12 was preformed rather than generated de novo. In prior work, 1 μM cycloheximide was sufficient to inhibit lipid body formation in response to platelet-activating factor by 68% (9). In contrast, release of IL-12 after engagement of LIR7 or CD9 was blocked by pretreatment of the cells with brefeldin A (Table 2), which disrupts the formation of vesicles. Brefeldin A had no effect on the release of IL-12 from A23187-stimulated eosinophils. These data are consistent with release of IL-12 in response to engagement of LIR7 or CD9 by a process of vesicular transport. The response was remarkably selective with no release of IL-4 (Fig. 5), which contrasts with the selective release of IL-4 but not IL-12 in response to eotaxin and the release of both mediators in response to the calcium ionophore. It is notable that stimulation of eosinophils with plate-bound secretory IgA elicited IL-10 release, whereas cross-linking of CD28 elicited release of IFNγ and IL-2, supporting the concept of stimulus-dependent, selective release of cytokines (36). IL-12 favors the development of Th1 responses, whereas IL-4 favors the development of Th2 responses (37). The release of IL-4 but not IL-12 in response to eotaxin is consistent with the proposed role of that chemokine in Th2-dependent eosinophilic inflammation (38, 39). On the other hand, the specific release of IL-12 but not IL-4 from eosinophils in response to engagement of LIR7 suggests a possible function for LIR7 in tempering Th2 cell-dependent inflammatory responses and indicates that the counterregulatory function of the LIRs may not be confined to the inhibitory members of this family of receptors.
Table 2.
Cross-linking of LIR7 elicits preformed IL-12 release from eosinophils by vesicular transport
| Condition | % eosinophils releasing IL-12 | IL-12 fluorescent intensity/cell |
|---|---|---|
| α-CD9 | 45.2 ± 8.3 | 0.70 ± 0.11 |
| +BFA (0.1 μg/ml) | 45.0 ± 4.2 | 0.50 ± 0.11* |
| +BFA (1 μg/ml) | 29 ± 1.7* | 0.10 ± 0.08** |
| +CHX (10 μM) | 55.5 ± 4.9 | 0.63 ± 0.18 |
| α-LIR7 | 39.6 ± 5.4 | 0.83 ± 0.14 |
| +BFA (0.1 μg/ml) | 37.5 ± 3.0 | 0.51 ± 0.12* |
| +BFA (1 μg/ml) | 20.3 ± 4.3* | 0.20 ± 0.10** |
| +CHX (10 μM) | 47.5 ± 0.7 | 0.85 ± 0.08 |
| A23187 | 96.9 ± 0.8 | 1.78 ± 0.56 |
| +BFA (0.1 μg/ml) | 98.1 ± 3.0 | 1.60 ± 0.39 |
| +BFA (1 μg/ml) | 96.8 ± 3.0 | 1.91 ± 0.21 |
| +CHX (10 μM) | 96.4 ± 3.0 | 1.73 ± 0.10 |
Agarose-embedded eosinophils were stimulated for 3 h with 0.5 μM calcium ionophore or 10 μg/ml mAbs to CD9 or LIR7. EliCell assay for IL-12 release was performed as described in Materials and Methods. The values of IL-12 release are expressed either as the percentages of eosinophils exhibiting red extracellular immunolabeling or as the average of electronically measured immunofluorescent intensities of extracellular IL-12. Control values for eosinophils stimulated with mAb to MHC class I (10 μg/ml) were 3.3 ± 3.0% and 0.00 ± 0.00 units, respectively. Results are means ± SD from at least three donors. BFA, brefeldin A; CHX, cycloheximide. *, P < 0.05, and **, P < 0.01, compared with cells stimulated in the absence of inhibitors.
Acknowledgments
We thank Maaya Yasuda for excellent technical assistance. This work was supported by National Institutes of Health Grants AI31599, AI20241, AI22571, AI41995, HL70270, and HL07718, an Arthritis Foundation postdoctoral fellowship, and National Science Foundation Research Experience for Undergraduates Grant DBI-9322334.
Abbreviations
- EDN
eosinophil-derived neurotoxin
- LT
leukotriene
- LIR
leukocyte Ig-like receptor
- Fcγ
Fc receptor γ chain
- 5-LO
5-lipoxygenase
- Th
T helper
References
- 1.Gleich G. J Allergy Clin Immunol. 2000;105:651–663. doi: 10.1067/mai.2000.105712. [DOI] [PubMed] [Google Scholar]
- 2.Popken-Harris P, Thomas L, Oxvig C, Sottrup-Jensen L, Kubo H, Klein J S, Gleich G J. J Allergy Clin Immunol. 1994;94:1282–1289. doi: 10.1016/0091-6749(94)90343-3. [DOI] [PubMed] [Google Scholar]
- 3.Lacy P, Moqbel R. Chem Immunol. 2000;76:134–155. doi: 10.1159/000058782. [DOI] [PubMed] [Google Scholar]
- 4.Six A D, Dennis E A. Biochim Biophys Acta. 2000;1488:1–19. doi: 10.1016/s1388-1981(00)00105-0. [DOI] [PubMed] [Google Scholar]
- 5.Owen W F, Jr, Soberman R J, Yoshimoto T, Sheffer A L, Lewis R A, Austen K F. J Immunol. 1987;138:532–538. [PubMed] [Google Scholar]
- 6.Drazen J M, Israel E, O'Byrne P M. N Engl J Med. 1999;340:197–206. doi: 10.1056/NEJM199901213400306. [DOI] [PubMed] [Google Scholar]
- 7.Brock T G, Anderson J A, Fries F P, Peters-Golden M, Sporn P H S. J Immunol. 1999;162:1669–1676. [PubMed] [Google Scholar]
- 8.Bozza P T, Yu W, Penrose J F, Morgan E S, Dvorak A M, Weller P F. J Exp Med. 1997;186:909–920. doi: 10.1084/jem.186.6.909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ravetch J V, Lanier L L. Science. 2000;290:84–89. doi: 10.1126/science.290.5489.84. [DOI] [PubMed] [Google Scholar]
- 10.Borges L, Hsu M L, Fanger N, Kubin M, Cosman D. J Immunol. 1997;159:5192–5196. [PubMed] [Google Scholar]
- 11.Samaridis J, Colonna M. Eur J Immunol. 1997;27:660–665. doi: 10.1002/eji.1830270313. [DOI] [PubMed] [Google Scholar]
- 12.Arm J P, Nwankwo C, Austen K F. J Immunol. 1997;159:2342–2349. [PubMed] [Google Scholar]
- 13.Wagtmann N, Rojo S, Eichler E, Mohrenweiser H, Long E O. Curr Biol. 1997;7:615–618. doi: 10.1016/s0960-9822(06)00263-6. [DOI] [PubMed] [Google Scholar]
- 14.Fanger N A, Cosman D, Peterson L, Braddy S C, Maliszewski C R, Borges L. Eur J Immunol. 1998;28:3423–3434. doi: 10.1002/(SICI)1521-4141(199811)28:11<3423::AID-IMMU3423>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 15.Nakajima H, Samaridis J, Angman L, Colonna M. J Immunol. 1999;162:5–8. [PubMed] [Google Scholar]
- 16.Cosman D, Fanger N, Borges L, Kubin M, Chin W, Peterson L, Hsu M L. Immunity. 1997;7:273–282. doi: 10.1016/s1074-7613(00)80529-4. [DOI] [PubMed] [Google Scholar]
- 17.Colonna M, Samaridis J, Cella M, Angman L, Allen R L, O'Callaghan C A, Dunbar R, Ogg G S, Cerundolo V, Rolink A. J Immunol. 1998;160:3096–3100. [PubMed] [Google Scholar]
- 18.Navarro F, Llano M, Bellon T, Colonna M, Geraghty D E, Lopez-Botet M. Eur J Immunol. 1999;29:277–283. doi: 10.1002/(SICI)1521-4141(199901)29:01<277::AID-IMMU277>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 19.Cella M, Dohring C, Samaridis J, Dessing M, Brockhaus M, Lanzavecchia A, Colonna M. J Exp Med. 1997;185:1743–1751. doi: 10.1084/jem.185.10.1743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Colonna M, Nakajima H, Cella M. Semin Immunol. 2000;12:121–127. doi: 10.1006/smim.2000.0214. [DOI] [PubMed] [Google Scholar]
- 21.Tedla N, Gibson K, McNeil H P, Cosman D, Borges L, Arm J P. Am J Pathol. 2002;160:425–431. doi: 10.1016/S0002-9440(10)64861-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim J T, Gleich G J, Kita H. J Immunol. 1997;159:926–933. [PubMed] [Google Scholar]
- 23.Bandeira-Melo C, Phoofolo M, Weller P F. J Biol Chem. 2001;276:22779–22787. doi: 10.1074/jbc.M101436200. [DOI] [PubMed] [Google Scholar]
- 24.Bandeira-Melo C, Gillard G, Ghiran I, Weller P F. J Immunol Methods. 2000;244:105–115. doi: 10.1016/s0022-1759(00)00264-7. [DOI] [PubMed] [Google Scholar]
- 25.Bandeira-Melo C, Sugiyama K, Woods L J, Weller P F. J Immunol. 2001;166:4813–4817. doi: 10.4049/jimmunol.166.8.4813. [DOI] [PubMed] [Google Scholar]
- 26.Bozza P T, Payne J L, Morham S G, Langenbach R, Smithies O, Weller P F. Proc Natl Acad Sci USA. 1996;93:11091–11096. doi: 10.1073/pnas.93.20.11091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Daheshia M, Friend D S, Grusby M J, Austen K F, Katz H R. J Exp Med. 2001;194:227–234. doi: 10.1084/jem.194.2.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Walsh G M, Kay A B. Clin Exp Immunol. 1986;63:466–472. [PMC free article] [PubMed] [Google Scholar]
- 29.Schievella A R, Regier M K, Smith W L, Lin L L. J Biol Chem. 1995;270:30749–30754. doi: 10.1074/jbc.270.51.30749. [DOI] [PubMed] [Google Scholar]
- 30.Brock T G, McNish R W, Peters-Golden M. J Biol Chem. 1995;270:21652–21658. doi: 10.1074/jbc.270.37.21652. [DOI] [PubMed] [Google Scholar]
- 31.Woods J W, Evans J F, Ethier D, Scott S, Vickers P J, Hearn L, Heibein J A, Charleson S, Singer I I. J Exp Med. 1993;178:1935–1946. doi: 10.1084/jem.178.6.1935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Penrose J F, Spector J, Lam B K, Friend D S, Xu K, Jack R M, Austen K F. Am J Respir Cell Mol Biol. 1995;152:283–289. doi: 10.1164/ajrccm.152.1.7599836. [DOI] [PubMed] [Google Scholar]
- 33.Yu W, Bozza P T, Tzizik D M, Gray J P, Cassara J, Dvorak A M, Weller P F. Am J Pathol. 1998;152:759–769. [PMC free article] [PubMed] [Google Scholar]
- 34.Weller P F, Bozza P T, Yu W, Dvorak A M. Int Arch Allergy Appl Immunol. 1999;118:450–452. doi: 10.1159/000024161. [DOI] [PubMed] [Google Scholar]
- 35.Bartemes K R, McKinney S, Gleich G J, Kita H. J Immunol. 1999;162:2982–2989. [PubMed] [Google Scholar]
- 36.Woerly G, Roger N, Loiseau S, Dombrowicz D, Capron A, Capron M. J Exp Med. 1999;190:487–496. doi: 10.1084/jem.190.4.487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Romagnani S. Immunol Today. 1997;18:263–266. doi: 10.1016/s0167-5699(97)80019-9. [DOI] [PubMed] [Google Scholar]
- 38.Hogan S P, Mishra A, Brandt E B, Foster P S, Rothenberg M E. Proc Natl Acad Sci USA. 2000;97:6681–6686. doi: 10.1073/pnas.97.12.6681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rothenberg M E. Am J Respir Cell Mol Biol. 1999;21:291–295. doi: 10.1165/ajrcmb.21.3.f160. [DOI] [PubMed] [Google Scholar]