Abstract
Cells typically respond quickly to stress, altering their metabolism to compensate. In mammalian cells, stress signaling usually leads to either cell-cycle arrest or apoptosis, depending on the severity of the insult and the ability of the cell to recover. Stress also often leads to reorganization of nuclear architecture, reflecting the simultaneous inhibition of major nuclear pathways (e.g., replication and transcription) and activation of specific stress responses (e.g., DNA repair). In this review, we focus on how two nuclear organelles, the nucleolus and the Cajal body, respond to stress. The nucleolus senses stress and is a central hub for coordinating the stress response. We review nucleolar function in the stress-induced regulation of p53 and the specific changes in nucleolar morphology and composition that occur upon stress. Crosstalk between nucleoli and CBs is also discussed in the context of stress responses.
Main Text
Nucleolar Dynamics under Stress Conditions
The main function of the nucleolus is the rapid production of small and large ribosome subunits, a process that must be highly regulated to achieve proper cellular proliferation and cell growth (Lempiainen and Shore, 2009). Many aspects of nucleolar organization and function are conserved within eukaryotic organisms, from yeast to human (Kressler et al., 2010). This review focuses on how stress responses in mammalian cells affect the nucleolus and Cajal bodies (CBs), and we introduce this topic by giving a brief overview of ribosome subunit biogenesis in mammalian cells. For an overview of the related processes of ribosome subunit biogenesis in yeast, we refer the reader to the following reviews: Henras et al. (2008) and Tschochner and Hurt (2003).
Nucleoli in mammalian cells disassemble when cells divide and reform at the end of mitosis around the tandemly repeated clusters of rDNA genes known as nucleolar organizing regions (NORs). This results in a subnuclear compartment that concentrates the factors involved in ribosomal RNA (rRNA) transcription and processing, as well as ribosome subunit assembly (for detailed review, see Kressler et al., 2010). Transcription of rDNA genes by RNA polymerase I (RNA Pol I) leads to the synthesis of a 47S precursor ribosomal RNA transcript (pre-rRNA). The pre-rRNA is either co- or posttranscriptionally processed and modified by snoRNPs (small nucleolar ribonucleoproteins) to generate the 28S, 18S, and 5.8S rRNAs. These snoRNP-mediated modifications include 2′-O-methylation and pseudouridine formation (Matera et al., 2007). The 28S, 18S, and 5.8S rRNAs are assembled with ribosomal proteins (RPs) to form the small and large preribosome subunits, which are each exported separately to the cytoplasm and undergo final processing steps to become the mature 40S and 60S ribosome subunits.
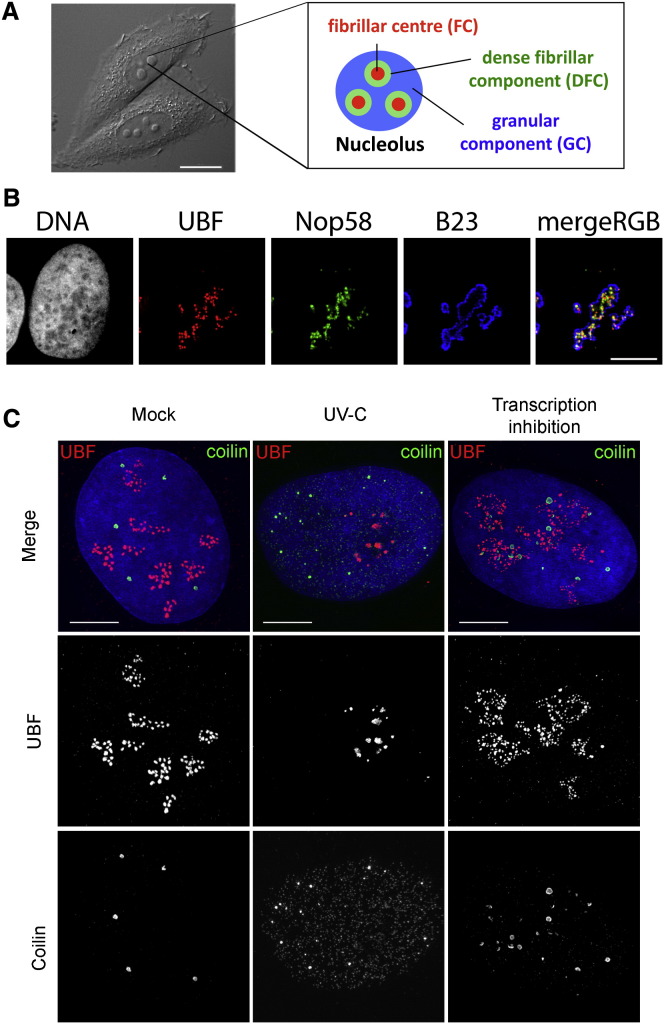
The three main events that occur within the nucleolus—pre-rRNA transcription, processing, and ribosomal RNP assembly—are reflected in its “tripartite” internal structure. These events, or at least the molecules that mediate them, are concentrated in three distinct subnucleolar compartments called the fibrillar center (FC), the dense fibrillar component (DFC), and the granular component (GC), as summarized in Figure 1A. It is generally accepted that pre-rRNA is transcribed from rDNA either in the FC or at the border between the FC and DFC. FCs are enriched in components of the RNA Pol I machinery, such as UBF, whereas the DFC harbors pre-rRNA processing factors, such as the snoRNAs and snoRNP proteins, fibrillarin and Nop58. Both the FC and the DFC are enclosed by the GC, where preribosome subunit assembly takes place (reviewed in Boisvert et al., 2007, Sirri et al., 2008) (Figure 1B). The morphology and size of nucleoli are linked to nucleolar activity, which in turn depends on cell growth and metabolism.
Figure 1.
Overview of Nucleolar Organization under Physiological Conditions in the Mammalian Cell Nucleus, and Visualization by Immunofluorescence of Stress-Induced Changes to Nucleolar and Cajal Body Organization
(A) Differential interference contrast (DIC) image of live HeLa cells: nucleoli are readily observed as phase-dense structures. Scale bar, 15 μm (left panel). Schematic representation of nucleolar tripartite internal organization, formed by the fibrillar center (FC), the dense fibrillar component (DFC), and the granular component (GC) (right panel).
(B) Fluorescence microscopy images showing the three subnucleolar compartments in human U2OS cells. FC is visualized using antibodies against UBF, DFC using antibodies against Nop58, and GC using antibodies against B23/NPM. Scale bar, 10 μm.
(C) Examples of stress-induced changes in nucleolar and CB organization in U2OS cells. (Left panel) Untreated cells. (Middle panel) UV-C-treated cells (6 hr postirradiation, 30 J/m2). (Right panel) DRB-treated cells (3 hr, 25 μg/mL). All images show UBF (nucleolar fibrillar center) in red and coilin (CB) in green. UV-C treatment induces nucleolar segregation and relocalization of coilin to nucleoplasmic microfoci. In contrast, DRB treatment induces nucleolar fragmentation and unravelling of the FC, as well as CB disruption and association of coilin with the nucleolus in cap-like structures. Scale bar, 5 μm.
Reorganization of the Nucleolus under Stress
The varied effects on ribosome subunit production and cell growth induced by different types of cellular stress are often accompanied by dramatic changes in the organization and composition of the nucleolus (Table 1 ). A well-described phenomenon is the nucleolar segregation caused by DNA damage (e.g., following UV irradiation or inhibition of topoisomerase II by drugs such as etoposide) and/or transcriptional inhibition (e.g., by actinomycin D) (Al-Baker et al., 2005, Govoni et al., 1994). Segregation is characterized by the condensation and subsequent separation of the FC and GC, together with the formation of “nucleolar caps” around the nucleolar remnant (also called central body) (Shav-Tal et al., 2005). Different types of caps are formed by nucleolar proteins such as UBF (Figure 1C), nucleoplasmic proteins (mostly RNA-binding proteins), and the CB marker, coilin. Importantly, nucleolar segregation is different from the process of nucleolar fragmentation, which occurs following inhibition of either RNA Pol II (but not I) or protein kinases (David-Pfeuty, 1999, Haaf and Ward, 1996) and leads to unravelling of the FC (Figure 1C). Viral infections can also cause specific changes in nucleolar morphology, such as an increase in nucleolar and/or FC size following corona virus infection (reviewed in Greco, 2009, Hiscox, 2007).
Table 1.
Summary of the Effects of Different Stress Types on Nucleolar and CB Organization
| Stress Type | Trigger | p53 Stabilization | Effects on Nucleolus | Effects on CBs | References |
|---|---|---|---|---|---|
| DNA damage/genotoxic stress |
UV-C | ✓ | Nucleolar segregation, delocalization of Ki-67 | CB disruption and coilin in nucleoplasmic microfoci | Rubbi and Milner, 2003, Al-Baker et al., 2005, Cioce et al., 2006 |
| IR (DSB) | ✓ | Nucleolar disruption, ATM-dependent inhibition of RNA pol I activity | No major effect on coilin distribution | Rubbi and Milner, 2003, Kruhlak et al., 2007 | |
| Camptothecin Bleomycin | ✓ | Nucleolar disruption | N/A | Rubbi and Milner, 2003 | |
| Temperature change | Heat shock | ✓ | Nucleolar disruption | CBs smaller at 39°C; micro-CBs in Xenopus | Rubbi and Milner, 2003, Carmo-Fonseca et al., 1993, Handwerger et al., 2002 |
| Cold shock | CBs bigger at 32°C | Carmo-Fonseca et al., 1993 | |||
| Hypoxia | – | ✓ | Nucleolar disruption, VHL-dependent reduction of rRNA transcription | N/A | Rubbi and Milner, 2003, Mekhail et al., 2006 |
| Osmotic stress | N/A | N/A | Disruption of CBs | Cioce et al., 2006 | |
| Viral infection | Adenovirus, Coronavirus, HCV, HIV, HPV, HSV-1, Poliovirus, West Nile virus | N/A | Changes in nucleolar morphology and proteome | Coilin in nucleoplasmic microfoci and rosettes (adenovirus); ICP0-induced accumulation of coilin at damaged centromeres (HSV-1) | Greco, 2009, Rebelo et al., 1996, James et al., 2010, Morency et al., 2007 |
| Nutrient stress | Serum starvation | N/A | Reduction in ribosomal biogenesis | CB number decreases | Mayer and Grummt, 2006, Murayama et al., 2008, Hoppe et al., 2009, Zhou et al., 2009, Tanaka et al., 2010, Andrade et al., 1993 |
| Inhibition of RNA polymerase I and/or II |
Actinomycin D | ✓ | Nucleolar disruption, release of RPs into the nucleoplasm | Coilin in nucleolar caps | Lindstrom, 2009, Warner and McIntosh, 2009, Zhang and Lu, 2009, Carmo-Fonseca et al., 1992, Shav-Tal et al., 2005 |
| DRB | ✓ | Nucleolar disruption | Nucleolar association of coilin | Rubbi and Milner, 2003 | |
| α-Amanitin | ✓ | Nucleolar disruption | Coilin in cap-like structures associated with the nucleolus | Rubbi and Milner, 2003, Carmo-Fonseca et al., 1992 | |
| Inhibition of nuclear export | Leptomycin B (LMB) | ✓ | No disruption of nucleolar integrity | Nucleolar association of coilin | Rubbi and Milner, 2003, Sleeman et al., 2001 |
| Inhibition of phosphatases | Okadaic acid | N/A | N/A | Accumulation of coilin in the nucleolus | Lyon et al., 1997 |
| Inhibition of DNA and RNA synthesis | 5-Fluorouracil | ✓ | Nucleolar disruption, release of RPs into the nucleoplasm and p53 stabilisation. rRNA processing disrupted in the nucleolus | N/A | Lindstrom, 2009, Warner and McIntosh, 2009, Zhang and Lu, 2009, Burger et al., 2010 |
| Alteration of proteasome activity |
MG132 | ✓ | No disruption of nucleolar integrity, inhibition of late rRNA processing | No disruption of CBs | Rubbi and Milner, 2003, Burger et al., 2010; personal observation |
| Overexpression of PA28γ | N/A | N/A | Disruption of CBs | Cioce et al., 2006 | |
|
Alteration of snRNP biogenesis |
Depletion of SMN, PHAX, TGS1 | N/A | N/A | Disruption of CBs and nucleolar localization of coilin | Lemm et al., 2006 |
| SmB overexpression | N/A | Increase in CB number | Sleeman et al., 2001 | ||
| Oncogenic stress | c-myc or Ras activation | ✓ | Up regulation of nucleolar proteins p14ARF and B23/NPM | N/A | Kruse and Gu, 2009, Lee and Gu, 2010, Chen et al., 2010 |
|
Alteration of ribosome subunit biogenesis |
Malfunction of nucleolar proteins (e.g., Bop1, B23/NPM, nucleostemin) | ✓ | Release of RPs into the nucleoplasm following, in most cases, nucleolar disruption. | N/A | Fumagalli et al., 2009, Lindstrom, 2009, Warner and McIntosh, 2009, Zhang and Lu, 2009 |
| RP knockdown | |||||
IR, gamma irradiation; DSB, double-strand breaks; HCV, hepatitis C virus; HIV, human immunodeficiency virus; HSV-1, herpes simplex virus type 1; RP, ribosomal proteins; N/A, not available.
Cellular Stress and Ribosome Subunit Biogenesis
Efficient ribosome subunit biogenesis is central to gene expression and is also a highly energy-consuming process that is tightly coupled to cell growth, i.e., the ability of a cell to achieve a certain cell mass before cell division can occur. A high transcription rate of rDNA genes and the activity of all three RNA polymerases is required in animal cells undergoing rapid proliferation to meet the cellular demand for ribosomes. Under stress conditions that affect cell-cycle progression and/or intracellular energy status, such as nutrient deprivation, alteration of ribosome subunit biosynthesis is one strategy that can preserve cellular energy homeostasis (Sengupta et al., 2010). This can occur at several different levels, which include regulation of RNA Pol I transcription and/or rRNA processing (reviewed in Chedin et al., 2007, Grummt and Voit, 2010).
Only ∼50% of the ∼400 rDNA repeats in the human diploid genome are transcriptionally active. Interestingly, a change in growth conditions predominantly triggers a change in the transcriptional efficiency of already active genes, rather than the activation of silent genes (reviewed in McStay and Grummt, 2008, Moss et al., 2007). The basal transcription factors, TIF-1A, SL1, and UBF, are essential for transcription by RNA Pol I and appear to be modulated by different signaling pathways in response to changes in environmental conditions (reviewed in Grummt, 2003). A key player for the regulation of ribosomal synthesis in response to extracellular conditions is the kinase mammalian target of rapamycin (mTOR), which promotes pre-rRNA synthesis by regulating the localization and/or activity of TIF-1A, SL1, and UBF, as well as translation of RPs (Mayer and Grummt, 2006, Xiao and Grove, 2009).
mTOR inactivation either by nutrient deprivation or treatment of cells with the specific mTOR inhibitor rapamycin leads to reduced pre-rRNA transcription and thereby decreased ribosome subunit production. Although the mechanisms for starvation-induced inactivation of mTOR are not completely understood, mTOR activity is inhibited by an increase in the cellular AMP/ATP ratio upon nutrient deprivation, via activation of the LKB1-AMPK pathway (Hardie, 2005, Sengupta et al., 2010). Overall, a complex signaling network that integrates mTOR, PI3K (phosphatidylinositol 3-kinase), and MAPK (mitogen-activated protein kinase) pathways is involved in the regulation of ribosome subunit production in response either to changes in nutrient levels, or to growth factor signaling such as IGF-1 (insulin-like growth factor 1) signaling (James and Zomerdijk, 2004).
DNA damage, such as chromosomal double-strand breaks, transiently reduces RNA Pol I transcription in MEFs in an ATM-dependent manner, by interfering with initiation complex assembly and impairing efficient transcription elongation (Kruhlak et al., 2007). rRNA synthesis is also decreased during hypoxia in a process requiring the interaction of the von Hippel-Lindau (vHL) tumor suppressor protein with the rDNA promoter (Mekhail et al., 2006). Interestingly, viral infection can either inhibit or promote the host's pre-RNA synthesis. For example, poliovirus inhibits RNA Pol I activity by inducing SL1 cleavage and UBF posttranslational modification (Banerjee et al., 2005), while hepatitis C virus stimulates RNA Pol I activity, thereby promoting liver carcinogenesis (Kao et al., 2004).
Despite the consensus that stress-dependent regulation of pre-rRNA synthesis mainly occurs by influencing the transcriptional rate of already active genes, recent findings also point toward additional regulatory pathways, such as epigenetic regulation of rRNA transcription. For example, protein complexes whose activities are responsive to intracellular energy status, including eNoSC and NoRC, as well as the JmjC histone demethylase KDM2A, can induce the formation of transcriptionally silent heterochromatin in the nucleolus by triggering either H3K9 dimethylation, acetylation of H4K16, or demethylation of H3K36 in the rDNA locus, thereby repressing rRNA transcription in response to energy deprivation (Murayama et al., 2008, Tanaka et al., 2010, Zhou et al., 2009).
Although most examples of mechanisms for modulating ribosome subunit biogenesis upon cellular stress involve control of RNA Pol I activity, alternative pathways may also exist. For instance, it has been shown recently that infection of human cells with herpes simplex virus type 1 (HSV-1) affects rRNA processing independently of rRNA transcription (Belin et al., 2010).
The Multifunctional Nucleolus: A Central Hub in the Stress Response
The nucleolus appears to be involved in additional cellular functions that may not be directly related to ribosome subunit biogenesis (Pederson and Tsai, 2009, Warner and McIntosh, 2009). The diverse functions of over 4500 nucleolus-associated proteins identified through proteomic experiments are consistent with additional roles for the nucleolus (Ahmad et al., 2009). Classification of the molecular functions of nucleolar proteins shows that only ∼30% have a function clearly related to the production of ribosome subunits. The other functions include biogenesis of multiple RNPs, cell-cycle control, apoptosis, viral infection, DNA replication, and repair, consistent with a role for the nucleolus in providing a link between ribosome subunit biosynthesis, cell-cycle progression, and stress signaling.
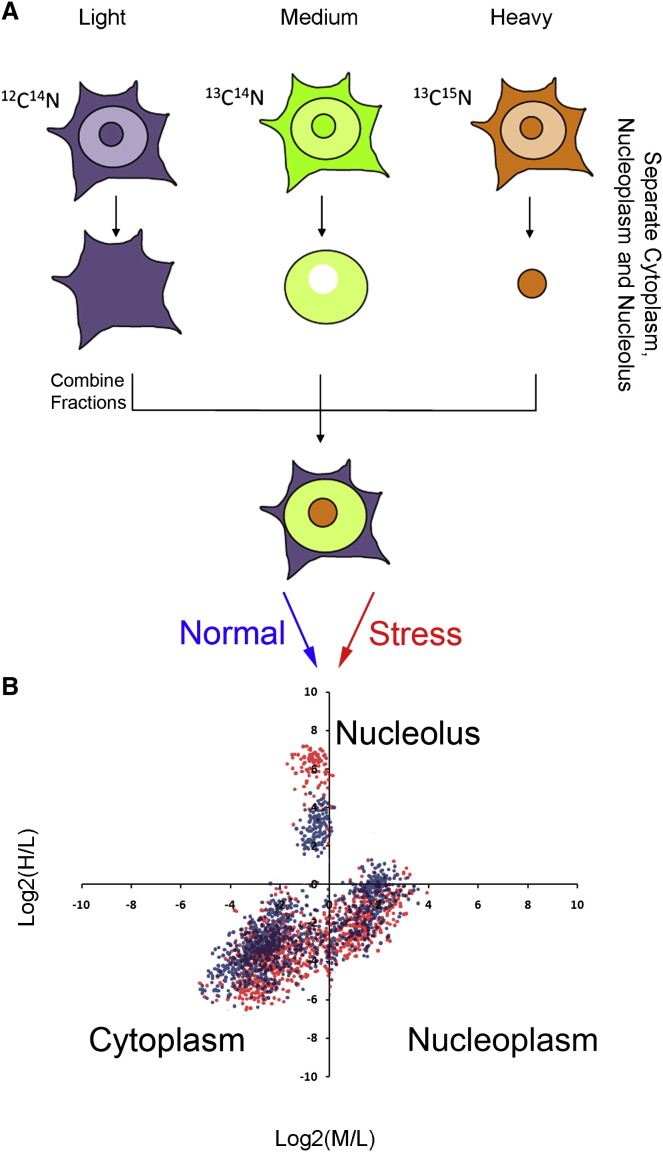
Interestingly, the protein content of the nucleolus is dynamic and alters under stress conditions. Proteomic studies have analyzed the dynamics of the nucleolar proteome in response to stress stimuli, including actinomycin D-mediated inhibition of transcription, viral infection, and DNA damage (Andersen et al., 2005, Boisvert et al., 2010, Boisvert and Lamond, 2010, Emmott et al., 2010, Lam et al., 2010), revealing a complex reorganization of the nucleolus during the stress response. In these studies, mass spectrometry (MS)-based SILAC (stable isotope labeling by amino acids in cell culture) quantitative proteomics (Ong et al., 2002) provide an unbiased and high-throughput approach for characterizing the nucleolar proteome and its dynamics under stress conditions. Rather than defining a nucleolar proteome without reference to the rest of the cell, the “spatial proteomics” approach measures the relative distribution of cellular proteins between different cell compartments. For example, measurement of protein levels between cytoplasm, nucleus, and nucleolus was used to characterize dynamic changes in protein localization following etoposide-induced DNA damage (Boisvert et al., 2010, Boisvert and Lamond, 2010) (Figure 2 ). Furthermore, spatial proteomics performed in parallel on two HCT116 cell lines, either p53 +/+ or p53 −/−, showed that etoposide-induced redistribution of RPs from the nucleolus to the nucleoplasm is p53 dependent (Boisvert and Lamond, 2010).
Figure 2.
“Spatial Proteomics” Approach to Study Nucleolar Proteome Dynamics in Response to Stress
(A) Typical “spatial proteomics” protocol. Cells are labeled with heavy-isotope containing amino acids (SILAC-based labeling) prior to fractionation into cytoplasm, nucleoplasm, and nucleolus. A whole-cell extract is created by recombining differentially labeled subcellular fractions and analyzed by LC-MS/MS. This can be used to measure the relative distribution of cellular proteins between the three compartments (medium/light [M/L] ratio, nucleoplasm/cytoplasm; heavy/medium [H/L] ratio, nucleolus/cytoplasm ratio) and analyze the stress-induced changes in protein localization, as visualized in (B).
Quantitative MS-based proteomics and live-cell fluorescence microscopy can be used as a “dual strategy” to study intracellular protein dynamics and generate quantitative time-lapse measurements of changes in protein levels in different cellular compartments. A complementary fluorescence imaging approach has measured the levels and locations of more than 1000 endogenously tagged proteins in individual living cells before and after CPT (camptothecin) treatment, a specific topoisomerase I inhibitor. This revealed rapid changes in concentration and localization of proteins involved in nucleolar stress, DNA damage, and oxidative stress pathways (Cohen et al., 2008). Interestingly, most proteins that translocated in response to CPT are nucleolar proteins. For example, nucleolar intensity of tagged topoisomerase I, the drug target, decreases in less than 2 min after treatment while other nucleolar proteins, including RPs, show a decrease in nucleolar intensity in less than 1 hr following the addition of the drug. This suggests that nucleolar stress is an immediate effect of CPT and that the nucleolus is involved in early steps of the cellular stress response.
In addition to high-throughput proteomics and imaging methods, many individual studies have characterized the stress-induced reorganization of the nucleolus and analyzed the underlying mechanisms. For example, the NPM/B23-p14ARF-p53 pathway has been well described and represents a good example of nucleolar proteins that are translocated to the nucleoplasm under stress conditions, leading to the stabilization of p53 via interaction of p14ARF with Hdm2, a major player in the negative regulation of p53 (described in detail below) (Gjerset and Bandyopadhyay, 2006). In yeast, detailed molecular mechanisms that link cdc14B translocation from the nucleolus to the nucleoplasm upon DNA damage with the activation of the G2 checkpoint have been deciphered, showing that nucleoplasmic accumulation of cdc14B stabilizes both Claspin, an activator of the DNA damage checkpoint, and Wee1, an inhibitor of cell-cycle progression (Bassermann et al., 2008). In contrast, RelA(p65), a subunit of NF-kB, is targeted to the nucleolus in a stress-dependent manner and triggers apoptosis (Thoms et al., 2010). Nucleolar stress signaling pathways rely on the dynamic sequestration and/or release of proteins in response to stress stimuli, which raises the question of the molecular mechanisms involved in protein nucleolar targeting (discussed by Emmott and Hiscox, 2009). It is anticipated that posttranslational modifications (PTMs) of nucleolar proteins may also play a major role in stress-induced changes in protein localization.
Recent analysis of the nucleolar proteome has uncovered a network comprising chaperones, cochaperones, and multitasking proteins within the nucleolus (Banski et al., 2010a). For example, viral infections induce the nucleolar accumulation of chaperones such as HSP70 (Greco et al., 2010), while other proteins, such as Hsc70s (heat shock cognate proteins 70), can accumulate in the nucleolus when cells are recovering from stress (Banski et al., 2010b). This relocalization results from a combination of a constitutive nucleolar targeting signal along with an autoinhibitory motif indicating that this is a tightly controlled mechanism and not just an indirect effect of stress (Banski et al., 2010b). These observations suggest that the cellular stress response requires the coordinated action of these chaperones and associated proteins in the nucleolus, possibly because essential cellular activities that are particularly sensitive to stress require special attention from chaperones.
Nucleolar Signaling of p53 in the Stress Response
Mechanisms underlying the nucleolar stress response are complex and intertwined. We discuss below the essential role of the nucleolus in the activation of p53 under stress conditions. Several p53-independent pathways that require nucleolar proteins, such as ARF and/or NPM, have also been well studied (reviewed in Li and Hann, 2009, and Sherr, 2006) but are not discussed here due to space limitations.
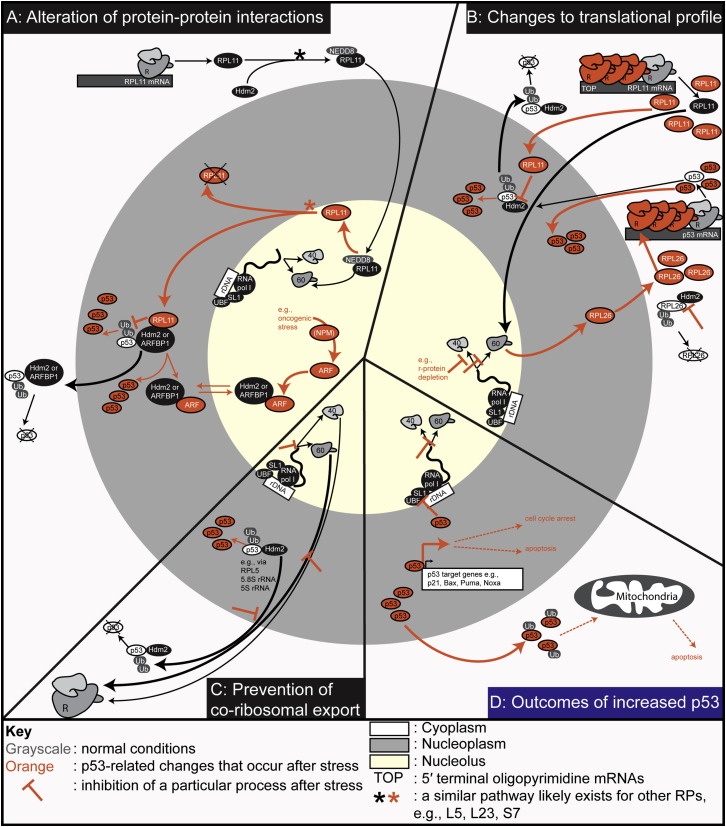
Many of the pathways that convert stress signals into a cellular response link the nucleolus to the stabilization and activation of the tumor suppressor and so-called “guardian of the genome,” p53 (Olson, 2004, Pederson and Tsai, 2009, Rubbi and Milner, 2003). p53-nucleolar signaling pathways rely on different mechanisms that can mediate the increase of cellular p53 levels (illustrated as separate pathways in Figure 3 ). However, it is likely that in reality these pathways are interconnected, depending on the particular stress and cellular conditions.
Figure 3.
Overview of the Nucleolar-Related, Stress-Induced Mechanisms that Result in Increased p53 Activity
The different mechanisms have been separated into three broad categories, namely those that primarily involve either (A) alterations of protein-protein interactions, (B) changes to the translational profile, or (C) prevention of coribosomal export of p53 and Hdmd2. The size of each quadrant represents their relative importance according to current literature. Grayscale text and objects indicate processes that occur during normal physiological conditions, whereas orange text and objects show the changes that occur following stress that result in increased p53 levels and activity. The outcomes of increases p53 are summarized in (D), namely the upregulation of p53 target genes which ultimately leads to either cell-cycle arrest or apoptosis, and the inhibition of ribosome biogenesis via the inhibition of rRNA transcription. Refer to text for detailed explanations of these pathways and for references.
The main nuclear role of p53 is to upregulate transcription of RNA polymerase II-transcribed genes such as p21, Bax, Puma, and Noxa, whereas in the cytoplasm p53 activates the mitochondrial apoptotic pathway (Lee and Gu, 2010). p53 can also function as an inhibitor of RNA Pol I transcription in the nucleolus by disrupting SL1-UBF interactions, which results in a decrease in ribosome subunit biogenesis (Figure 3D; Zhai and Comai, 2000).
p53 levels are kept low under normal physiological conditions due to its interactions with E3 ubiquitin ligases, the best-studied of which is Hdm2 (also called Mdm2 in mouse), resulting in p53 nuclear export (following monoubiquitinylation), degradation (following polyubiquitinylation), and/or inhibition of its transactivation domain (Kruse and Gu, 2009, Lee and Gu, 2010, Olson, 2004, Vousden and Prives, 2009). p53 activation is essential for both stress-induced cell-cycle arrest and apoptosis. Indeed, failure of a cell to properly regulate p53 activity following disruption of ribosome biogenesis (ribosomal stress) has been linked to human disorders such as Diamond Blackfan Anaemia and TCOF1 (Treacher Collins-Franceschetti syndrome 1) (reviewed in Narla and Ebert, 2010). In mice, mutations in genes encoding RPs Rps19 and Rps20 cause p53-dependent pigmentation defects (McGowan et al., 2008).
Nucleolar-related mechanisms to activate p53 can be broadly separated into three categories: alterations to protein-protein interactions, prevention of coribosomal export of p53-Hdm2, and changes to the cellular translational profile (Figures 3A–3C). All three mechanisms involve the relocalization of proteins, and in many ways, the nucleolus can be seen as a “barrier” to promote or inhibit particular p53-based interactions. It has been observed that most types of cellular stress that result in an increase in p53 stability also involve the disruption of nucleolar integrity, leading to the hypothesis of the nucleolus being a stress sensor responsible for the suppression of p53 levels (Table 1; Rubbi and Milner, 2003). This model provides an explanation for the ability of disparate stress pathways, including DNA damage, transcriptional inhibition, depletion of nucleotide pools, oncogene expression, viral infection, and heat shock, to converge and result in a similar outcome. However, some stress pathways, such as those activated following bleomycin treatment, or disruption of 40S ribosome subunit biogenesis, lead to the activation of p53 with minimal disruption of nucleolar integrity (Fumagalli et al., 2009, Olson, 2004), suggesting that Rubbi and Milner's nucleolar disruption model explains many, but not all, of the mechanisms that activate p53 following stress.
Stress-Induced p53 Stabilization Mechanisms that Mainly Involve Changes to Protein-Protein Interactions
The first example of a mechanism that regulates p53 stability at the level of protein-protein interactions involves the upregulation of the tumor suppressor protein, p14ARF. This occurs following oncogenic or genotoxic stresses, such as hyperproliferative signals emanating from oncogenic Ras and overexpressed Myc. p14ARF, like p53, is maintained at low levels at steady state in most cell types due to ubiquitin-mediated degradation. Upregulation of p14ARF prevents the proliferation of cells bearing damaged DNA via the stabilization of p53, and consistent with this, p14ARF is often mutated or silenced in tumor cells (Montanaro et al., 2008). p14ARF binds Hdm2 and blocks its interaction with p53. Although p14ARF is a nucleolar protein, it is not clear if relocalization of Hdm2 to the nucleolus is necessary to prevent its interactions with p53, or if it is sufficient for p14ARF to inhibit the ubiquitin ligase activity of Hdm2 in the nucleoplasm (Kruse and Gu, 2009, Lee and Gu, 2010). Indeed, the C-terminal region of ARF, which includes the nucleolar localization sequence, is not essential for the regulation of p53 ubiquitinylation by Hdm2 (Zhang and Xiong, 1999).
While long-term upregulation of p14ARF involves increased synthesis at the transcriptional level (Gil and Peters, 2006), short-term p14ARF stabilization is achieved by promoting its nucleolar localization via interactions with B23/NPM, which in turn prevents its association with ubiquitin ligases, such as ULF, in the nucleoplasm. B23/NPM is also upregulated following oncogenic stress (Chen et al., 2010). Failure to activate these p14ARF-B23/NPM-p53 pathways responsible for p14ARF stabilization is observed in many forms of cancer; for example, a high-mutation rate of B23/NPM is observed in primary acute myeloid leukemia, and the overexpression of ULF is detected in many human tumors (Chen et al., 2010). These pathways are also subjected to a negative feedback loop, since p14ARF promotes the ubiquitinylation and degradation of B23/NPM, leading to inhibition of pre-rRNA processing and subsequent disruption of ribosome subunit biogenesis (Olson, 2004).
Another example of changes to protein-protein interactions resulting in increased p53 activity is the release of RPs from the nucleolus following ribosomal stress. This stress can be triggered by serum starvation, cell contact inhibition, depletion of nucleotides such as GTP, treatment with low doses of actinomycin D (1−5 nM) or 5-Fluorouracil, or the malfunction of nucleolar proteins involved in ribosome subunit biogenesis (such as expression of a Bop1 dominant-negative mutant, inhibition of B23/NPM activity by p14ARF, overexpression of nucleostemin, or the reduction of particular RPs) (Deisenroth and Zhang, 2010, Lindstrom, 2009, Warner and McIntosh, 2009, Zhang and Lu, 2009). Nucleoplasmic RPs such as L5, L11, L23, and S7 have been shown to directly interact with Hdm2 and thus prevent the ubiquitin-mediated degradation of p53, which delays proliferation under unfavorable conditions (Zhang and Lu, 2009).
RPL11 is a well-studied example of a RP that participates in a p53-nucleolar stress response pathway. RPL11 is normally modified by the ubiquitin-like modifier NEDD8 following its translation in the cytoplasm. NEDD8 modification is promoted by Hdm2 and is required for RPL11 stabilization and nucleolar localization. Following stress, RPL11 is de-NEDDylated, resulting in its nucleoplasmic accumulation. Although this leads to RPL11 degradation, it can first interact with Hdm2 and mediate p53 stabilization (Sundqvist et al., 2009). The release of RPL11 into the nucleoplasm appears to be a common link between a number of pathways that survey the maturation of the small and large ribosomal subunits (e.g., 18S and 28S rRNA production) and regulate p53 stability in response to defects in these processes (Holzel et al., 2010).
Translation-Mediated Mechanisms to Increase p53 Levels
In addition to the nucleolar deNEDDylation discussed above, an increase in nucleoplasmic RPL11 levels and subsequent inhibition of Hdm2-mediated p53 degradation can also result from increased translation of RPL11 mRNA. This can be triggered by stress resulting from disruption of 40S ribosome subunit biogenesis (Fumagalli et al., 2009). The RPL11-encoding mRNA (along with many other RP-encoding mRNAs) contains a TOP (5′-terminal oligopyrimidine) sequence, which allows a subset of mRNAs to be translated under conditions in which global protein synthesis is otherwise inhibited (Caldarola et al., 2009).
RPL26 also increases the translation of p53 mRNA by binding to its 5′ untranslated region. Under normal conditions, this is prevented by Hdm2-mediated ubiquitinylation and degradation of RPL26. However, following genotoxic or ribosomal stress, both a release of RPL26 from the 60S ribosomal subunit and a decrease in Hdm2 activity (possibly via PTMs) result in an increase of free RPL26, which upregulates p53 translation (Ofir-Rosenfeld et al., 2008, Zhang and Lu, 2009).
Prevention of Coribosomal Export of p53/Hdm2
Based on the observation that p53 and/or Hdm2 can interact with a number of RPs (such as RPL5 and RPL11), as well as with the 5.8S and 5S rRNAs (Fontoura et al., 1992, Riley and Maher, 2007, Zhang and Lu, 2009), it has been suggested that p53/Hdm2 are cotransported with the ribosomal subunits during their export from the nucleolus to the cytoplasm. This may either prevent p53 interacting with its target genes in the nucleoplasm or promote its ubiquitin-mediated degradation in the cytoplasm, or both. Following stress, both production and export of the ribosome subunits are decreased and p53/Hdm2 are no longer transported to the cytoplasm, thus increasing the opportunity for p53 to activate transcription of its target genes in the nucleoplasm.
The Role of p53-Related Posttranslational Modifications in the Nucleolar Stress Response
There is evidence that PTMs of p53 play a role in nucleolar stress responses that result in the activation of p53. As already discussed, under normal conditions, ubiquitination of p53 via E3 ligases such as Hdm2 is associated with the suppression of p53 activity. Many stress-responses pathways lead to a decrease in p53 ubiquitination, via mechanisms that involve either increases, such as for p14ARF and B23/NPM, or decreases, such as for RPs, in the nucleolar levels of particular proteins (see above; Hock and Vousden, 2010, Lee and Gu, 2010). Furthermore, following cellular stresses/oncogenic activation that upregulate p14ARF, there is evidence that SUMOylation of p53 is upregulated in an Hdm2-dependent manner, which may result in an increase in the nucleolar localization of p53 and thus suppress nucleoplasmic functions of p53. However, the precise role of SUMOylation in p53 function is yet to be clearly elucidated (Carter and Vousden, 2009, Stehmeier and Muller, 2009).
Acetylation of lysine residues in p53 also appears to be important for the accumulation of p53 following nucleolar stress. Under normal conditions, p53 acetylation is actively suppressed by Hdm2. However, ribosomal stress induced by actinomycin D treatment specifically leads to p300/CBP-mediated acetylation of both p53 and Hdm2, which in turn upregulates p53 transcriptional activity and inhibits Hdm2 E3 ubiquitin ligase activity (Carter and Vousden, 2009, Lee and Gu, 2010).
The mechanistic role of p53 PTMs in the nucleolar stress response is not yet fully understood, given the plethora of p53 PTMs that have been observed during both normal homeostasis and following stress. For example, more than 36 different amino acids within p53 have been shown to be modified (Kruse and Gu, 2008, Kruse and Gu, 2009). These modifications include phosphorylation, methylation, NEDDylation, O-linked N-acetylglucosamine, ADP-ribosylation, prolyl isomerization, and oxidation of methionine. Many of the p53 PTMs are targeted to overlapping residues, suggesting that some are in direct competition (Carter and Vousden, 2009). Moving forward, it will be important to understand how such a diverse range of stress types can result in so many differently modified forms of p53 and yet elicit similar cellular responses.
Crosstalk between Cajal Bodies and the Nucleolus in the Stress Response
CBs are distinct nuclear bodies that function in coordinating maturation of certain nuclear RNAs, including small nuclear RNA (snRNA), small nucleolar RNA (snoRNA), and histone mRNA processing (reviewed in Kiss et al., 2006, Matera et al., 2007, Nizami et al., 2010, Stanek and Neugebauer, 2006). CBs were first identified in human neurons in 1903 by the Spanish neurocytologist Santiago Ramon y Cajal but have since been described in many other organisms and cell types (Matera, 2006). They are characterized by the p80-coilin protein, a main structural component of CBs, which is commonly used as a signature marker. In addition to coilin, CBs contain an ever-growing list of residents that are involved in distinct cellular pathways, including snRNP and snoRNP biogenesis, as well as histone mRNA processing (reviewed in Cioce and Lamond, 2005). Recent studies performed in zebrafish and mice showed that depletion of p80-coilin leads to CB dispersal, defects in snRNP biogenesis and splicing, and developmental arrest (Strzelecka et al., 2010, Walker et al., 2009), indicating that the formation of CBs plays an essential role in snRNP processing during embryogenesis. In addition, CBs are linked to the regulation of cell-cycle progression. CB number and composition vary throughout the cell cycle, with CB size being maximal at G1/S phase transition (Andrade et al., 1993). In S phase, the recruitment of the cyclinE/cdk2 complex to CBs leads to the phosphorylation of NPAT and the subsequent activation of histone mRNA transcription (Marzluff et al., 2008). The observation that CBs contain factors that are functionally related to cell proliferation, such as ZPR1 and FGF-2 (Cioce and Lamond, 2005), suggests an additional role for CBs in the transduction of proliferative signals to the nucleus.
CBs are intimately linked with the nucleolus, both on physical and functional levels. CBs were originally called “nucleolar accessory bodies” because of their close association with nucleoli in neurons (Gall, 2000, Lafarga et al., 2009). CBs are even sometimes found within the nucleolus and a budding yeast counterpart of the CB, termed the “nucleolar body,” is part of the nucleolus (Cioce and Lamond, 2005, Verheggen et al., 2002). In addition, CBs are involved in the maturation of snoRNPs, which are subsequently transported to the nucleoli, where they participate in rRNA processing. In general, both nucleoli and CBs are involved in the production of non-poly(A)-tailed RNAs that are tightly connected to cell growth, including histone mRNAs, snRNAs, and snoRNAs in CBs, and rRNAs in nucleoli. Consistent with this, both structures are prominent in cells that are transcriptionally and metabolically active, such as neuronal and cancer cells (Berciano et al., 2007).
Altogether, these findings suggest an intimate link between CBs and nucleoli. Given that the nucleolus acts as a major hub in coordinating the stress response, it is not surprising that the related CBs are also involved in the cellular response to stress.
Dynamics of CBs upon Cellular Stress
The structure of CBs is altered by different types of stress, such as UV irradiation, heat shock, transcriptional inhibition, osmotic stress, starvation, and viral infection, all of which correlate with a decrease in cellular metabolism and RNA processing. However, the redistribution of coilin, and other CB components, varies according to the type of stress, as summarized in Table 1. Nutrient deprivation results in a decreased number of CBs (Andrade et al., 1993), whereas UV-C irradiation, osmotic stress, and heat shock reversibly disrupt CBs, as visualized by the redistribution of coilin to nucleoplasmic microfoci (Cioce et al., 2006, Handwerger et al., 2002) (Figure 1C). A subset of UV-C-irradiated cells also forms coilin-containing nucleolar caps, similar to those observed upon the inhibition of RNA Pol I and II by actinomycin D (Andersen et al., 2005, Shav-Tal et al., 2005). In cells treated with DRB (5,6-dichloro-1-β-D-ribobenzimidazole), a kinase inhibitor that inhibits RNA polymerase II transcription, coilin relocalizes to cap-like structures associated with the nucleolus (Figure 1C). Following viral infection, different types of CB-like structures have been observed. In HSV-1-infected cells, ICP0 (viral protein infected cell protein 0) expression induces the destabilization of centromeres and the subsequent redistribution of coilin, SMN, and fibrillarin to the damaged centromeres (Morency et al., 2007). In contrast, adenovirus-induced “rosettes” reflect the recruitment of coilin and other CB components to the periphery of viral replication centers, where they are involved in processing of late-phase viral transcripts (James et al., 2010). Similarly, in plants, groundnut rosette virus (GRV) infection leads to the reorganization of CBs into CB-like structures containing viral protein ORF3, which fuse with the nucleolus and facilitate the formation of viral RNP particles that are efficient for systemic infection (Kim et al., 2007). The redistribution of coilin observed upon viral infection may reflect that the CB RNA processing machinery is “hijacked” by the virus.
Most stress signals that induce CB disruption are accompanied by transcription inhibition. In particular, snRNA transcription is inhibited upon UV-C irradiation, in a p53-dependent manner (Gridasova and Henry, 2005), as well as histone mRNA transcription (Bongiorno-Borbone et al., 2010). This supports the idea that UV-C-induced CB disruption is associated with CB activity being, at least partially, shut down. However, is the inhibition of CB activity the cause or the consequence of CB disruption? In fact, the inhibition of either transcription, or snRNP biogenesis, can induce CB disruption (Carmo-Fonseca et al., 1992, Lemm et al., 2006). Conversely, CB disruption induced by coilin depletion induces defects in snRNP biogenesis (Strzelecka et al., 2010). It is therefore likely that mechanisms involved in snRNA processing inhibition and CB disruption are intimately interconnected.
Given the functional link between CBs and nucleoli, it would not be surprising that common mechanisms are involved in the stress-induced rearrangements of these two organelles. Interestingly, p53 has been detected in CBs upon UV-C treatment (Young et al., 2002). In addition, different types of stress that affect CB integrity, such as the inhibition of transcription and/or snRNA processing, induce the redistribution of coilin to the nucleolus (Figure 1C and Table 1). An important question is whether the stress-induced redistribution of coilin to the nucleolus is associated with a specific role of coilin, and other CB components, in the stress response. Given that coilin is required for efficient viral infection (James et al., 2010, Kim et al., 2007) and that coilin is sometimes found associated with remnants of nucleolar transcription upon UV-C irradiation and with the unravelled rDNA transcription units upon α-amanitin treatment (Cioce et al., 2006, Haaf and Ward, 1996), it is possible that CB-like structures are involved in the processing of specific subsets of RNA transcripts produced under stress conditions. Altogether, although underlying mechanisms remain to be deciphered, these observations suggest a possible crosstalk between the nucleolus and CBs during the stress response.
Mechanisms Involved in CB Disassembly upon Stress
Kaiser and colleagues showed that CB assembly exhibits hallmarks of a self-organizing structure (Kaiser et al., 2008). Therefore, it is likely that stress-induced disassembly of CBs results from specific mechanisms that affect interactions between CB components, and/or provoke the degradation of one or several CB components. Changes in the CB protein interaction network could result from posttranslational modification of key CB components. In particular, SUMOylation of CB components has been proposed to increase upon stress (Navascues et al., 2008). Other modifications of coilin, such as phosphorylation and methylation, could play a role in stress-induced coilin redistribution. Indeed, coilin phosphorylation, as well as coilin demethylation, affects its self-association properties and results in its nucleolar accumulation (Hebert, 2010, Tapia et al., 2010).
UV-C-induced coilin redistribution is mediated, at least in part, by the proteasome activator subunit PA28γ (Cioce et al., 2006). This suggests that proteasomal activity is required for CB disruption and that active mechanisms are responsible for stress-induced CB fragmentation. Although coilin levels show little or no change upon UV-C irradiation (Cioce et al., 2006), a recent study demonstrated that FLASH, an essential component of CBs, is degraded by the proteasome upon UV-C treatment (Bongiorno-Borbone et al., 2010). Since PA28γ and FLASH interact in a yeast two-hybrid assay (Mao et al., 2008), it will be interesting to analyze whether this degradation is PA28γ dependent, and whether other CB components are also specifically degraded upon stress. More generally, the role of the proteasome in the stress-induced reorganization of nuclear architecture also needs to be deciphered. At this stage, it is tempting to draw a parallel with the proteasome impairment observed in several neurodegenerative diseases, such as Huntington's or Parkinson's, which leads to the accumulation of misfolded proteins in nuclear aggregates (Rubinsztein, 2006). Could these aggregates be linked to a defect in the proteasome-mediated reorganization of the nucleus upon stress? Future studies to analyze the role of the proteasome and PA28γ in stress-induced reorganization of nuclear architecture may provide further insights into our understanding of these diseases.
Perspectives
The nucleolus and related CBs are morphologically distinct subnuclear organelles that are involved in coordinating major RNA-protein assembly and modification processes in proliferating cells. It is therefore not surprising that the structure and function of both entities are affected by cellular stress. Both the nucleolus and CBs appear to be major targets of signaling pathways that are activated by the cellular stress response, resulting in a complex range of changes in the organization, size, and protein content of these two nuclear organelles. Nucleolar function in coupling ribosome subunit biogenesis and cell-cycle progression, by controlling the activity of the tumor suppressor protein p53, places the nucleolus as a central hub in coordinating the cellular stress response. Although the mechanisms controlling these stress signaling pathways have been analyzed extensively, and a wide range of stress-responsive protein complexes have been identified, many details remain uncharacterized. In particular, the molecular mechanisms that affect nucleolar and CB structure and function under stress conditions need to be elucidated. Hence, valuable new insights should be provided by studying the dynamics of large-scale protein-protein interaction networks and the changes in PTMs of key nuclear factors under stress conditions. Given the essential role of ribosome subunit biogenesis in cell growth, further characterization of nucleolar stress signaling pathways and the subsequent identification of new biomarkers and molecular targets will be important for defining and manipulating mechanisms involved in cancer and neurodegenerative diseases.
Acknowledgments
We are grateful to Laura Trinkle-Mulcahy for providing DIC images of live HeLa cells. We thank Dimitris Xirodimas for critical reading of the manuscript and members of the Lamond laboratory for helpful discussions and advice. This work was funded by a Wellcome Trust Programme Grant (083524/Z/07/Z), with additional support from the EU networks “PROSPECTS” (HEALTH-F4-2008-201648) and “EURASNET” (LSHG-CT-2005-518238), and from an interdisciplinary RASOR (Radical Solutions for Researching the Proteome) initiative. A.I.L. is a Wellcome Trust Principal Research Fellow. S.B. is a Human Frontier Science Program long-term fellow. B.J.W. is a Marie Curie International Incoming Fellow. S.H. is a Leopoldina Research Fellow. F.-M.B. is supported by a fellowship from the Caledonian Research Foundation. We apologize to the many authors whose primary research articles could not all be cited due to space limitations.
References
- Ahmad Y., Boisvert F.M., Gregor P., Cobley A., Lamond A.I. NOPdb: Nucleolar Proteome Database—2008 update. Nucleic Acids Res. 2009;37:D181–D184. doi: 10.1093/nar/gkn804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Baker E.A., Oshin M., Hutchison C.J., Kill I.R. Analysis of UV-induced damage and repair in young and senescent human dermal fibroblasts using the comet assay. Mech. Ageing Dev. 2005;126:664–672. doi: 10.1016/j.mad.2004.12.002. [DOI] [PubMed] [Google Scholar]
- Andersen J.S., Lam Y.W., Leung A.K., Ong S.E., Lyon C.E., Lamond A.I., Mann M. Nucleolar proteome dynamics. Nature. 2005;433:77–83. doi: 10.1038/nature03207. [DOI] [PubMed] [Google Scholar]
- Andrade L.E., Tan E.M., Chan E.K. Immunocytochemical analysis of the coiled body in the cell cycle and during cell proliferation. Proc. Natl. Acad. Sci. USA. 1993;90:1947–1951. doi: 10.1073/pnas.90.5.1947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee R., Weidman M.K., Navarro S., Comai L., Dasgupta A. Modifications of both selectivity factor and upstream binding factor contribute to poliovirus-mediated inhibition of RNA polymerase I transcription. J. Gen. Virol. 2005;86:2315–2322. doi: 10.1099/vir.0.80817-0. [DOI] [PubMed] [Google Scholar]
- Banski P., Kodiha M., Stochaj U. Chaperones and multitasking proteins in the nucleolus: networking together for survival? Trends Biochem. Sci. 2010;35:361–367. doi: 10.1016/j.tibs.2010.02.010. [DOI] [PubMed] [Google Scholar]
- Banski P., Mahboubi H., Kodiha M., Shrivastava S., Kanagaratham C., Stochaj U. Nucleolar targeting of the chaperone HSC70 is regulated by stress, cell signaling and a composite targeting signal which is controlled by autoinhibition. J. Biol. Chem. 2010;285:21858–21867. doi: 10.1074/jbc.M110.117291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassermann F., Frescas D., Guardavaccaro D., Busino L., Peschiaroli A., Pagano M. The Cdc14B-Cdh1-Plk1 axis controls the G2 DNA-damage-response checkpoint. Cell. 2008;134:256–267. doi: 10.1016/j.cell.2008.05.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belin S., Kindbeiter K., Hacot S., Albaret M.A., Roca-Martinez J.X., Therizols G., Grosso O., Diaz J.J. Uncoupling ribosome biogenesis regulation from RNA polymerase I activity during herpes simplex virus type 1 infection. RNA. 2010;16:131–140. doi: 10.1261/rna.1935610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berciano M.T., Novell M., Villagra N.T., Casafont I., Bengoechea R., Val-Bernal J.F., Lafarga M. Cajal body number and nucleolar size correlate with the cell body mass in human sensory ganglia neurons. J. Struct. Biol. 2007;158:410–420. doi: 10.1016/j.jsb.2006.12.008. [DOI] [PubMed] [Google Scholar]
- Boisvert F.M., Lamond A.I. P53-dependent subcellular proteome localization following DNA damage. Proteomics. 2010 doi: 10.1002/pmic.201000213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boisvert F.M., van Koningsbruggen S., Navascues J., Lamond A.I. The multifunctional nucleolus. Nat. Rev. Mol. Cell Biol. 2007;8:574–585. doi: 10.1038/nrm2184. [DOI] [PubMed] [Google Scholar]
- Boisvert F.M., Lam Y.W., Lamont D., Lamond A.I. A quantitative proteomics analysis of subcellular proteome localization and changes induced by DNA damage. Mol. Cell. Proteomics. 2010;9:457–470. doi: 10.1074/mcp.M900429-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bongiorno-Borbone L., De Cola A., Barcaroli D., Knight R.A., Di Ilio C., Melino G., De Laurenzi V. FLASH degradation in response to UV-C results in histone locus bodies disruption and cell-cycle arrest. Oncogene. 2010;29:802–810. doi: 10.1038/onc.2009.388. [DOI] [PubMed] [Google Scholar]
- Burger K., Muhl B., Harasim T., Rohrmoser M., Malamoussi A., Orban M., Kellner M., Gruber-Eber A., Kremmer E., Holzel M., Eick D. Chemotherapeutic drugs inhibit ribosome biogenesis at various levels. J. Biol. Chem. 2010;285:12416–12425. doi: 10.1074/jbc.M109.074211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caldarola S., De Stefano M.C., Amaldi F., Loreni F. Synthesis and function of ribosomal proteins–fading models and new perspectives. FEBS J. 2009;276:3199–3210. doi: 10.1111/j.1742-4658.2009.07036.x. [DOI] [PubMed] [Google Scholar]
- Carmo-Fonseca M., Pepperkok R., Carvalho M.T., Lamond A.I. Transcription-dependent colocalization of the U1, U2, U4/U6, and U5 snRNPs in coiled bodies. J. Cell Biol. 1992;117:1–14. doi: 10.1083/jcb.117.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmo-Fonseca M., Ferreira J., Lamond A.I. Assembly of snRNP-containing coiled bodies is regulated in interphase and mitosis—evidence that the coiled body is a kinetic nuclear structure. J. Cell Biol. 1993;120:841–852. doi: 10.1083/jcb.120.4.841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter S., Vousden K.H. Modifications of p53: competing for the lysines. Curr. Opin. Genet. Dev. 2009;19:18–24. doi: 10.1016/j.gde.2008.11.010. [DOI] [PubMed] [Google Scholar]
- Chedin S., Laferte A., Hoang T., Lafontaine D.L., Riva M., Carles C. Is ribosome synthesis controlled by pol I transcription? Cell Cycle. 2007;6:11–15. doi: 10.4161/cc.6.1.3649. [DOI] [PubMed] [Google Scholar]
- Chen D., Shan J., Zhu W.G., Qin J., Gu W. Transcription-independent ARF regulation in oncogenic stress-mediated p53 responses. Nature. 2010;464:624–627. doi: 10.1038/nature08820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cioce M., Lamond A.I. Cajal bodies: a long history of discovery. Annu. Rev. Cell Dev. Biol. 2005;21:105–131. doi: 10.1146/annurev.cellbio.20.010403.103738. [DOI] [PubMed] [Google Scholar]
- Cioce M., Boulon S., Matera A.G., Lamond A.I. UV-induced fragmentation of Cajal bodies. J. Cell Biol. 2006;175:401–413. doi: 10.1083/jcb.200604099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen A.A., Geva-Zatorsky N., Eden E., Frenkel-Morgenstern M., Issaeva I., Sigal A., Milo R., Cohen-Saidon C., Liron Y., Kam Z. Dynamic proteomics of individual cancer cells in response to a drug. Science. 2008;322:1511–1516. doi: 10.1126/science.1160165. [DOI] [PubMed] [Google Scholar]
- David-Pfeuty T. Potent inhibitors of cyclin-dependent kinase 2 induce nuclear accumulation of wild-type p53 and nucleolar fragmentation in human untransformed and tumor-derived cells. Oncogene. 1999;18:7409–7422. doi: 10.1038/sj.onc.1203103. [DOI] [PubMed] [Google Scholar]
- Deisenroth C., Zhang Y. Ribosome biogenesis surveillance: probing the ribosomal protein-Mdm2-p53 pathway. Oncogene. 2010;29:4253–4260. doi: 10.1038/onc.2010.189. [DOI] [PubMed] [Google Scholar]
- Emmott E., Hiscox J.A. Nucleolar targeting: the hub of the matter. EMBO Rep. 2009;10:231–238. doi: 10.1038/embor.2009.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emmott E., Rodgers M., Macdonald A., McCrory S., Ajuh P., Hiscox J.A. Quantitative proteomics using stable isotope labeling with amino acids in cell culture (SILAC) reveals changes in the cytoplasmic, nuclear and nucleolar proteomes in Vero cells infected with the coronavirus infectious bronchitis virus. Mol. Cell. Proteomics. 2010;9:5335–5345. doi: 10.1074/mcp.M900345-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontoura B.M., Sorokina E.A., David E., Carroll R.B. p53 is covalently linked to 5.8S rRNA. Mol. Cell. Biol. 1992;12:5145–5151. doi: 10.1128/mcb.12.11.5145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fumagalli S., Di Cara A., Neb-Gulati A., Natt F., Schwemberger S., Hall J., Babcock G.F., Bernardi R., Pandolfi P.P., Thomas G. Absence of nucleolar disruption after impairment of 40S ribosome biogenesis reveals an rpL11-translation-dependent mechanism of p53 induction. Nat. Cell Biol. 2009;11:501–508. doi: 10.1038/ncb1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gall J.G. Cajal bodies: the first 100 years. Annu. Rev. Cell Dev. Biol. 2000;16:273–300. doi: 10.1146/annurev.cellbio.16.1.273. [DOI] [PubMed] [Google Scholar]
- Gil J., Peters G. Regulation of the INK4b-ARF-INK4a tumour suppressor locus: all for one or one for all. Nat. Rev. Mol. Cell Biol. 2006;7:667–677. doi: 10.1038/nrm1987. [DOI] [PubMed] [Google Scholar]
- Gjerset R.A., Bandyopadhyay K. Regulation of p14ARF through subnuclear compartmentalization. Cell Cycle. 2006;5:686–690. doi: 10.4161/cc.5.7.2623. [DOI] [PubMed] [Google Scholar]
- Govoni M., Farabegoli F., Pession A., Novello F. Inhibition of topoisomerase II activity and its effect on nucleolar structure and function. Exp. Cell Res. 1994;211:36–41. doi: 10.1006/excr.1994.1055. [DOI] [PubMed] [Google Scholar]
- Greco A. Involvement of the nucleolus in replication of human viruses. Rev. Med. Virol. 2009;19:201–214. doi: 10.1002/rmv.614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greco A., Di Benedetto A., Howard C.M., Kelly S., Nande R., Dementieva Y., Miranda M., Brunetti A., Salvatore M., Claudio L. Eradication of therapy-resistant human prostate tumors using an ultrasound-guided site-specific cancer terminator virus delivery approach. Mol. Ther. 2010;18:295–306. doi: 10.1038/mt.2009.252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gridasova A.A., Henry R.W. The p53 tumor suppressor protein represses human snRNA gene transcription by RNA polymerases II and III independently of sequence-specific DNA binding. Mol. Cell. Biol. 2005;25:3247–3260. doi: 10.1128/MCB.25.8.3247-3260.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grummt I. Life on a planet of its own: regulation of RNA polymerase I transcription in the nucleolus. Genes Dev. 2003;17:1691–1702. doi: 10.1101/gad.1098503R. [DOI] [PubMed] [Google Scholar]
- Grummt I., Voit R. Linking rDNA transcription to the cellular energy supply. Cell Cycle. 2010;9:225–226. doi: 10.4161/cc.9.2.10614. [DOI] [PubMed] [Google Scholar]
- Haaf T., Ward D.C. Inhibition of RNA polymerase II transcription causes chromatin decondensation, loss of nucleolar structure, and dispersion of chromosomal domains. Exp. Cell Res. 1996;224:163–173. doi: 10.1006/excr.1996.0124. [DOI] [PubMed] [Google Scholar]
- Handwerger K.E., Wu Z., Murphy C., Gall J.G. Heat shock induces mini-Cajal bodies in the Xenopus germinal vesicle. J. Cell Sci. 2002;115:2011–2020. doi: 10.1242/jcs.115.10.2011. [DOI] [PubMed] [Google Scholar]
- Hardie D.G. New roles for the LKB1→AMPK pathway. Curr. Opin. Cell Biol. 2005;17:167–173. doi: 10.1016/j.ceb.2005.01.006. [DOI] [PubMed] [Google Scholar]
- Hebert M.D. Phosphorylation and the Cajal body: modification in search of function. Arch. Biochem. Biophys. 2010;496:69–76. doi: 10.1016/j.abb.2010.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henras A.K., Soudet J., Gerus M., Lebaron S., Caizergues-Ferrer M., Mougin A., Henry Y. The post-transcriptional steps of eukaryotic ribosome biogenesis. Cell. Mol. Life Sci. 2008;65:2334–2359. doi: 10.1007/s00018-008-8027-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiscox J.A. RNA viruses: hijacking the dynamic nucleolus. Nat. Rev. Microbiol. 2007;5:119–127. doi: 10.1038/nrmicro1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hock A., Vousden K.H. Regulation of the p53 pathway by ubiquitin and related proteins. Int. J. Biochem. Cell Biol. 2010;42:1618–1621. doi: 10.1016/j.biocel.2010.06.011. [DOI] [PubMed] [Google Scholar]
- Holzel M., Orban M., Hochstatter J., Rohrmoser M., Harasim T., Malamoussi A., Kremmer E., Langst G., Eick D. Defects in 18 S or 28 S rRNA processing activate the p53 pathway. J. Biol. Chem. 2010;285:6364–6370. doi: 10.1074/jbc.M109.054734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoppe S., Bierhoff H., Cado I., Weber A., Tiebe M., Grummt I., Voit R. AMP-activated protein kinase adapts rRNA synthesis to cellular energy supply. Proc. Natl. Acad. Sci. USA. 2009;106:17781–17786. doi: 10.1073/pnas.0909873106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James M.J., Zomerdijk J.C. Phosphatidylinositol 3-kinase and mTOR signaling pathways regulate RNA polymerase I transcription in response to IGF-1 and nutrients. J. Biol. Chem. 2004;279:8911–8918. doi: 10.1074/jbc.M307735200. [DOI] [PubMed] [Google Scholar]
- James N.J., Howell G.J., Walker J.H., Blair G.E. The role of Cajal bodies in the expression of late phase adenovirus proteins. Virology. 2010;399:299–311. doi: 10.1016/j.virol.2010.01.013. [DOI] [PubMed] [Google Scholar]
- Kaiser T.E., Intine R.V., Dundr M. De novo formation of a subnuclear body. Science. 2008;322:1713–1717. doi: 10.1126/science.1165216. [DOI] [PubMed] [Google Scholar]
- Kao C.F., Chen S.Y., Lee Y.H. Activation of RNA polymerase I transcription by hepatitis C virus core protein. J. Biomed. Sci. 2004;11:72–94. doi: 10.1007/BF02256551. [DOI] [PubMed] [Google Scholar]
- Kim S.H., Ryabov E.V., Kalinina N.O., Rakitina D.V., Gillespie T., MacFarlane S., Haupt S., Brown J.W., Taliansky M. Cajal bodies and the nucleolus are required for a plant virus systemic infection. EMBO J. 2007;26:2169–2179. doi: 10.1038/sj.emboj.7601674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiss T., Fayet E., Jady B.E., Richard P., Weber M. Biogenesis and intranuclear trafficking of human box C/D and H/ACA RNPs. Cold Spring Harb. Symp. Quant. Biol. 2006;71:407–417. doi: 10.1101/sqb.2006.71.025. [DOI] [PubMed] [Google Scholar]
- Kressler D., Hurt E., Bassler J. Driving ribosome assembly. Biochim. Biophys. Acta. 2010;1803:673–683. doi: 10.1016/j.bbamcr.2009.10.009. [DOI] [PubMed] [Google Scholar]
- Kruhlak M., Crouch E.E., Orlov M., Montano C., Gorski S.A., Nussenzweig A., Misteli T., Phair R.D., Casellas R. The ATM repair pathway inhibits RNA polymerase I transcription in response to chromosome breaks. Nature. 2007;447:730–734. doi: 10.1038/nature05842. [DOI] [PubMed] [Google Scholar]
- Kruse J.P., Gu W. SnapShot: p53 posttranslational modifications. Cell. 2008;133:930–930.e1. doi: 10.1016/j.cell.2008.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruse J.P., Gu W. Modes of p53 regulation. Cell. 2009;137:609–622. doi: 10.1016/j.cell.2009.04.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lafarga M., Casafont I., Bengoechea R., Tapia O., Berciano M.T. Cajal's contribution to the knowledge of the neuronal cell nucleus. Chromosoma. 2009;118:437–443. doi: 10.1007/s00412-009-0212-x. [DOI] [PubMed] [Google Scholar]
- Lam Y.W., Evans V.C., Heesom K.J., Lamond A.I., Matthews D.A. Proteomics analysis of the nucleolus in adenovirus-infected cells. Mol. Cell. Proteomics. 2010;9:117–130. doi: 10.1074/mcp.M900338-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J.T., Gu W. The multiple levels of regulation by p53 ubiquitination. Cell Death Differ. 2010;17:86–92. doi: 10.1038/cdd.2009.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemm I., Girard C., Kuhn A.N., Watkins N.J., Schneider M., Bordonne R., Luhrmann R. Ongoing U snRNP biogenesis is required for the integrity of Cajal bodies. Mol. Biol. Cell. 2006;17:3221–3231. doi: 10.1091/mbc.E06-03-0247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lempiainen H., Shore D. Growth control and ribosome biogenesis. Curr. Opin. Cell Biol. 2009;21:855–863. doi: 10.1016/j.ceb.2009.09.002. [DOI] [PubMed] [Google Scholar]
- Li Z., Hann S.R. The Myc-nucleophosmin-ARF network: a complex web unveiled. Cell Cycle. 2009;8:2703–2707. doi: 10.4161/cc.8.17.9418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindstrom M.S. Emerging functions of ribosomal proteins in gene-specific transcription and translation. Biochem. Biophys. Res. Commun. 2009;379:167–170. doi: 10.1016/j.bbrc.2008.12.083. [DOI] [PubMed] [Google Scholar]
- Lyon C.E., Bohmann K., Sleeman J., Lamond A.I. Inhibition of protein dephosphorylation results in the accumulation of splicing snRNPs and coiled bodies within the nucleolus. Exp. Cell Res. 1997;230:84–93. doi: 10.1006/excr.1996.3380. [DOI] [PubMed] [Google Scholar]
- Mao I., Liu J., Li X., Luo H. REGgamma, a proteasome activator and beyond? Cell. Mol. Life Sci. 2008;65:3971–3980. doi: 10.1007/s00018-008-8291-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marzluff W.F., Wagner E.J., Duronio R.J. Metabolism and regulation of canonical histone mRNAs: life without a poly(A) tail. Nat. Rev. Genet. 2008;9:843–854. doi: 10.1038/nrg2438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matera A.G. Drosophila Cajal bodies: accessories not included. J. Cell Biol. 2006;172:791–793. doi: 10.1083/jcb.200602002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matera A.G., Terns R.M., Terns M.P. Non-coding RNAs: lessons from the small nuclear and small nucleolar RNAs. Nat. Rev. Mol. Cell Biol. 2007;8:209–220. doi: 10.1038/nrm2124. [DOI] [PubMed] [Google Scholar]
- Mayer C., Grummt I. Ribosome biogenesis and cell growth: mTOR coordinates transcription by all three classes of nuclear RNA polymerases. Oncogene. 2006;25:6384–6391. doi: 10.1038/sj.onc.1209883. [DOI] [PubMed] [Google Scholar]
- McGowan K.A., Li J.Z., Park C.Y., Beaudry V., Tabor H.K., Sabnis A.J., Zhang W., Fuchs H., de Angelis M.H., Myers R.M. Ribosomal mutations cause p53-mediated dark skin and pleiotropic effects. Nat. Genet. 2008;40:963–970. doi: 10.1038/ng.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McStay B., Grummt I. The epigenetics of rRNA genes: from molecular to chromosome biology. Annu. Rev. Cell Dev. Biol. 2008;24:131–157. doi: 10.1146/annurev.cellbio.24.110707.175259. [DOI] [PubMed] [Google Scholar]
- Mekhail K., Rivero-Lopez L., Khacho M., Lee S. Restriction of rRNA synthesis by VHL maintains energy equilibrium under hypoxia. Cell Cycle. 2006;5:2401–2413. doi: 10.4161/cc.5.20.3387. [DOI] [PubMed] [Google Scholar]
- Montanaro L., Trere D., Derenzini M. Nucleolus, ribosomes, and cancer. Am. J. Pathol. 2008;173:301–310. doi: 10.2353/ajpath.2008.070752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morency E., Sabra M., Catez F., Texier P., Lomonte P. A novel cell response triggered by interphase centromere structural instability. J. Cell Biol. 2007;177:757–768. doi: 10.1083/jcb.200612107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moss T., Langlois F., Gagnon-Kugler T., Stefanovsky V. A housekeeper with power of attorney: the rRNA genes in ribosome biogenesis. Cell. Mol. Life Sci. 2007;64:29–49. doi: 10.1007/s00018-006-6278-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murayama A., Ohmori K., Fujimura A., Minami H., Yasuzawa-Tanaka K., Kuroda T., Oie S., Daitoku H., Okuwaki M., Nagata K. Epigenetic control of rDNA loci in response to intracellular energy status. Cell. 2008;133:627–639. doi: 10.1016/j.cell.2008.03.030. [DOI] [PubMed] [Google Scholar]
- Narla A., Ebert B.L. Ribosomopathies: human disorders of ribosome dysfunction. Blood. 2010;115:3196–3205. doi: 10.1182/blood-2009-10-178129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navascues J., Bengoechea R., Tapia O., Casafont I., Berciano M.T., Lafarga M. SUMO-1 transiently localizes to Cajal bodies in mammalian neurons. J. Struct. Biol. 2008;163:137–146. doi: 10.1016/j.jsb.2008.04.013. [DOI] [PubMed] [Google Scholar]
- Nizami Z., Deryusheva S., Gall J.G. The Cajal body and histone locus body. Cold Spring Harb. Perspect. Biol. 2010;2 doi: 10.1101/cshperspect.a000653. Published online May 26, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ofir-Rosenfeld Y., Boggs K., Michael D., Kastan M.B., Oren M. Mdm2 regulates p53 mRNA translation through inhibitory interactions with ribosomal protein L26. Mol. Cell. 2008;32:180–189. doi: 10.1016/j.molcel.2008.08.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson M.O. Sensing cellular stress: another new function for the nucleolus? Sci. STKE. 2004;2004:pe10. doi: 10.1126/stke.2242004pe10. [DOI] [PubMed] [Google Scholar]
- Ong S.E., Blagoev B., Kratchmarova I., Kristensen D.B., Steen H., Pandey A., Mann M. Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol. Cell. Proteomics. 2002;1:376–386. doi: 10.1074/mcp.m200025-mcp200. [DOI] [PubMed] [Google Scholar]
- Pederson T., Tsai R.Y. In search of nonribosomal nucleolar protein function and regulation. J. Cell Biol. 2009;184:771–776. doi: 10.1083/jcb.200812014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebelo L., Almeida F., Ramos C., Bohmann K., Lamond A.I., Carmo-Fonseca M. The dynamics of coiled bodies in the nucleus of adenovirus-infected cells. Mol. Biol. Cell. 1996;7:1137–1151. doi: 10.1091/mbc.7.7.1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riley K.J., Maher L.J., 3rd p53 RNA interactions: new clues in an old mystery. RNA. 2007;13:1825–1833. doi: 10.1261/rna.673407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubbi C.P., Milner J. Disruption of the nucleolus mediates stabilization of p53 in response to DNA damage and other stresses. EMBO J. 2003;22:6068–6077. doi: 10.1093/emboj/cdg579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubinsztein D.C. The roles of intracellular protein-degradation pathways in neurodegeneration. Nature. 2006;443:780–786. doi: 10.1038/nature05291. [DOI] [PubMed] [Google Scholar]
- Sengupta S., Peterson T.R., Sabatini D.M. Regulation of the mTor complex 1 by nutrients, growth factors, and stress. Mol. Cell. 2010;40:310–322. doi: 10.1016/j.molcel.2010.09.026. this issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shav-Tal Y., Blechman J., Darzacq X., Montagna C., Dye B.T., Patton J.G., Singer R.H., Zipori D. Dynamic sorting of nuclear components into distinct nucleolar caps during transcriptional inhibition. Mol. Biol. Cell. 2005;16:2395–2413. doi: 10.1091/mbc.E04-11-0992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherr C.J. Divorcing ARF and p53: an unsettled case. Nat. Rev. Cancer. 2006;6:663–673. doi: 10.1038/nrc1954. [DOI] [PubMed] [Google Scholar]
- Sirri V., Urcuqui-Inchima S., Roussel P., Hernandez-Verdun D. Nucleolus: the fascinating nuclear body. Histochem. Cell Biol. 2008;129:13–31. doi: 10.1007/s00418-007-0359-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sleeman J.E., Ajuh P., Lamond A.I. snRNP protein expression enhances the formation of Cajal bodies containing p80-coilin and SMN. J. Cell Sci. 2001;114:4407–4419. doi: 10.1242/jcs.114.24.4407. [DOI] [PubMed] [Google Scholar]
- Stanek D., Neugebauer K.M. The Cajal body: a meeting place for spliceosomal snRNPs in the nuclear maze. Chromosoma. 2006;115:343–354. doi: 10.1007/s00412-006-0056-6. [DOI] [PubMed] [Google Scholar]
- Stehmeier P., Muller S. Regulation of p53 family members by the ubiquitin-like SUMO system. DNA Repair (Amst.) 2009;8:491–498. doi: 10.1016/j.dnarep.2009.01.002. [DOI] [PubMed] [Google Scholar]
- Strzelecka M., Trowitzsch S., Weber G., Luhrmann R., Oates A.C., Neugebauer K.M. Coilin-dependent snRNP assembly is essential for zebrafish embryogenesis. Nat. Struct. Mol. Biol. 2010;17:403–409. doi: 10.1038/nsmb.1783. [DOI] [PubMed] [Google Scholar]
- Sundqvist A., Liu G., Mirsaliotis A., Xirodimas D.P. Regulation of nucleolar signalling to p53 through NEDDylation of L11. EMBO Rep. 2009;10:1132–1139. doi: 10.1038/embor.2009.178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka Y., Okamoto K., Teye K., Umata T., Yamagiwa N., Suto Y., Zhang Y., Tsuneoka M. JmjC enzyme KDM2A is a regulator of rRNA transcription in response to starvation. EMBO J. 2010;29:1510–1522. doi: 10.1038/emboj.2010.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tapia O., Bengoechea R., Berciano M.T., Lafarga M. Nucleolar targeting of coilin is regulated by its hypomethylation state. Chromosoma. 2010;119:527–540. doi: 10.1007/s00412-010-0276-7. [DOI] [PubMed] [Google Scholar]
- Thoms H.C., Loveridge C.J., Simpson J., Clipson A., Reinhardt K., Dunlop M.G., Stark L.A. Nucleolar targeting of RelA(p65) is regulated by COMMD1-dependent ubiquitination. Cancer Res. 2010;70:139–149. doi: 10.1158/0008-5472.CAN-09-1397. [DOI] [PubMed] [Google Scholar]
- Tschochner H., Hurt E. Pre-ribosomes on the road from the nucleolus to the cytoplasm. Trends Cell Biol. 2003;13:255–263. doi: 10.1016/s0962-8924(03)00054-0. [DOI] [PubMed] [Google Scholar]
- Verheggen C., Lafontaine D.L., Samarsky D., Mouaikel J., Blanchard J.M., Bordonne R., Bertrand E. Mammalian and yeast U3 snoRNPs are matured in specific and related nuclear compartments. EMBO J. 2002;21:2736–2745. doi: 10.1093/emboj/21.11.2736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vousden K.H., Prives C. Blinded by the light: the growing complexity of p53. Cell. 2009;137:413–431. doi: 10.1016/j.cell.2009.04.037. [DOI] [PubMed] [Google Scholar]
- Walker M.P., Tian L., Matera A.G. Reduced viability, fertility and fecundity in mice lacking the cajal body marker protein, coilin. PLoS ONE. 2009;4:e6171. doi: 10.1371/journal.pone.0006171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warner J.R., McIntosh K.B. How common are extraribosomal functions of ribosomal proteins? Mol. Cell. 2009;34:3–11. doi: 10.1016/j.molcel.2009.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao L., Grove A. Coordination of ribosomal protein and ribosomal RNA gene expression in response to TOR signaling. Curr. Genomics. 2009;10:198–205. doi: 10.2174/138920209788185261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young P.J., Day P.M., Zhou J., Androphy E.J., Morris G.E., Lorson C.L. A direct interaction between the survival motor neuron protein and p53 and its relationship to spinal muscular atrophy. J. Biol. Chem. 2002;277:2852–2859. doi: 10.1074/jbc.M108769200. [DOI] [PubMed] [Google Scholar]
- Zhai W., Comai L. Repression of RNA polymerase I transcription by the tumor suppressor p53. Mol. Cell. Biol. 2000;20:5930–5938. doi: 10.1128/mcb.20.16.5930-5938.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Xiong Y. Mutations in human ARF exon 2 disrupt its nucleolar localization and impair its ability to block nuclear export of MDM2 and p53. Mol. Cell. 1999;3:579–591. doi: 10.1016/s1097-2765(00)80351-2. [DOI] [PubMed] [Google Scholar]
- Zhang Y., Lu H. Signaling to p53: ribosomal proteins find their way. Cancer Cell. 2009;16:369–377. doi: 10.1016/j.ccr.2009.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y., Schmitz K.M., Mayer C., Yuan X., Akhtar A., Grummt I. Reversible acetylation of the chromatin remodelling complex NoRC is required for non-coding RNA-dependent silencing. Nat. Cell Biol. 2009;11:1010–1016. doi: 10.1038/ncb1914. [DOI] [PubMed] [Google Scholar]